虹鳟免疫诱导型基因Cathelicidin2启动子功能分析*
赵紫霞 许 建 江炎亮 白庆利 蒋立坤 陈葆华 徐 鹏,3
虹鳟免疫诱导型基因启动子功能分析*
赵紫霞1①许 建1江炎亮1白庆利2蒋立坤1陈葆华1徐 鹏1,3
(1. 中国水产科学研究院 农业农村部水生动物基因组学重点实验室 渔业生物技术北京市重点实验室 北京 100141;2. 中国水产科学研究院黑龙江水产研究所 哈尔滨 150070; 3. 厦门大学海洋与地球学院 厦门 361102)
本研究通过实时荧光定量PCR实验对虹鳟()()基因的转录模式进行了分析。结果显示,该基因在鳃、头肾等与机体免疫防御功能密切相关的组织内转录,在细菌和病毒感染后,转录水平均显著升高。对基因上游调控序列进行启动子和转录因子结合位点预测,发现该启动子具有真核生物典型的TATA盒和CAAT盒结构,基因上游直至第一内含子区域内,密集存在多个免疫相关转录因子结合位点,其中,2个核因子κB(Nuclear factor kappa B,NFκB)预测结合位点均位于核心启动子正链区域。在草鱼()肾组织细胞系内,绿色荧光蛋白和萤火虫荧光素酶基因都能够在该启动子驱动下表达,表明其具有启动子活性,且启动子活性在受到免疫诱导后增强,包括细菌脂多糖(Lipopolysaccharides, LPS)模拟的细菌感染和聚肌胞苷酸(Polyinosinic polycytidylic acid, Poly I:C)模拟的病毒感染。双荧光素酶报告基因检测显示,启动子活性在与NFκB转录因子表达时,增强至4.39倍,证明基因受NFκB通路调控。研究表明,虹鳟基因能够在多种免疫刺激诱导下表达,其启动子可以应用为免疫诱导型的基因工程元件,驱动外源免疫基因在鱼体内适时表达,抵御外界病原感染,同时,避免非必要条件下的过度表达形成生长负担。
虹鳟;抗菌肽;;启动子;转录调控
抗菌肽(Antimicrobial peptide, AMP)又称为宿主防御肽(Host defense peptide, HDP),是非特异性免疫系统的关键组分之一,在生物体对抗病原微生物感染中发挥着重要的分子屏障作用。Cathelicidin是已知最大的抗菌肽家族之一,在脊椎动物中广泛分布,包括哺乳动物、鸟类、爬行动物、两栖动物和鱼类中都有报道(Maier, 2008; Hao, 2012; Cheng, 2015),其中,哺乳动物Cathelicidin的结构和功能研究较为深入(Hilchie, 2013; 王晨等, 2017)。
第1个硬骨鱼类基因由Chang等(2005)在虹鳟()中发现,此后,在大西洋鲑()等多种鲑科(Salmonidae鱼类(D'Este, 2016)、大西洋鳕() (Shewring, 2011)、香鱼() (Nsrelden, 2017)、日本鳗鲡() (张东玲等, 2015)等物种中陆续发现,并因其具广谱抗菌能力而吸引了广泛的注意,但另一些已完成的硬骨鱼类基因组图谱中却无法筛查到该家族基因(Shewring, 2011),如斑马鱼()、青鳉()、河豚()等。因此,Cathelicidin可能为部分硬骨鱼类群所特有,其基因功能和表达调控的研究将有助于加深对鱼类,特别是对鲑科鱼类非特异性免疫机制的理解,并促进鱼类抗病育种研究进展。
Cathelicidin成熟肽为带有正电荷的短肽,主要通过插入并破坏细胞膜结构杀灭病原微生物。体外实验表明,虹鳟4个基因的各种可变剪切表达产物均能够抑制多种革兰氏阳性和阴性菌的生长(Zhang, 2015),基因产物还具有一定的抗真菌效果,能够延缓水霉()孢子形成(de Bruijn, 2012)。而在大西洋鲑的研究中则发现,基因表达呈现出广泛的免疫应答特征,不仅能够被细菌所诱导,也能够被不具有细胞结构的病毒模拟物所诱导(Shewring, 2011; Broekman, 2013),其转录调控机制尚不明确。本研究以虹鳟基因为例,通过实时荧光定量PCR(Quantitative Real Time PCR, qRT-PCR)实验验证了该基因的免疫诱导转录模式,并开展了启动子克隆与功能分析,对基因转录调控机制进行了初步探索。
1 材料与方法
1.1 实验材料
无特定病原虹鳟5尾,采自中国水产科学研究院黑龙江水产研究所渤海冷水性鱼试验站;自然感染鳗弧菌()发病的虹鳟5尾,采自山西朔州;自然感染传染性造血器官坏死病毒(, IHNV)发病的虹鳟5尾,采自北京怀柔。草鱼肾组织细胞系(Kidney, CIK)保藏于国家水产种质资源平台。
1.2 基因表达分析
使用Trizol试剂(Life technologies)分别提取鳃、头肾组织总RNA,DNase I(Sigma-Aldrich)处理以避免基因组DNA污染。使用ReverTra Ace-α-RT-PCR cDNA第一链合成试剂盒(TOYOBO)进行mRNA反转录。使用SYBR Green RT-PCR试剂盒(TOYOBO)在Applied Biosystems®7500 Real-Time PCR系统(Life technologies)上进行qRT-PCR实验,分析各样本内基因(NCBI:NM_001124463.1)的表达量差异。使用(NCBI:NM_001124235.1)作为内参基因,2–DDCt法(Schmittgen2008)计算基因相对表达量。定量引物Cath2-qPCR-F:CAACACCCTCAACACTGACCG;Cath2-qPCR-R:GAATCTTTTCTACCCATCTTAGG;定量引物actin-F:ACAGGTCATCACCATCGGCA;actin-R:GGTCTCGTGGATACCGCAAG。
1.3 启动子测序分析
