水稻抗稻瘟病天然免疫机制及抗病育种新策略
何峰,张浩,刘金灵,王志龙,王国梁
1. 湖南农业大学农学院,长沙 410128;
2. 中国农业科学院植物保护研究所,北京100193
水稻是最重要的粮食作物之一,是全球 50%左右人口的主食。稻瘟病是由子囊菌(Magnaporthe oryzae)引起的一种严重的水稻病害,为植物十大真菌性病害之一[1]。全球每年由于稻瘟病危害造成的产量损失可达水稻总产的 10%~30%[2]。近 10中(2004~2013)我国稻瘟病年平均危害面积在 8000万亩左右(数据来源于全国农业技术推广服务中心网站,http:// www.moa.gov.cn/sydw/njzx/),水稻平均单产约430公斤/亩(数据来源于国家统计局网站,http://www.stats. gov.cn/),按每亩减产5%计算,年造成产量损失超过15亿公斤。尽管利用抗病品种是防治稻瘟病最经济有效的措施,但是由于田间稻瘟菌群体频繁变异,往往导致抗病品种在应用数年后就丧失抗性。此外,我国近年育成水稻品种整体抗性水平并不高。例如,2004~2008年国审 174个品种稻瘟病平均抗病指数为 5.5,属于中感级别(数据来源于国家水稻数据中心网站,http://www.ricedata.cn/)。因此,发掘新抗病种质资源并发展新的高效抗病育种技术一直是水稻抗病育种的迫切需求。
植物在与病原菌长期协同进化过程中形成了两种天然免疫机制[3],即病原菌相关分子模式(Pathogenassociated molecular patterns,PAMPs)诱导的抗病反应机制(PAMP-triggered immunity,PTI)和病原菌效应蛋白(Effector)诱导的抗病反应机制(Effector-triggered immunity,ETI)。PTI通常由植物细胞表面模式识别受体(Pattern recognition receptors,PRRs)识别保守的病原菌PAMPs分子,然后激活植物体内相对较弱的基础防御反应。ETI则依赖于植物抗病蛋白(R proteins)直接或间接识别病原菌分泌的效应蛋白,进而激发更为强烈的抗性反应,以抑制病原菌侵染,通常表现为植物过敏反应(Hypersensitive response,HR)。目前,植物PTI和ETI防卫反应分子机制在拟南芥(Arabidopsis thaliana)中已研究得比较深入[4]。
近年来,水稻与稻瘟菌互作已发展成植物与病原真菌分子互作研究的模式系统之一,人们在认识水稻稻瘟病PTI和ETI抗性机制及稻瘟菌致病性机制等方面取得了重要研究成果,并发展了一些具有重要应用前景的育种新技术。本文总结了近年水稻抗稻瘟病PTI和ETI天然免疫分子机制研究的最新进展,并探讨了水稻抗病育种改良的新技术策略,同时对当前水稻抗稻瘟病机制研究与抗病育种应用的问题和挑战进行了展望。
1 稻瘟病PTI防御机制
PTI是由PRR蛋白识别PAMPs而激发的抗性反应。PAMPs是病原菌中一类保守的结构性分子,如细菌鞭毛蛋白(Flagellin,flg22)、延伸因子(Elongation factor Tu,EF-Tu)、Ax21(Sulfated peptide Ax21)、肽聚糖(Peptidoglycan,PGN)、脂多糖(Lipopolysaccharides,LPS)、真菌细胞壁多糖、几丁质、葡聚糖等[5~8]。在拟南芥中,PTI信号激活分子机制已经有了较系统地研究[9~11]。近年来,水稻 PTI信号响应机制也取得了重要的进展。
1.1 flg22与flg22受体OsFLS2及其介导的信号通路
flg22是细菌鞭毛蛋白 N 端一段含 22 个氨基酸的保守多肽,该肽段是鞭毛蛋白诱导植物抗性反应的关键功能结构[12]。拟南芥中,flg22能够被PRR受体FLS2(Flagellinsensing 2)识别与结合,并激活下游抗病信号。FLS2编码一个丝氨酸/苏氨酸类受体蛋白激酶(Receptor-like kinase,RLK)。研究表明,水稻中FLS2同源基因OsFLS2也具有类似的保守功能。OsFLS2能够与flg22结合,并能互补拟南芥fls2突变体的表型[13]。过表达OsFLS2增强了水稻对flg22的应答,并激活了细胞死亡等抗性反应[13]。
最近,晶体结构研究发现flg22能够引起FLS2和蛋白激酶BAK1(BRI1-associated receptor kinase 1)形成异源二聚体,并导致 FLS2和 BAK1相互磷酸化[14]。FLS2-BAK1二聚化后,通过磷酸化蛋白激酶BIK1(Botrytis-induced kinase 1),而激活下游信号通路[15]。bik1突变体降低了拟南芥对flg22的敏感性,并增强了对假单胞菌 DC3000的感病性[15]。最新研究发现,BIK1通过磷酸化NADPH氧化酶RbohD,而诱导ROS的产生[16,17]。OsBAK1过表达水稻出现叶夹角小、发育不良和矮化等表型[18]。过表达OsBIK1增强了水稻对稻瘟病的抗性,且OsBIK1能够互补拟南芥bik1突变体的功能[19],但水稻中OsFLS2与 OsBAK1是否具有与拟南芥类似的保守功能,还有待深入研究。