以虹鳟基因组图谱序列(Berthelot2014)为参考,通过基因组步移法测定基因序列,使用CLC Genomics Workbench 9.5.4数据分析平台拼接,并结合在线分析软件Neural Network Promoter Prediction(Reese, 2001)(http://www.fruitfly.org/seq_tools/promoter. html),JASPAR2016 (Mathelier2016)(http://jaspar. genereg.net/),AliBaba2.1(http://gene-regulation.com/pub/ programs/alibaba2/index.html),PROMO (Messeguer2002)(http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3),开展启动子位置和转录因子结合位点分析。
1.4 载体构建
以虹鳟基因组DNA为模板,使用Cath2启动子引物Cath2-promoter-F:TCTCACGCGTGGCAGACAGGCATGGGAATA;Cath2-promoter-R: TCTCAAGCTTCTGACCTCTGACCAGCAAGC进行PCR扩增。将纯化后的扩增产物进行MluⅠ、HindⅢ双酶切处理,获取带有黏性末端的启动子片段。分别将pGL3- EGFP和pGL3-basic载体(Promega)进行MluⅠ、HindⅢ双酶切处理,获取带有黏性末端的载体质粒。将纯化后的启动子和载体质粒按照物质的量比例3∶1混合,T4 DNA连接酶16℃处理12 h,转化DH5α感受态细菌,涂布含有氨苄青霉素的平板,37℃倒置培养12 h。挑取单克隆,碱裂解法提取质粒,测序鉴定,获得成功插入虹鳟Cath2启动子完整序列的pGL3- EGFP-Cath2和pGL3-Cath2启动子载体。按照同样步骤构建插入核因子κB (Nuclear factor kappa B,NFκB)编码序列(NCBI:XM_019064772.1)的pEGFP-NFκB表达载体,酶切位点为EcoRⅠ和BamHⅠ。
1.5 细胞转染和免疫刺激
使用含5% 胎牛血清的杜氏改良伊格尔培养基(Dulbecco’s Modified Eagle Medium,DMEM),初始接种浓度为每孔2×105个细胞,5% CO2培养箱25℃培养12 h,待单层细胞贴壁超过60%后,用pH为7.4的PBS缓冲液洗涤细胞3次,每孔加入无血清DMEM培养基900 μl,1 h后进行转染。
转染实验使用Lipofectamine®2000(Life technologies)体系进行脂质体转染,分为7组,每组6孔,各孔分别加入(1)2000 ng pGL3-EGFP空载体+6 μl Lipofectamine® 2000转染试剂+994 μl无血清DMEM培养基;(2)、(3)、(4)2000 ng pGL3-EGFP-Cath2启动子载体+6 μl Lipofectamine® 2000转染试剂+994 μl无血清DMEM培养基;(5)1200 ng pEGFP空载体+600 ng pGL3-basic载体+60 ng PRL-CMV海肾荧光素酶内参载体+6 μl Lipofectamine® 2000转染试剂+994 μl无血清DMEM培养基;(6)1200 ng pEGFP空载体+600 ng pGL3-Cath2启动子载体+60 ng PRL-CMV海肾荧光素酶内参载体+6 μl Lipofectamine® 2000转染试剂+998 μl无血清DMEM培养基;(7)1200 ng pEGFP- NFκB表达载体+600 ng pGL3-Cath2启动子载体+60 ngPRL-CMV海肾荧光素酶内参载体+6 μl Lipofectamine® 2000转染试剂+994 μl无血清DMEM培养基。轻轻摇动平皿混匀,5% CO2培养箱25℃培养。
转染后6 h,换半液,换液使用含有5%胎牛血清的DMEM培养基,继续5% CO2培养箱25℃培养。转染后24 h,在荧光显微镜下观察各组细胞形态和绿色荧光蛋白荧光信号,计数并计算转染效率,转染效率=表达绿色荧光蛋白的细胞数/活细胞总数。向(3)组各孔中加入1 μg/μl的细菌脂多糖(Lipopolysaccharides, LPS)10 μl,向(4)组各孔中加入10 μg/μl的聚肌胞苷酸(Polyinosinic polycytidylic acid,Poly I:C)10 μl,轻轻混匀,继续5% CO2培养箱25℃培养6 h。在荧光显微镜下观察(1)~(4)组细胞形态和绿色荧光蛋白荧光信号。
1.6 双荧光素酶报告基因检测
使用Dual-Luciferase® Reporter Assay System双荧光素酶报告基因检测试剂盒(Promega),按说明书完成检测步骤。将(5)~(7)组各孔内细胞充分裂解后,分别测定萤火虫荧光素酶和海肾荧光素酶活力。使用海肾荧光素酶活力值作为内参,使萤火虫荧光素酶活力值正态化,荧光素酶活性=萤火虫荧光素酶活力/海肾荧光素酶活力。再以只转染对照载体和内参载体的第(5)组数值为对照,将该组荧光素酶活性归一化,荧光素酶相对活性=实验组荧光素酶活性/对照组荧光素酶活性(Pannier, 2007)。
2 结果
2.1 虹鳟Cath2基因转录模式分析
通过qRT-PCR对虹鳟基因转录特征进行了研究。结果表明,该基因转录呈现出显著的免疫应答特征,细菌和病毒感染均能增强该基因转录。基因在健康个体的鳃、头肾组织内均有表达,其中,头肾表达量较高;在鳗弧菌感染个体中,基因表达量显著上调,上升幅度达到10.7倍(鳃)和13.5倍(头肾);在IHNV感染个体中,基因表达量也有明显上调,但上升幅度小于鳗弧菌感染个体(图1),分别为6.98倍(鳃)和6.84倍(头肾)。

图1 虹鳟Cath2基因在感染条件下mRNA表达量变化
1:健康个体;2:鳗弧菌感染个体;3:IHNV感染个体
1: Healthy control; 2:infected; 3: IHNV infected
图中实验组相对表达量以健康对照组鳃组织基因表达量为基准计算,误差线为相对标准差(=5),星号为检验显著性水平(**,< 0.01),下同