1.2 几丁质与几丁质受体 OsCEBiP及其介导的信号通路
几丁质,即β-(1,4)-N-乙酰氨基-2-脱氧-D-葡聚糖,是真菌细胞壁的重要组分,也是诱导植物 PTI反应的一类 PAMPs。拟南芥中,几丁质能够被含LysM结构域的类受体蛋白激酶CERK1(Chitin elicitor receptor kinase 1)识别、结合并激活 PTI反应[20~22]。但不同的是,水稻中几丁质不能直接结合 CERK1的同源蛋白 OsCERK1,而是先与跨膜蛋白OsCEBiP(Chitin elicitor binding protein)结合,再由OsCEBiP与 OsCERK1形成异源二聚体,进而传导对几丁质的响应,激活下游信号[23,24]。OsCEBiP也编码含LysM结构的类受体蛋白(RLP),但无胞内的激酶结构域,为非典型的PRR蛋白。生物学功能分析表明,水稻OsCEBiP敲除突变体显著抑制了几丁质诱导的 PTI反应,并增强了对稻瘟病菌的感病性[23]。OsCERK1RNAi转基因水稻也显著抑制几丁质引起的防卫反应[24]。
最新研究发现,OsCERK1能够磷酸化激活PRONE型鸟苷酸交换因子 OsRacGEF1,促进 Rho型小G蛋白OsRac1与GTP结合,变为活性状态[25]。OsRac1为水稻PTI和ETI防御信号中一个关键性集成调控因子[26]。活性态的 OsRac1能够激活下游多样性的抗病信号途径。例如,OsRac1与NBS-LRR 蛋白Pit直接互作,激活ETI信号;OsRac1能够激活膜上的呼吸爆发氧化酶 OsRbohB,氧化 NADPH,促进ROS的产生。此外,OsRac1还能调节MAPK级联反应介导的抗病信号[25~27]。
1.3 几丁质、肽聚糖与受体蛋白LYP4和LYP6介导的抗病信号途径
近来研究表明,水稻中OsCEBiP并不是几丁质的唯一受体,另外两个 LysM结构域蛋白 LYP4和LYP6(Lysin motif-containing proteins)也参与了对几丁质的识别[28]。在细菌中LYPs识别细菌肽聚糖,而在植物中 LYPs能与肽聚糖相关的几丁质和结瘤因子(Nod factor)结合[29,30]。拟南芥中,LysM蛋白LYM1(At-LYP3)和 LYM3(At-LYP2)能够识别结合肽聚糖而激活PTI反应,但LYM1和LYM3是否也能识别几丁质,还有待进一步研究[31~33]。而水稻 LYP4和LYP6具有双重功能,能够结合肽聚糖和几丁质。LYP4和LYP6是两个细胞膜定位蛋白,具有N端的信号肽序列、2个LysMs结构以及C端的糖基磷脂酰肌醇锚钩。研究发现,细菌性病原菌及其他多种PAMPs,如肽聚糖、几丁质、脂多糖和flg22等,都可以诱导LYP4和LYP6的表达。生物学功能鉴定表明,LYP4或LYP6RNAi水稻均降低了肽聚糖或几丁质介导的防卫反应,并增强了对白叶枯菌(Xanthomonas oryzaepv.oryzae)和稻瘟菌的感病性[28]。
2 稻瘟病ETI防御机制
2.1 稻瘟病抗性基因与无毒基因的克隆
克隆抗稻瘟病基因与其对应的无毒基因,并阐明它们之间的互作关系是认识稻瘟病ETI免疫机制的关键。目前,水稻中已定位了超过 100个稻瘟病抗性基因,其中报道克隆了 22个(表 1)。这些已克隆的基因编码包括核苷酸结合位点(Nucleotide binding sites,NBS)/富亮氨酸重复(Leucine rich repeat,LRR)蛋白(NBS-LRR),如Pib、Pi-ta、Pi9、Pi2、Piz-t、Pi36、Pi37、Pikm、Pit、Pi5、Pid3、Pish、Pb1、Pi54、Pik、Pik-p、Pia、Pi25、Pi1和Pi35;受体蛋白激酶类(Receptor-like kinase protein,RLK),如Pi-d2[34];富脯氨酸类蛋白,如Pi21[35]等 3种类型的蛋白。此外,已克隆了 10个稻瘟菌无毒基因PWL1、PWL2、AvrPi-ta、AvrPiz-t、AvrPia、AvrPii、AvrPik/km/kp、Avr1-CO39、ACE1和AvrPi9(表 2)。

表1 已克隆的稻瘟病抗性基因(R)信息
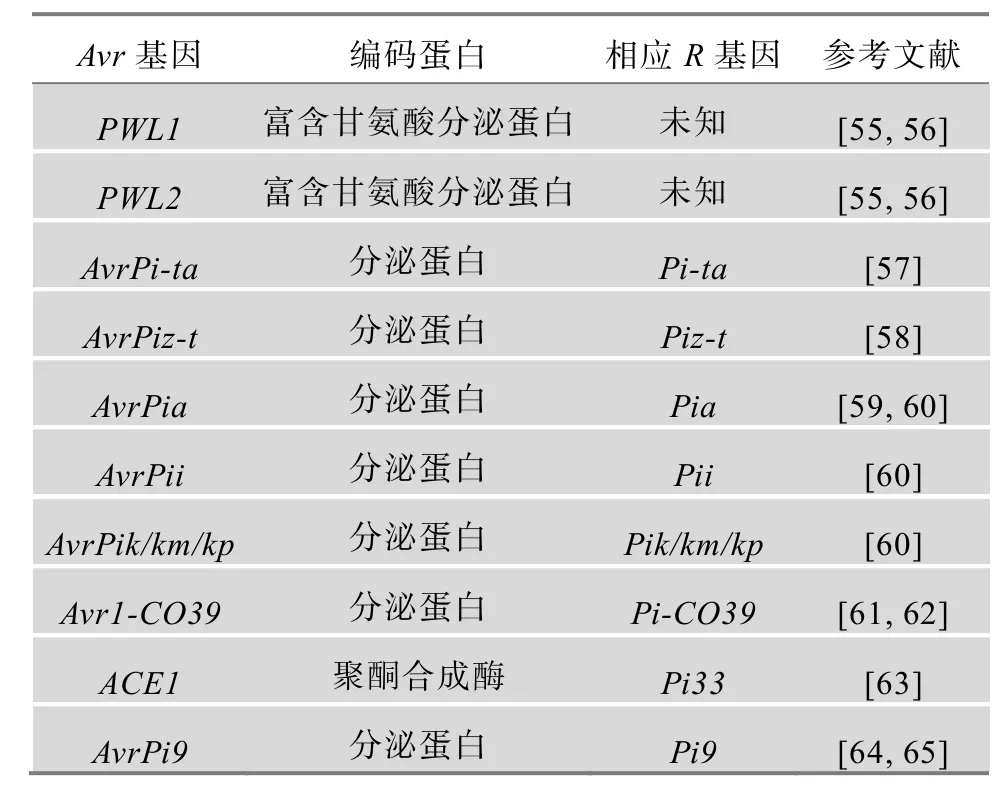
表2 已克隆的稻瘟菌无毒基因(Avr)信息
2.2 R和Avr蛋白直接互作介导的ETI激活模式