The relative expression level in each group was calculated by comparing with the value in gill of healthy control. Error bars represented standard error of the mean (=5). Statistical differences relative to unaffected tissues were calculated by using Student’s t test (**,< 0.01), the same as below
2.2 虹鳟Cath2基因启动子序列分析
将虹鳟基因(NCBI:NM_001124463.1)序列在虹鳟基因组图谱中进行Blat比对,该基因被定位于ChrUn_8重叠群,使用该区段序列为参考,通过基因组步移法对基因上游调控序列进行Sanger测序和拼接,获得起始密码子前1000 bp序列。对该区域进行启动子功能分析,推测基因转录起始位点自–7 bp开始,在–30 bp位置有TATA盒,在–69 bp位置有CAAT盒,具有典型的真核生物启动子结构。

图2 虹鳟Cath2基因上游调控序列功能预测示意
预测该基因核心启动子区为–1~–351 bp区间,为更完整地保留特征性转录调控区域,设计PCR引物扩增–927 bp~76 bp间的1003 bp序列,将其克隆进入启动子载体用于转录活性分析实验。
在虹鳟基因–1000~312 bp区间范围内,密集存在着多个顺式作用元件(图2),其中,既包括免疫应答转录因子结合位点,如核因子κB(Nuclear factor kappa B, NFκB)、Ccaat增强子结合蛋白(Ccaat /Enhancer Binding Protein,CEBP)、cAMP应答元件结合蛋白(cAMP-responsive element binding proteins,CREBP)、巨噬细胞活化因子相关转录因子(Macrophage activating factor related factor,MAFF)、干扰素调节因子(Interferon regulatory factors, IRF)等,也包括高频出现的干扰素增强子GAAA基序以及数个环境压力应答元件,如低氧应答元件(Hypoxia response element, HRE)。
2.3 虹鳟Cath2基因启动子活性及免疫应答
对预测得到的虹鳟基因启动子区域进行PCR扩增,将该启动子连接进入pGL3-EGFP质粒载体,使虹鳟启动子驱动绿色荧光蛋白(Green fluorescent protein, GFP)基因表达,构建成功pGL3- EGFP-Cath2质粒载体。
使用pGL3-EGFP-Cath2质粒转染CIK细胞,转染后细胞生长状况良好(图3A),荧光显微镜下可见GFP绿色荧光(图3B),表明转入的DNA片段能够成功驱动GFP基因表达,具有启动子活性,转染效率为(34.7±4.6)%。向细胞培养液中加入免疫刺激剂LPS (图3C、图3D)或Poly I:C(图3E、图3F),分别模拟细菌感染和病毒感染,细胞的荧光强度显著强于无免疫刺激对照组,表明启动子活性在免疫激活后显著增强,为免疫诱导型启动子。
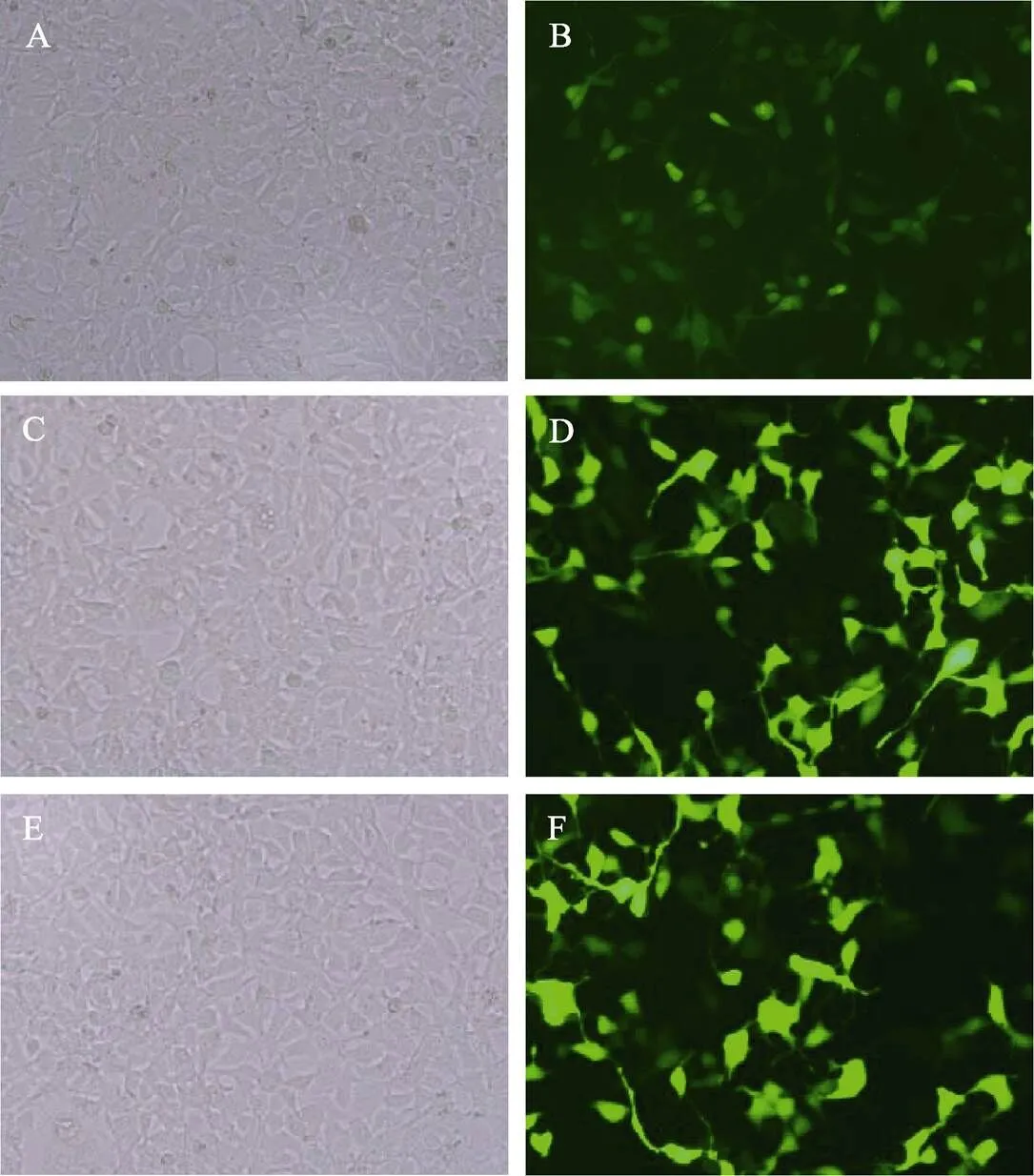
图3 转染pGL3-EGFP-Cath2质粒的CIK细胞
A:无免疫刺激,明场显微镜照片;B:无免疫刺激,荧光显微镜照片;C:LPS模拟细菌感染,明场显微镜照片; D:LPS模拟细菌感染,荧光显微镜照片;E:Poly I:C模拟病毒感染,明场显微镜照片;F:Poly I:C模拟病毒感染,荧光显微镜照片
A: Control cells without immune stimulus, by bright field microscope; B: Control cells without immune stimulus, by fluorescence microscope; C: Immune stimulated cells with LPS, by bright field microscope; D: Immune stimulated cells with LPS, by fluorescence microscope; E: Immune stimulated cells with Poly I:C, by bright field microscope; F: Immune stimulated cells with Poly I:C, by fluorescence microscope
2.4 转录因子NFκB调控虹鳟Cath2基因启动子活性
将虹鳟基因启动子序列连接进入pGL3- basic质粒载体,使虹鳟启动子驱动萤火虫荧光素酶基因表达,构建成功pGL3-Cath2质粒载体;将虹鳟基因cDNA序列连接进入pEGFP载体,使虹鳟基因与GFP基因共表达,构建成功pEGFP-NFκB质粒载体。

图4 双荧光素酶报告基因实验测定虹鳟Cath2基因启动子活性
使用pGL3-Cath2启动子载体质粒、pEGFP-NFκB转录因子表达载体质粒单独或共同转染CIK细胞,所有细胞中均有PRL-CMV海肾荧光素酶内参质粒共转染,使用未负载启动子的pGL3-basic空质粒和未负载转录因子的pEGFP空质粒作为对照,使各组转染DNA总量均等
CIK cells were transfected by pGL3-Cath2 promoter vector plasmid and pEGFP-NFκB transcription factors expression vector plasmid, independently or simultaneously. All cells were co-transfected by PRL-CMV renilla luciferase plasmids as internal control. Blank vectors, pGL3-basic and pEGFP plasmids were also used for co-transfection, to keep equal quantity of transfected DNA among different groups
使用pGL3-Cath2启动子载体、pEGFP-NFκB表达载体,与PRL-CMV海肾荧光素酶内参载体质粒共转染CIK细胞,转染效率为(28.9 ± 3.5)%。定量测定启动子活性,结果见图4。从图4可以看出,转染pGL3- Cath2实验组测定的萤光素酶活性比空载体对照组显著增强,表明基因启动子在该体系中表现出明显的启动子活性;pGL3-Cath2和pEGFP-NFκB共转染实验组测定的萤光素酶活性显著高于单独转染pGL3-Cath2实验组,前者约为后者的4.39倍,表明外源NFκB转录因子的增强性表达能够提高启动子活性,证实了NFκB为促进基因启动子表达的反式作用因子。
3 讨论
3.1 病毒诱导虹鳟Cath2基因转录
多项不同来源的转录组数据分析表明,虹鳟基因转录水平不仅在细菌感染或LPS模拟刺激后大幅度上升(MacKenzie, 2008; Langevin, 2012; Bridle, 2012),也在病毒或模拟物刺激后有不同程度的上升,如IHNV BLK94和WA毒株(Purcell, 2011),IHNV灭活病毒(MacKenzie, 2008),病毒性出血败血症病毒(Viral hemorrhagic septicemia virus, VHSV)(Aquilino, 2014)以及病毒双链RNA模拟物Poly I:C(Pacitti, 2016)诱导后的表达谱数据中,基因均呈现上调表达。
为在实验层面确认以上生物信息学分析数据,本研究通过qRT-PCR检测,证实自然感染鳗弧菌或IHNV的虹鳟鳃、头肾组织中,基因转录水平都有显著升高(图1),进一步的体外转录研究结果(图3)显示,细菌模拟物LPS和病毒模拟物Poly I:C都能够诱导基因启动子表达增强。基因表达能够被细菌感染所诱导,与其抗菌生理功能相吻合,而该基因同样参与病毒感染免疫应答,该基因可能不局限于通常所知的生物学功能,即破坏病原生物膜结构,还可能以其他的方式发挥作用,以抵御不具有细胞结构的病毒感染。
D’Este等(2016)对鲑科鱼类Cathelicidin成熟多肽结构进行了分析,并推测其富含丝氨酸和甘氨酸的分子,可通过刺激吞噬细胞的吞噬功能来发挥免疫调节作用,而非直接杀伤病原体。本研究从转录特征方面支持了Cathelicidin的非直接杀菌功能,这种受病毒诱导表达的转录模式在鱼类抗菌肽中并非个例,在大西洋鳕中也曾有报道,包括Hepcidin (Solstad, 2008)和Cathelicidin(Shewring, 2011; Broekman, 2013),其抗病毒机制仍需进一步研究验证。
3.2 虹鳟Cath2基因转录调控分析
为解析虹鳟基因转录调控机制,本研究对基因上游调控序列进行了测序和分析,成功获得了长度为351 bp的核心启动子序列。序列相似性比对结果表明,虹鳟基因起始密码子前的上游调控序列与人()、大西洋鳕基因以及虹鳟基因对应序列的碱基一致性较低,但具有一些共同的结构特征,各基因启动子均具有真核生物启动子典型的TATA盒和CAAT盒结构,在基因上游直至第一内含子区域,密集存在多个免疫相关转录因子结合位点(Chang, 2005; Chakraborty, 2009; Shewring, 2011;Dhawan, 2015)。
虹鳟基因上游调控区共有3个NFκB预测结合位点,其中,2个都在核心启动子正链上,分别位于–78 bp和–180 bp位置,靠近CAAT盒。对已报道的鱼类其他基因序列进行了NFκB结合位点预测,包括虹鳟、大西洋鳕研究发现,NFκB结合特征性序列普遍存在于基因上游,但此前尚无NFκB调控基因转录的研究结果。本研究通过双荧光素酶报告基因研究,将虹鳟启动子连入pGL3质粒载体,转染草鱼CIK细胞系,萤火虫荧光素酶基因能够在该启动子驱动下表达,当与NFκB表达载体共转染时,萤火虫荧光素酶活性显著增强,表明NFκB是增强基因转录的反式作用因子。
NFκB是经典的炎症和免疫调控核转录因子,该通路既能被细胞表面病原识别受体直接激活,也能被多种诱导磷酸激酶活化的胞内信号所激活,在接受免疫刺激后,数分钟即可迅速活化,参与包括细菌和病毒在内的多种病原感染应答(Ghosh, 1998;Varejckova, 2017)。虹鳟基因转录受NFκB调控,因此,能够在不同病原感染个体中迅速大幅上调表达,呈现出免疫诱导型转录特征。
3.3 免疫诱导型启动子的应用
启动子常根据作用方式及功能分为3类:组成型启动子,在全部或多数组织中保持持续的转录活性;特异型启动子,在特定的组织或发育时期具有转录活性;诱导型启动子,受特定物理或化学信号调控 (李圣彦等, 2014)。本研究中获得的虹鳟启动子即为免疫诱导型启动子,在基因工程抗病育种中具有较高的应用价值。
利用基因工程技术,将免疫基因,如免疫球蛋白或抗菌肽等转入养殖鱼类体内,培育疾病抗性品种,是防控养殖病害的有效途径之一(叶鼎等, 2014)。启动子是各种转基因技术方案中必备的转基因元件,转基因载体中负载的外源目的基因必须置于有效的启动子介导之下,才能够在宿主细胞内成功转录表达。
目前,已报道的用作转基因元件的鱼类启动子多为组成型,如鲤()β-肌动蛋白、美洲大绵鳚()抗冻蛋白启动子等(叶星等, 2011; Ledford, 2015; 汪亚平等, 2016; 胡炜等, 2016)。但对抗病转基因鱼而言,使用组成型启动子,将使未接触病原的鱼体内长期大量表达外源免疫基因,不仅没有必要,还可能因消耗能量而造成生长负担。如果使用免疫诱导型启动子,则可介导外源基因仅在鱼体遭受免疫胁迫时出现高表达,合理利用能量并抵御病害。