在已克隆的R基因与Avr基因中,Pi-ta与Avr-Pi-ta[66]、Pik 与 AvrPik[67]、Pia 与 AvrPia[62]、PiCO39与Avr1-CO39[62]4对蛋白具有直接相互作用(图1)。Pi-ta与AvrPi-ta最早被报道具有直接相互作用[66]。与Pi-ta/AvrPi-ta不同的是,Pik、Pia、PiCO39功能的实现需要两个NBS-LRR蛋白共同作用,相应的无毒蛋白 AvrPik、AvrPia、Avr1-CO39只与其中一个NBS-LRR成员互作。例如,Pik由 NBS-LRR基因Pik-1和Pik-2组成[49],Pik-1被证明与AvrPik直接互作[67]。研究发现,Avr-Pik有Avr-Pik-A、B、C、D和E5个等位基因,同样Pik也有 5个等位基因Pik*、Pikp、Pikm、Piks和Pikh。不同的Pik等位基因只识别特定的AvrPik等位基因。例如,Pikp只识别Avr-Pik-D,而不识别其他几个等位基因,揭示了抗病基因与无毒基因之间的协同进化机制[67]。此外,Pia由两个反向串联的NBS-LRR基因RGA4和RGA5组成[51]。AvrPia也只与 RGA5直接互作,且只与RGA5两个转录本RGA5-A和RGA5-B中的RGA5-A互作[62]。RGA4和RGA5也被证明能够识别无毒蛋白Avr1-CO39,并且RGA5-A也能与Avr1-CO39互作[62]。这些结果揭示了同一抗病基因能够识别多个不同无毒基因的新机制,至于形成这种识别机制的原因和进化动力是什么,目前尚不清楚。
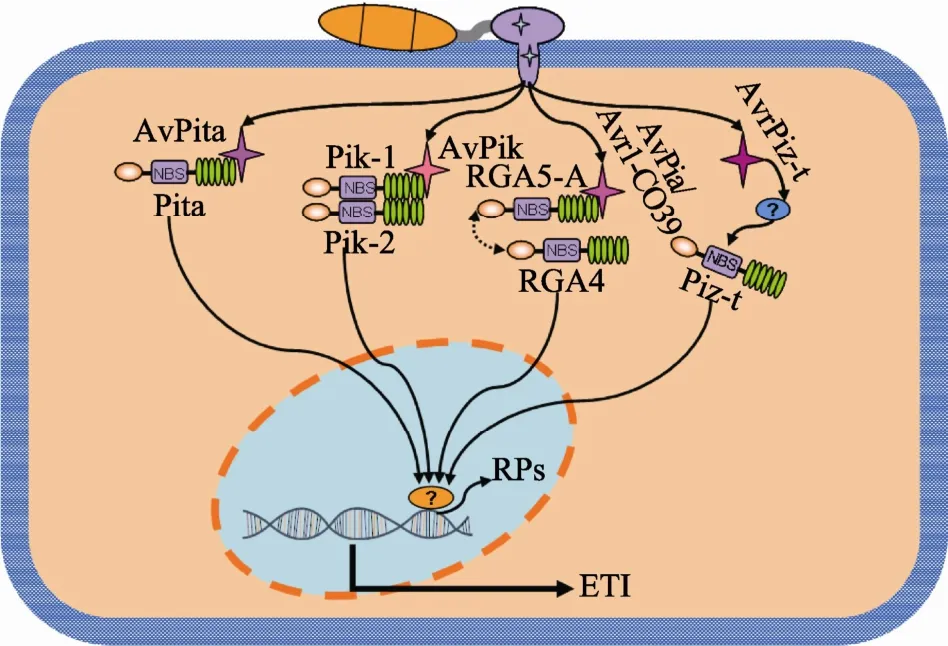
图1 水稻稻瘟病抗性基因与相应无毒基因互作模式
2.3 R-Avr基因非直接互作介导的ETI激活模式
AvrPiz-t是广谱抗稻瘟病基因Piz-t对应的无毒基因。研究表明,AvrPiz-t与Piz-t无直接相互作用。AvrPiz-t编码一个 N端含信号肽、长度为 108个氨基酸的新分泌蛋白[58]。生物学功能研究发现,AvrPiz-t具有毒性,能够抑制 BAX诱导的烟草(N. benthamiana)细胞死亡。水稻中异源表达AvrPiz-t则增强了水稻对稻瘟病的感病性,并显著抑制flg22和几丁质诱导的PTI反应[68]。此外,利用酵母双杂交技术,以AvrPiz-t为诱饵,筛选水稻cDNA文库,鉴定了 12个 AvrPiz-t互作蛋白,命名为 APIPs(Avr-Piz-t-interacting proteins);对其中RING finger型泛素连接酶基因APIP6的功能进行研究,发现AvrPiz-t通过干扰APIP6的泛素连接酶活性而抑制APIP6介导的 PTI反应,反之 APIP6能够泛素化 AvrPiz-t;生物学功能研究表明,APIP6RNAi水稻增强了对稻瘟菌的感病性[68]。这些结果揭示了稻瘟菌效应蛋白通过干扰植物泛素蛋白降解系统而抑制植物抗病性的新机制。
3 抗稻瘟病育种新策略
3.1 全基因组选择育种
随着新一代高通量测序技术的发展,开展大规模水稻重测序、发掘覆盖全基因组的高密度SNP标记已成为可能。最近,全基因组关联分析(Genome wide association studies,GWAS)技术在水稻、玉米(Zea mays)等作物基因定位中已成功应用[69~72]。例如,Huang等[73]利用二代测序技术对517份我国水稻种质材料进行重测序,利用识别的全基因组 SNP标记对14个性状进行了 GWAS分析,定位了与这些性状相关的 80个基因位点。最近,Huang等[69]又对100份我国粳稻品种和来自33个国家的330份水稻材料进行了重测序,结合之前 517份材料的数据,对共 947份材料的抽穗期和产量性状进行了GWAS分析,发现了32个新的与抽穗、产量性状相关位点。此外,Zhao等[74]利用基于水稻重测序开发的44M的SNP芯片对413份来自82个国家的水稻材料的34个性状进行 GWAS分析,鉴定了大量水稻性状相关基因。这些研究极大地推动了以全基因组高密度分子标记和大规模群体表型数据为基础的全基因组选择(Genome-wide selection,GWS)育种技术的应用[75,76]。在稻瘟病抗性基因GWAS研究中,Zhao等[74]也鉴定了5个抗性基因位点。最近,Kang等[77]利用来自 5个国家的 5个稻瘟菌小种对 Zhao等[74]研究中413份水稻材料进行了GWAS分析,鉴定了66个稻瘟病抗性位点,其中53个为新鉴定的位点。这些结果表明,利用GWAS 进行水稻抗病分子育种具备可行性。
3.2 靶基因调控技术育种应用
3.2.1 TALEN和CRISPR技术
TALEN(Transcription activator-like effector nucleases)是新近发展的利用 TAL效应子识别靶基因特定序列,并结合核酸内切酶对靶DNA进行切割的一种基因编辑技术[78,79]。目前已在人类细胞、酵母(Saccharomyces)、斑马鱼(Danio rerio)、大鼠(Rattusnorvegicus)、小鼠(Musculus)、拟南芥和水稻等生物中成功应用。最近,Li等[80]运用TALEN技术准确敲除了水稻白叶枯病感病基因Os11N3,成功获得了抗白叶枯病水稻。与TALEN技术繁琐的载体构建和组装过程相比,最新发展的 CRISPR(Clustered regularly interspaced short palindromic repeats /CRISPR –associated 9,CRISPR/Cas9)技术则弥补了这一缺点[81]。它是一种RNA介导的基因组定点编辑技术,具有构建简单、基因编辑准确率高的特点[82~84]。该技术也已在人类细胞、小鼠、斑马鱼、果蝇(Drosophila)、线虫(Nematoda)、拟南芥及水稻等多个物种中成功应用。TALEN和CRISPR技术的应用将极大地推动水稻基因靶向改良分子育种的发展。