目前,关于免疫诱导型启动子的报道仍较少 (杜小溪等, 2013; Fu, 2016),本研究即提供了一种具有快速免疫响应特征的鱼类启动子。虹鳟基因的表达模式较好地符合了抗病转基因鱼对外源免疫基因转录特征的需求,将该基因启动子用作抗病转基因载体构建的启动子元件,可能驱动外源免疫基因保持类似的表达模式。
鳃和头肾组织都在鱼类免疫防御中具有重要地位。鳃是鱼类的呼吸器官,比表面积大,表面密布毛细血管,随时与外界水流接触,因而易受环境中的病原菌侵袭。头肾是硬骨鱼类特有的淋巴器官,在鱼体内发挥重要的免疫功能。将虹鳟基因启动子用作转基因元件,可能使转基因鱼在受到免疫刺激时,鳃和头肾大量表达转入的免疫基因,及时抵御外界病原感染,同时,避免在不必要的组织和健康状态下过多表达外源基因,在鱼类基因工程抗病育种领域具有广阔的应用前景。
Aquilino C, Castro R, Fischer U,. Transcriptomic responses in rainbow trout gills upon infection with viral hemorrhagic septicemia virus (VHSV). Developmental and Comparative Immunology, 2014, 44(1): 12–20
Berthelot C, Brunet F, Chalopin D,. The rainbow trout genome provides novel insights into evolution after whole- genome duplication in vertebrates. Nature Communication, 2014, 5(4): 3657
Bridle AR, Koop BF, Nowak BF. Identification of surrogates of protection against yersiniosis in immersion vaccinated Atlantic salmon. PLoS One, 2012, 7(7): e40841
Broekman DC, Guðmundsson GH, Maier VH. Differential regulation of cathelicidin in salmon and cod. Fish and Shellfish Immunology, 2013, 35(2): 532–538
Chakraborty K, Maity PC, Sil AK,. cAMP stringently regulates human cathelicidin antimicrobial peptide expression in the mucosal epithelial cells by activating cAMP-response element-binding protein, AP-1, and inducible cAMP early repressor. Journal of Biological Chemistry, 2009, 284(33): 21810–21827
Chang CI, Pleguezuelos O, Zhang YA,. Identification of a novel cathelicidin gene in the rainbow trout,. Infection and Immunity, 2005, 73(8): 5053–5064
Cheng Y, Prickett MD, Gutowska W,. Evolution of the avian β-defensin and cathelicidin genes. BMC Evolutionary Biology, 2015, 15(1): 188
de Bruijn I, Belmonte R, Anderson VL ,. Immune gene expression in trout cell lines infected with the fish pathogenic oomycete. Developmental and Comparative Immunology, 2012, 38(1): 44–54
D'Este F, Benincasa M, Cannone G,. Antimicrobial and host cell-directed activities of Gly/Ser-rich peptides from salmonid cathelicidins. Fish and Shellfish Immunology, 2016(59): 456–468
Dhawan P, Wei R, Sun C,. C/EBPα and the vitamin D receptor cooperate in the regulation of cathelicidin in lung epithelial cells. Journal of Cellular Physiology, 2015, 230(2): 464–472
Du XX, Gao XG, Chen PH,. Research on gene promoter of fish. Biotechnology Bulletin, 2013(8): 12–16 [杜小溪, 高祥刚, 陈潘海,等. 鱼类基因启动子的研究进展. 生物技术通报, 2013(8): 12–16]
Fu XQ, Ding ZJ, Fan J,. Characterization, promoter analysis and expression of the interleukin-6 gene in blunt snout bream,. Fish Physiology and Biochemistry, 2016, 42(6): 1527–1540
Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: Evolutionarily conserved mediators of immune responses. Annual Review of Immunology, 1998, 16(1): 225–260
Hao X, Yang H, Wei L,. Amphibian cathelicidin fills the evolutionary gap of cathelicidin in vertebrate. Amino Acids, 2012, 43(2): 677–685
Hilchie AL, Wuerth K, Hancock RE. Immune modulation by multifaceted cationic host defense (Antimicrobial) peptides. Nature Chemical Biology, 2013, 9(12): 761–768
Hu W, Zhu ZY. Enlightenments for China from the industrialization of the transgenic atlantic salmon in the US. Engineering Sciences, 2016, 18(3): 105–109 [胡炜, 朱作言. 美国转基因大西洋鲑产业化对我国的启示. 中国工程科学, 2016, 18(3): 105–109]
Langevin C, Blanco M, Martin SA,Transcriptional responses of resistant and susceptible fish clones to the bacterial pathogen. PLoS One, 2012, 7(6): 39126
Ledford H. Salmon approval heralds rethink of transgenic animals. Nature, 2015, 527(7579): 417–418
Li SY, Lang ZH, Huang DF. Research progress on eukaryotic promoter. Current Biotechnology, 2014, 4(3): 158–164 [李圣彦, 郎志宏, 黄大昉. 真核生物启动子研究概述. 生物技术进展, 2014, 4(3): 158–164]
MacKenzie S, Balasch JC, Novoa B,. Comparative analysis of the acute response of the trout,head kidney tochallenge with virulent and attenuated infectious hematopoietic necrosis virus and LPS-induced inflammation. BMC Genomics, 2008, 9(1): 141
Maier VH, Dorn KV, Gudmundsdottir BK,. Characterisation of cathelicidin gene family members in divergent fish species. Molecular Immunology, 2008, 45(14): 3723–3730
Mathelier A, Fornes O, Arenillas DJ,. JASPAR 2016: A major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Research, 2016, 44(D1): 110–115
Messeguer X, Escudero R, Farré D,. PROMO: Detection of known transcription regulatory elements using species- tailored searches. Bioinformatics, 2002, 18(2): 333–334
Nsrelden RM, Horiuchi H, Furusawa S. Expression of ayu antimicrobial peptide genes after LPS stimulation. Journal of Veterinary Medical Science, 2017, 79(6): 1072–1080
Pacitti D, Lawan MM, Feldmann J,. Impact of selenium supplementation on fish antiviral responses: A whole transcriptomic analysis in rainbow trout () fed supranutritional levels of Sel-Plex®. BMC Genomics, 2016, 17(1): 116
Pannier AK, Ariazi EA, Bellis AD,. Bioluminescence imaging for assessment and normalization in transfected cell arrays. Biotechnology and Bioengineering, 2007, 98(2): 468–497