3.2.2 宿主诱导的基因沉默技术
宿主诱导的基因沉默(Host Induced Gene Silence,HIGS)技术是新近发现的一种利用RNAi策略抑制病原菌侵染的技术。该技术是将病原菌致病相关基因的RNAi沉默片段异源表达于寄主植物体内,通过诱导入侵病原菌中靶基因的 RNAi沉默,而抑制病原菌的侵染的[85,86]。目前已在大麦、小麦抗白粉病等研究中获得成功[85,86]。最近,Wang等[87]在水稻中利用 HIGS技术对稻瘟菌相关致病基因进行了初步研究,并获得了 HIGS抗病性增强的转基因水稻,表明HIGS技术可以用于水稻抗病育种。
3.2.3 病原菌诱导的基因表达调控(PIGR)技术
很多抗病相关基因过表达和 RNAi沉默后,除了增强抗病性外,还引起植株矮化、不育或细胞死亡等生长发育连锁负效应,使得转基因植物失去育种利用价值。近来,研究者发现一些受病原菌诱导表达的启动子,如在接种稻瘟菌12 h后显著诱导表达的OsQ16水稻基因启动子[88]。我们可以利用这类启动子获得转基因植物,正常条件下,靶基因不表达或者低水平表达,植株正常生长发育。当病原菌侵染时,靶基因被瞬时过表达或 RNAi沉默,并迅速激活抗病信号,以抑制病菌侵染,而后表达恢复正常,不影响正常生长发育。这种病原菌诱导的基因表达调控(Pathogen induced gene regulation,PIGR)技术,即病原菌诱导的基因过表达(Pathogen induced gene overexpression,PIGO)和病原菌诱导的基因沉默(Pathogen induced gene silencing,PIGS),将在植物抗病育种中具有重要应用前景。
4 展 望
近年来,人们在认识水稻抗稻瘟病分子机制方面取得了一系列重要的进展,特别是克隆和鉴定了一批PTI和ETI信号调控的关键基因。此外,GWAS、TALEN、CRISPR、HIGS等技术的发展,为水稻抗病分子育种提供了新的重要方法,但也带来了一些新的问题和挑战。
(1) 水稻抗稻瘟病持久抗性分子机制是什么?尽管NBS-LRR基因Pb1和Pi35的克隆,在认识水稻持久抗性机制方面取得了重要的突破[47,54]。但对于持久抗稻瘟病分子机制的认识仍有限。此外,Pi35是已克隆主效基因Pish的复等位基因,这为 Wang等[89]揭示的“水稻持久抗病性需要主效基因和微效基因的共同参与”提供了更为直接的证据。但持久抗病调控中主效基因与微效基因介导的信号途径是否具有共性?还有待深入探讨。
(2) R蛋白与Avr蛋白互作后激活的下游信号途径是什么?尽管目前已克隆了22个R基因和10个Avr基因。但是,R蛋白与相应的Avr蛋白识别后激活下游信号途径是什么?R蛋白,特别是NBS-LRR蛋白,直接调控的下游靶标是什么?仍知之甚少。最近研究发现,NBS-LRR蛋白 Pb1与转录因子OsWRKY45直接互作,而激活防卫反应[90],为揭示R蛋白调控的信号途径提供了新的重要证据。但要系统认识水稻R蛋白介导的抗病信号途径,有待深入研究。
(3)Avr基因在水稻中的靶调控基因是什么,它是怎样调控这些靶基因介导的抗病信号的?尽管已有少数研究表明,稻瘟菌Avr基因能够干扰水稻抗病信号,如AvrPiz-t通过干扰E3泛素连接酶APIP6,而抑制水稻PTI反应[68]。但其他Avr基因是否也具有类似功能?其在水稻中的靶标是什么?是怎样调控水稻抗病信号的?都有待系统研究。
(4)Avr基因检测技术能否应用到水稻抗病品种选育及布局上?目前,已定位了超过 40个稻瘟菌Avr基因[91],其中10个已克隆。因此,是否可以利用分子检测技术适时检测田间稻瘟菌群Avr基因频率和变化动态,为地区性抗病品种选育、布局提供科学指导?将成为一项值得开发利用的技术。
(5) 如何将新的分子育种技术与常规育种技术有效结合,以提高抗病品种选择效率,加速育种进程?尽管新发展的GWAS、TALEN、CRISPR、HIGS等为开发新的分子育种技术提供了重要信息。但如何将它们与常规育种程序科学有序结合,发展一套系统、高效、低成本而便于育种家利用的技术体系,仍是一个难题。
[1]Dean R, Van Kan JA, Pretorius ZA, Hammond-Kosack KE,Di Pietro A, Spanu PD, Rudd JJ, Dickman M, Kahmann R,Ellis J, Foster GD. The Top 10 fungal pathogens in molecular plant pathology.Mol Plant Pathol, 2012, 13(4):414–430.