Purcell MK, Marjara IS, Batts W,. Transcriptome analysis of rainbow trout infected with high and low virulence strains of infectious hematopoietic necrosis virus. Fish and Shellfish Immunology, 2011, 30(1): 84–93
Reese MG. Application of a time-delay neural network to promoter annotation in thegenome. Computers and Chemistry, 2001, 26(1): 51–56
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nature Protocols, 2008, 3(6): 1101–1108
Shewring DM, Zou J, Corripio-Miyar Y,. Analysis of the cathelicidin 1 gene locus in Atlantic cod (). Molecular Immunology, 2011, 48(5): 782–787
Solstad T, Larsen AN, Seppola M,. Identification, cloning and expression analysis of a hepcidin cDNA of the Atlantic cod (L.). Fish and Shellfish Immunology, 2008, 25(3): 298–310
Varejckova M, Gallardo-Vara E, Vicen M,Soluble endoglin modulates the pro-inflammatory mediators NF-κB and IL-6 in cultured human endothelial cells. Life Sciences, 2017, 175: 52–60
Wang C, Feng L, Yu HN,. Relationship between structure and function of Cathelicidins and their molecular design: A review. Chinese Journal of Biotechnology, 2017, 33(1): 27–35 [王晨, 冯兰, 于海宁,等. Cathelicidins 结构与功能的关系及其分子设计研究进展. 生物工程学报, 2017, 33(1): 27–35]
Wang YP, He LB. Retrospect and prospect of transgenic fish breeding in China. Chinese Journal of Biotechnology, 2016, 32(7): 851–860 [汪亚平, 何利波. 我国转基因鱼研制的历史回顾与展望. 生物工程学报, 2016, 32(7): 851–860]
Ye D, Zhu ZY, Sun YH. Fish genome manipulation and directional breeding. Science China: Life Sciences, 2014, 44(12): 1253–1261 [叶鼎, 朱作言, 孙永华. 鱼类基因组操作与定向育种. 中国科学: 生命科学, 2014, 44(12): 1253–1261]
Ye X, Tian YY, Gao FY. Progress in transgenic fish techniques and application. Hereditas (Beijing), 2011, 33(5): 494–503 [叶星, 田园园, 高风英. 转基因鱼的研究进展与商业化前景. 遗传, 2011, 33(5): 494–503]
Zhang DL, Yu DH. Cloning and prokaryotic expression of Cathelicidin gene from Japanese Eel I,. Biotechnology Bulletin,2015, 31(7): 124–131 [张东玲, 喻达辉. 日本鳗鲡I型Cathelicidin基因的克隆与原核表达. 生物技术通报, 2015, 31(7): 124–131]
Zhang XJ, Zhang XY, Zhang N,. Distinctive structural hallmarks and biological activities of the multiple cathelicidin antimicrobial peptides in a primitive teleost fish. Journal of Immunology, 2015, 194(10): 4974–4987
(编辑 陈严)
Functional Analysis of the Immune-Induced Promoter of the Rainbow TroutGene
ZHAO Zixia1①, XU Jian1, JIANG Yanliang1, BAI Qingli2, JIANG Likun1, CHEN Baohua1, XU Peng1,3
(1. Key Laboratory of Aquatic Genomics, Ministry of Agriculture and Rural Affairs, Beijing Key Laboratory of Fishery Biotechnology, Chinese Academy of Fishery Sciences, Beijing 100141; 2. Heilongjiang River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Harbin 150070;3. College of Ocean and Earth Sciences, Xiamen University, Xiamen 361102)