[2]Skamnioti P, Gurr SJ. Against the grain: safeguarding rice from rice blast disease.Trends Biotechnol, 2009, 27(3):141–150.
[3]Jones JDG, Dangl JL. The plant immune system.Nature,2006, 444(7117): 323–329.
[4]Boller T, He SY. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens.Science, 2009,324(5928): 742–744.
[5]Akerley BJ, Cotter PA, Miller JF. Ectopic expression of the flagellar regulon alters development of the Bordetella-host interaction.Cell, 1995, 80(4): 611–620.
[6]Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G. The N terminus of bacterial elongation factor Tu elicits innate immunity inArabidopsisplants.Plant Cell, 2004,16(12): 3496–3507.
[7]Dow M, Newman MA, von Roepenack E. The induction and modulation of plant defense responses by bacterial lipopolysaccharides.Annu Rev Phytopathol, 2000, 38:241–261.
[8]Erbs G, Silipo A, Aslam S, De Castro C, Liparoti V, Flagiello A, Pucci P, Lanzetta R, Parrilli M, Molinaro A. Peptidoglycan and muropeptides from pathogens agrobacterium and xanthomonas elicit plant innate immunity:structure and activity.Chem Biol, 2008, 15(5): 438–448.
[9]Asai T, Tena G, Plotnikova J, Willmann MR, Chiu W-L,Gomez-Gomez L, Boller T, Ausubel FM, Sheen J. MAP kinase signalling cascade inArabidopsisinnate immunity.Nature, 2002, 415(6875): 977–983.
[10]Qi Y, Tsuda K, Glazebrook J, Katagiri F. Physical association of pattern-triggered immunity (PTI) and effector-triggered immunity (ETI) immune receptors in Arabidopsis.Mol Plant Pathol, 2011, 12(7): 702–708.
[11]Thomma BPHJ, Nürnberger T, Joosten MHAJ. Of PAMPs and effectors: the blurred PTI-ETI dichotomy.Plant Cell,2011, 23(1): 4–15.
[12]Felix G, Duran JD, Volko S, Boller T. Plants have a sensitive perception system for the most conserved domain of bacterial flagellin.Plant J, 1999, 18(3): 265–276.
[13]Takai R, Isogai A, Takayama S, Che FS. Analysis of flagellin perception mediated by flg22 receptor OsFLS2 in rice.Mol Plant Microbe Interact, 2008, 21(12): 1635–1642.
[14]Sun Y, Li L, Macho AP, Han Z, Hu Z, Zipfel C, Zhou JM,Chai J. Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex.Science, 2013,342(6158): 624–628.
[15]Lu D, Wu S, Gao X, Zhang Y, Shan L, He P. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity.Proc Natl Acad Sci USA, 2010, 107(1): 496–501.
[16]Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S,Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A. Direct regulation of the NADPH oxidase RBOHD by the PRR-Associated kinase BIK1 during plant immunity.Mol Cell, 2014, 54(1): 43–55.
[17]Li L, Li M, Yu LP, Zhou ZY, Liang XX, Liu ZX, Cai GH,Gao LY, Zhang XJ, Wang YC, Chen S, Zhou JM. The FLS2-Associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity.Cell Host Microbe, 2014, 15(3): 329–338.
[18]Li D, Wang L, Wang M, Xu YY, Luo W, Liu YJ, Xu ZH,Li J, Chong K. Engineering OsBAK1 gene as a molecular tool to improve rice architecture for high yield.Plant Biotechnol J, 2009, 7(8): 791–806.
[19]张慧娟. 磷酸-1-鞘氨醇在植物抗病反应中的作用及水稻和拟南芥 BIK1 在逆境反应中的功能分析[学位论文]. 杭州: 浙江大学, 2009.
[20]Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. CERK1,a LysM receptor kinase, is essential for chitin elicitor signaling inArabidopsis.Proc Natl Acad Sci USA, 2007,104(49): 19613–19618.
[21]Iizasa Ei, Mitsutomi M, Nagano Y. Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1,to chitin in vitro.J Biol Chem, 2010, 285(5): 2996–3004.
[22]Petutschnig EK, Jones AM, Serazetdinova L, Lipka U,Lipka V. The lysin motif receptor-like kinase (LysM-RLK)CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation.J Biol Chem, 2010, 285(37): 28902–28911.
[23]Kaku H, Nishizawa Y, Ishii-Minami N, Akimoto-Tomiyama C, Dohmae N, Takio K, Minami E, Shibuya N.Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor.Proc Natl Acad Sci USA, 2006, 103(29): 11086–11091.
[24]Shimizu T, Nakano T, Takamizawa D, Desaki Y, Ishii-Minami N, Nishizawa Y, Minami E, Okada K, Yamane H,Kaku H, Shibuya N. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice.Plant J, 2010, 64(2): 204–214.
[25]Akamatsu A, Wong HL, Fujiwara M, Okuda J, Nishide K,Uno K, Imai K, Umemura K, Kawasaki T, Kawano Y,Shimamoto K. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity.Cell Host Microbe, 2013, 13(4): 465–476.
[26]Kawano Y, Akamatsu A, Hayashi K, Housen Y, Okuda J,Yao A, Nakashima A, Takahashi H, Yoshida H, Wong HL,Kawasaki T, Shimamoto K. Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity.Cell Host Microbe, 2010, 7(5): 362–375.
[27]Kawano Y, Shimamoto K. Early signaling network in rice PRR-mediated and R-mediated immunity.Curr Opin Plant Biol, 2013, 16(4): 496–504.
[28][28]Liu B, Li JF, Ao Y, Qu J, Li Z, Su J, Zhang Y, Liu J,Feng D, Qi KB, He YM, Wang JF, Wang HB. Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity.Plant Cell, 2012, 24(8): 3406–3419.
[29]Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD).J Mol Biol, 2000, 299(4): 1113–1119.
[30]Silipo A, Erbs G, Shinya T, Dow JM, Parrilli M, Lanzetta R, Shibuya N, Newman MA, Molinaro A. Glyco-conjugates as elicitors or suppressors of plant innate immunity.Glycobiology, 2010, 20(4): 406–419.
[31]Willmann R, Lajunen HM, Erbs G, Newman MA, Kolb D,Tsuda K, Katagiri F, Fliegmann J, Bono JJ, Cullimore JV,Jehle AK, Götz F, Kulik A, Molinaro A, Lipka V, Gust AA,Nürnberger T.Arabidopsislysin-motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection.Proc Natl Acad Sci USA, 2011, 108(49): 19824–19829.
[32]Shinya T, Motoyama N, Ikeda A, Wada M, Kamiya K,Hayafune M, Kaku H, Shibuya N. Functional characterization of CEBiP and CERK1 homologs inArabidopsisand rice reveals the presence of different chitin receptor systems in plants.Plant Cell Physiol, 2012, 53(10): 1696–1706.
[33]Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M,Robatzek S, Lipka V, Maule AJ. LYM2-dependent chitin perception limits molecular flux via plasmodesmata.Proc Natl Acad Sci USA, 2013, 110(22): 9166–9170.
[34]Chen XW, Shang JJ, Chen DX, Lei CL, Zou Y, Zhai WX,Liu GZ, Xu JC, Ling ZZ, Cao G, Ma BT, Wang YP, Zhao XF, Li SG, Zhu LH. AB ‐ lectin re ceptor kinase gene conferring rice blast resistance.Plant J, 2006, 46(5): 794–804.
[35]Fukuoka S, Saka N, Koga H, Ono K, Shimizu T, Ebana K,Hayashi N, Takahashi A, Hirochika H, Okuno K, Yano M.Loss of function of a proline-containing protein confers durable disease resistance in rice.Science, 2009, 325(5943):998–1001.
[36]Wang ZX, Yano M, Yamanouchi U, Iwamoto M, Monna L,Hayasaka H, Katayose Y, Sasaki T. The Pib gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes.Plant J, 1999, 19(1): 55–64.
[37]Bryan GT, Wu K-S, Farrall L, Jia Y, Hershey HP, McA-dams SA, Faulk KN, Donaldson GK, Tarchini R, Valent B.A single amino acid difference distinguishes resistant and susceptible alleles of the rice blast resistance gene Pi-ta.Plant Cell, 2000, 12(11): 2033–2046.
[38]Qu SH, Liu GF, Zhou B, Bellizzi M, Zeng LR, Dai LY,Han B, Wang GL. The broad-spectrum blast resistance gene Pi9 encodes a nucleotide-binding site–leucine-rich repeat protein and is a member of a multigene family in rice.Genetics, 2006, 172(3): 1901–1914.
[39]Zhou B, Qu SH, Liu GF, Dolan M, Sakai H, Lu GD, Bellizzi M, Wang GL. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity toMagnaporthe grisea.Mol Plant Microbe Interact, 2006,19(11): 1216–1228.
[40]Liu X, Lin F, Wang L, Pan Q. The in silico map-based cloning of Pi36, a rice coiled-coil-nucleotide-binding site- eucine-rich repeat gene that confers race-specific resistance to the blast fungus.Genetics, 2007, 176(4): 2541– 2549.
[41]Lin F, Chen S, Que Z, Wang L, Liu X, Pan Q. The blast resistance gene Pi37 encodes a nucleotide binding siteleucine-rich repeat protein and is a member of a resistance gene cluster on rice chromosome 1.Genetics, 2007,177(3): 1871–1880.
[42]Ashikawa I, Hayashi N, Yamane H, Kanamori H, Wu J,Matsumoto T, Ono K, Yano M. Two adjacent nucleotide-binding site-leucine-rich repeat class genes are required to confer Pikm-specific rice blast resistance.Genetics, 2008, 180(4): 2267–2276.
[43]Hayashi K, Yoshida H. Refunctionalization of the ancient rice blast disease resistance genePitby the recruitment of a retrotransposon as a promoter.Plant J, 2009, 57(3):413–425.
[44]Lee SK, Song MY, Seo YS, Kim HK, Ko S, Cao PJ, Suh JP,Yi G, Roh JH, Lee S,An G, Hahn TR, Wang GL, Ronald P,Jeon JS. Rice Pi5-mediated resistance to Magnaporthe oryzae requires the presence of two coiled-coil–nucleotide-binding–leucine-rich repeat genes.Genetics, 2009,181(4): 1627–1638.
[45]Shang J, Tao Y, Chen X, Zou Y, Lei C, Wang J, Li X, Zhao X, Zhang M, Lu Z, Xu J, Cheng Z, Wan J, Zhu L. Identification of a new rice blast resistance gene, Pid3, by genomewide comparison of paired nucleotide-binding site–leucine-rich repeat genes and their pseudogene alleles between the two sequenced rice genomes.Genetics, 2009, 182(4):1303–1311.
[46]Takahashi A, Hayashi N, Miyao A, Hirochika H. Unique features of the rice blast resistance Pish locus revealed by large scale retrotransposon-tagging.BMC Plant Biol, 2010,10(1): 175.
[47]Hayashi N, Inoue H, Kato T, Funao T, Shirota M, Shimizu T, Kanamori H, Yamane H, Hayano-Saito Y, Matsumoto T,Yano M, Takatsuji H. Durable panicle blast‐ resistance gene Pb1 encodes an atypical CC-NBS-LRR protein and was generated by acquiring a promoter through local genome duplication.Plant J, 2010, 64(3): 498–510.