This study aimed to investigate the transcriptional regulation of a teleost antimicrobial peptide. The expression pattern of the rainbow trout() gene was analyzed by quantitative real-time PCR (qRT-PCR).was expressed in tissues closely related to immune defense, including the gill and head kidney, and expression significantly increased after both bacterial and viral infections. Promoter and transcription factor binding sites were analyzed for the upstream regulatory sequence ofgene. The predicted promoter contained characteristic eukaryotic TATA and CAAT boxes, as well as multiple binding sites for immune-related transcription factors, including two candidate binding sites for nuclear factor kappa B (NFκB) on the positive strand of the core promoter. Inkidney cell lines, transcription of both green fluorescent protein and firefly luciferase genes was initiated by the predictedpromoter. Both the bacterial mimetic lipopolysaccharide and the viral mimetic polyinosinic polycytidylic acid up-regulated promoter activity. As shown in the dual luciferase reporter assay, promoter activity was enhanced by co-expression of the transcription factor NFκB, indicating thatis regulated by the NFκB pathway. These results suggest that expression of the rainbow troutgene could be induced by different immune stimuli, and its promoter might serve as an immune-inducible transgenic element. Thepromoter may initiate the transcription of heterologous immune genes against exogenous infection in a proper expression pattern, avoiding excessive transcription under unnecessary conditions; therefore, it has potential for genetic engineering approaches in the breeding of disease-resistant fish.
Rainbow trout; Antimicrobial peptide;; Promoter; Transcription regulation
ZHAO Zixia. E-mail: zhaozx@cafs.ac.cn
10.19663/j.issn2095-9869.20170614001
Q812, S917.4
A
2095-9869(2018)04-0037-09
* 国家科技支撑计划项目(2015BAD25B01)和中国水产科学研究院基本科研业务费专项(2015C007)共同资助 [This work was supported by the National Science and Technology Pillar Program (2015BAD25B01), and Central Public-interest Scientific Institution Basal Research Fund, CAFS (2015C007)]
赵紫霞, E-mail: zhaozx@cafs.ac.cn
2017-06-14,
2017-07-07
赵紫霞, 许建, 江炎亮, 白庆利, 蒋立坤, 陈葆华, 徐鹏. 虹鳟免疫诱导型基因启动子功能分析. 渔业科学进展, 2018, 39(4): 37–45
Zhao ZX, Xu J, Jiang YL, Bai QL, Jiang LK, Chen BH, Xu P. Functional analysis of the immune-induced promoter of the rainbow troutgene. Progress in Fishery Sciences, 2018, 39(4): 37–45
——一道江苏高考题的奥秘解读和拓展