[48]Rai AK, Kumar SP, Gupta SK, Gautam N, Singh NK,Sharma TR. Functional complementation of rice blast resistance gene Pi-k h (Pi54) conferring resistance to diverse strains ofMagnaporthe oryzae.J Plant Biochem Biotech,2011, 20(1): 55–65.
[49]Zhai C, Lin F, Dong ZQ, He XY, Yuan B, Zeng XS, Wang L, Pan QH. The isolation and characterization of Pik, a rice blast resistance gene which emerged after rice domestication.New Phytol, 2011, 189(1): 321–334.
[50]Yuan B, Zhai C, Wang WJ, Zeng XS, Xu XK, Hu HQ, Lin F, Wang L, Pan QH. The Pik-p resistance to Magnaporthe oryzae in rice is mediated by a pair of closely linked CC-NBS-LRR genes.Theor Appl Genet, 2011, 122(5):1017–1028.
[51]Okuyama Y, Kanzaki H, Abe A, Yoshida K, Tamiru M,Saitoh H, Fujibe T, Matsumura H, Shenton M, Galam DC,Undan J, Ito A, Sone T, Terauchi R. A multifaceted genomics approach allows the isolation of the rice Pia ‐blast resistance gene consisting of two adjacent NBS-LRR protein genes.Plant J, 2011, 66(3): 467–479.
[52]Chen J, Shi YF, Liu WZ, Chai RY, Fu YP, Zhuang JY, Wu JL. A Pid3 allele from rice cultivar Gumei2 confers resistance to Magnaporthe oryzae.J Genet Genomics, 2011,38(5): 209–216.
[53]Hua L, Wu JZ, Chen CX, Wu WH, He XY, Lin F, Wang L,Ashikawa I, Matsumoto T, Wang L, Pan QH. The isolation of Pi1, an allele at the Pik locus which confers broad spectrum resistance to rice blast.Theor Appl Genet, 2012,125(5): 1047–1055.
[54]Fukuoka S, Yamamoto SI, Mizobuchi R, Yamanouchi U,Ono K, Kitazawa N, Yasuda N, Fujita Y, Nguyen TTT,Koizumi S, Sugimoto K, Matsumoto T, Yano M. Multiple functional polymorphisms in a single disease resistance gene in rice enhance durable resistance to blast.Scientific Reports, 2014, 4,doi:10.1038/srep04550.
[55]Kang S, Sweigard JA, Valent B. The PWL host specificity gene family in the blast fungus Magnaporthe grisea.Mol Plant Microbe Interact, 1995, 8(6): 939–948.
[56]Sweigard JA, Carroll AM, Kang S, Farrall L, Chumley FG,Valent B. Identification, cloning, and characterization of PWL2, a gene for host species specificity in the rice blast fungus.Plant Cell, 1995, 7(8): 1221–1233.
[57]Orbach MJ, Farrall L, Sweigard JA, Chumley FG, Valent B.A telomeric avirulence gene determines efficacy for the rice blast resistance gene Pi-ta.Plant Cell, 2000, 12(11):2019–2032.
[58]Li W, Wang BH, Wu J, Lu GD, Hu YJ, Zhang X, Zhang ZG, Zhao Q, Feng Q, Zhang HY, Wang ZY, Wang GL,Han B, Wang ZH,Zhou B. The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t.Mol Plant Microbe Interact, 2009, 22(4):411–420.
[59]Miki S, Matsui K, Kito H, Otsuka K, Ashizawa T, Yasuda N, Fukiya S, Sato J, Hirayae K, Fujita Y, Nakajima T,Tomita F,Sone T. Molecular cloning and characterization of the AVR ‐Pia locus from a Japanese field isolate of Magnaporthe oryzae.Mol Plant Pathol, 2009, 10(3):361–374.
[60]Yoshida K, Saitoh H, Fujisawa S, Kanzaki H, Matsumura H, Yoshida K, Tosa Y, Chuma I, Takano Y, Win J, Kamoun S, Terauchi R. Association genetics reveals three novel avirulence genes from the rice blast fungal pathogen Magnaporthe oryzae.Plant Cell, 2009, 21(5): 1573–1591.
[61]Leong SA. The ins and outs of host recognition ofMagnaporthe oryzae// Genomics of Disease Stadler Genetics Symposia Series. New York: Springer, 2008: 199–216.
[62]Cesari S, Thilliez G, Ribot C, Chalvon V, Michel C, Jauneau A, Rivas S, Alaux L, Kanzaki H, Okuyama Y, Morel JB, Fournier E, Tharreau D, Terauchi R, Kroj T. The rice resistance protein pair RGA4/RGA5 recognizes the Magnaporthe oryzae effectors AVR-Pia and AVR1- CO39 by direct binding.Plant Cell, 2013, 25(4): 1463–1481.
[63]Böhnert HU, Fudal I, Dioh W, Tharreau D, Notteghem JL,Lebrun MH. A putative polyketide synthase/peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice.Plant Cell, 2004, 16(9): 2499–2513.
[64]Bao JW, Tang M, Zhu X, Wang H, Jeon JS, Han SS, Zhou B. Cloning of AvrPi9 by genome gomparison of a pair of putative wild and mutant strains: an important step toward the understanding of the mechanism underlying the broad-spectrum resistance mediated by the rice blast resistance gene Pi9. International Rice Blast Conference.Jeju, korea2013
[65]Kou YQ, Zhou B, Naqvi NI. A secreted effector confers avirulence towards Pi9-mediated broad-spectrum blast resistance.International Rice Blast Conference. Jeju, Korea2013.
[66]Jia YL, McAdams SA, Bryan GT, Hershey HP, Valent B.Direct interaction of resistance gene and avirulence gene products confers rice blast resistance.EMBO J, 2000,19(15): 4004–4014.
[67]Kanzaki H, Yoshida K, Saitoh H, Fujisaki K, Hirabuchi A,Alaux L, Fournier E, Tharreau D, Terauchi R. Arms race co-evolution of Magnaporthe oryzae AVR-Pik and rice Pik genes driven by their physical interactions.Plant J, 2012, 72(6): 894–907.
[68]Park CH, Chen S, Shirsekar G, Zhou B, Khang CH,Songkumarn P, Afzal AJ, Ning Y, Wang R, Bellizzi M,Valent B, Wang GL. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 Ubiquitin Ligase APIP6 to suppress pathogen-associated molecular pattern–triggered immunity in rice.Plant Cell, 2012, 24(11): 4748–4762.
[69]Huang XH, Zhao Y, Wei XH, Li CY, Wang AH, Zhao Q, Li WJ, Guo YN, Deng LW, Zhu CR, Fan DL, Lu YQ, Weng QJ, Liu KY, Zhou TY, Jing YF, Si LZ, Dong GJ, Huang T,Lu TT, Feng Q, Qian Q, Li JY, Han B. Genome-wide association study of flowering time and grain yield traits in a worldwide collection of rice germplasm.Nat Genet, 2012,44(1): 32–39.
[70]Huang XH, Lu TT, Han B. Resequencing rice genomes: an emerging new era of rice genomics.Trends Genet, 2013,29(4): 225–232.
[71]Kump KL, Bradbury PJ, Wisser RJ, Buckler ES, Belcher AR, Oropeza-Rosas MA, Zwonitzer JC, Kresovich S,McMullen MD, Ware D, Balint-Kurti PJ, Holland JB.Genome-wide association study of quantitative resistance to southern leaf blight in the maize nested association mapping population.Nat Genet, 2011, 43(2): 163–168.
[72]Tian F, Bradbury PJ, Brown PJ, Hung H, Sun Q,Flint-Garcia S, Rocheford TR, McMullen MD, Holland JB,Buckler ES. Genome-wide association study of leaf architecture in the maize nested association mapping population.Nat Genet, 2011, 43(2): 159–162.
[73]Huang XH, Wei XH, Sang T, Zhao Q, Feng Q, Zhao Y, Li CY, Zhu CR, Lu TT, Zhang ZW, Li M, Fan DL, Guo YL,Wang AH, Wang L, Deng LW, Li WJ, Lu YQ, Weng QJ,Liu KY, Huang Tao, Zhou TY, Jing YF, Li Wei, Lin Zhang,Buckler ES, Qian Q, Zhang QF, Li JY, Han B. Genome-wide association studies of 14 agronomic traits in rice landraces.Nat Genet, 2010, 42(11): 961–967.
[74]Zhao K, Tung CW, Eizenga GC, Wright MH, Ali ML,Price AH, Norton GJ, Islam MR, Reynolds A, Mezey J,McClung AM, Bustamante CD, McCouch SR. Genome-wide association mapping reveals a rich genetic architecture of complex traits inOryza sativa.Nat Commun,2011, 2: 467,doi: 10.1038/ncomms1467.
[75]Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps.Genetics, 2001, 157(4): 1819–1829.
[76]Daetwyler HD, Villanueva B, Bijma P, Woolliams JA. Inbreeding in genome‐ wide selection.J Anim Breed Genet,2007, 124(6): 369–376.
[77]Kang H, Zhang Y, Wang Y, Xiao Y, Wang D, Bellizzi M,Qu S, Korniliev P, Mezey JG, LiuW, Wang Z, Yan S, Li Z,Leung H, McCouch S, Wang GL. Molecular dissection of the complex genetic architecture of rice immunity to the blast fungusMagnaporthe oryzaeusing genome-wide association study. International Rice Blast Conference.Jeju, Korea2013.
[78]Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F,Hummel A, Bogdanove AJ, Voytas DF. Targeting DNA double-strand breaks with TAL effector nucleases.Genetics, 2010, 186(2): 757–761.
[79]Bogdanove AJ, Voytas DF. TAL effectors: customizable proteins for DNA targeting.Science, 2011, 333(6051):1843–1846.
[80]Li T, Liu B, Spalding MH, Weeks DP, Yang B.High-efficiency TALEN-based gene editing produces disease-resistant rice.Nat Biotechnol, 2012, 30(5): 390–392.
[81]Pennisi E. The CRISPR craze.Science, 2013, 341(6148):833–836.
[82]Gaj T, Gersbach CA, Barbas III CF. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering.Trends Biotechnol, 2013, 31(7): 397–405.
[83]Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW,Zhang F, Jaenisch R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering.Cell, 2013, 153(4): 910–918.
[84]Wei CX, Liu JY, Yu ZS, Zhang B, Gao GJ, Jiao RJ.TALEN or Cas9–rapid, efficient and specific choices for genome modifications.J Genet Genomics, 2013, 40(6):281–289.
[85]Nowara D, Gay A, Lacomme C, Shaw J, Ridout C, Douchkov D, Hensel G, Kumlehn J, Schweizer P. HIGS: hostinduced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis.Plant Cell, 2010, 22(9):3130–3141.
[86]Nunes CC, Dean RA. Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies.Mol Plant Pathol,2012, 13(5): 519–529.
[87]Wang MY, Wang XL, Wang GL. Development of a hostinduced gene silencing system to engineer resistant transgenic rice to the rice blast fungusMagnaporthe oryzae.International Rice Blast Conference. Jeju, Korea2013.
[88]王光, 吴智丹, 张磊, 刘凤权, 邵敏. 水稻稻瘟病菌诱导表达启动子 OsQ16p的克隆与功能分析. 作物学报,2012, 38(6): 980–987.
[89]Wang GL, Mackill DJ, Bonman JM, McCouch SR, Champoux MC, Nelson RJ. RFLP mapping of genes conferring complete and partial resistance to blast in a durably resistant rice cultivar.Genetics, 1994, 136(4): 1421–1434.
[90]Inoue H, Hayashi N, Matsushita A, Xinqiong L, Nakayama A, Sugano S, Jiang CJ, Takatsuji H. Blast resistance of CC-NB-LRR protein Pb1 is mediated by WRKY45 through protein–protein interaction.Proc Natl Acad Sci USA, 2013, 110(23): 9577–9582.
[91]Zhang S, Xu JR. Effectors and effector delivery inMagnaporthe oryzae.PLoS Pathog, 2014, 10(1): e1003826.

