谷类作物β-葡聚糖合成酶基因家族的研究进展
袁泓宇, 崔冬丽, John Seymour Heslop Harrison, 刘青
谷类作物-葡聚糖合成酶基因家族的研究进展
袁泓宇1,2,3, 崔冬丽1,2,3, John Seymour Heslop Harrison1,2,4, 刘青1,2,5*
(1. 中国科学院华南植物园,中国科学院植物资源保护与可持续利用重点实验室,广州 510650;2. 华南国家植物园,广州 510650;3. 中国科学院大学,北京 100049;4. 莱斯特大学遗传学和基因组生物学系,莱斯特 LE1 7RH,英国;5. 中国科学院核心植物园,广州 510650)
Beta-葡聚糖是由-(1,3)和-(1,4)糖苷键连接的非纤维素多糖,主要分布在谷类作物籽粒胚乳及糊粉层中,在高尔基体合成,经由囊泡运输到质膜,最终在细胞壁上沉积。通过增加胆汁酸排泄,延迟葡萄糖吸收,-葡聚糖可有效降低胆固醇及血糖水平。Beta-葡聚糖合成酶基因家族成员最早在水稻()中得到鉴定,后在其他作物中陆续被发现。该基因家族包括3个主要成员:、和亚基因家族,起源于不同分支,经过趋同演化,执行合成-葡聚糖的功能。Beta-葡聚糖基因家族成员均受到负选择压力,演化过程中序列高度保守。亚家族基因成员相对较多,常在染色体上形成基因簇,是介导-葡聚糖合成的主效基因。亚家族在叶基部等幼嫩组织中表达水平相对较高,且明显受到光照强度的影响;和亚家族成员较少,其中亚家族在叶尖等成熟组织中的表达水平高,而亚家族在籽粒中有较高的表达水平。该文综述了-葡聚糖合成酶基因家族成员的系统发育关系、表达模式,-葡聚糖合成酶的亚细胞定位,以及作物中的定向育种研究进展,提出-葡聚糖合成酶基因家族在染色体上的精准定位是未来的研究趋势,以期推动染色体工程在作物-葡聚糖定向育种中的应用。
Beta-葡聚糖;谷类作物定向育种;基因家族演化;染色体工程
Beta-葡聚糖是籽粒胚乳和糊粉层细胞壁的主要非纤维素多糖[1],在谷类作物及其他单子叶植物中均有分布[2]。Beta-葡聚糖可增加胆汁酸排泄,延迟葡萄糖吸收[3],降低胆固醇及血糖水平[4],在食品、药品等领域中具有较高的应用价值。最初在水稻()基因组中鉴定到-葡聚糖合成酶基因家族成员亚基因家族[5],并通过异源表达实验,证实亚基因家族编码-葡聚糖合成酶[6]。之后46种被子植物的研究表明,和亚基因家族也具有合成-葡聚糖的功能[7]。
由于对-葡聚糖合成的调控网络,以及物种间-葡聚糖含量差异的遗传因素缺乏精准认知, 作物-葡聚糖育种还停留在传统育种阶段。本文拟通过整理-葡聚糖合成酶基因家族研究进展, 提出未来研究的方向,为作物-葡聚糖定向育种提供理论支撑。
1 Beta-葡聚糖的化学特性和功能应用
1.1 化学结构和溶解度
Beta-葡聚糖是由d-吡喃葡萄糖苷单元通过-(1,3)和-(1,4)糖苷键连接组成的非纤维素多糖, 又名混合线性葡聚糖[Mixed-linkage (1,3;1,4)--d-glucan, MLG],无分支或取代基(图1)。Beta-葡聚糖链中,- (1,4)糖苷键赋予葡聚糖链刚性,而随机分布的-(1,3)糖苷键增加了葡聚糖链空间扭转的灵活性[8]。邻近的糖苷键相同时,葡聚糖链空间扭曲度减弱,使葡聚糖链趋向聚合,即溶解度降低,水溶液凝胶特性增加[9]。
Beta-葡聚糖的精细结构,表现为寡糖聚合度(degree of polymerization, DP)的比值。地衣酶能特异地断开-(1,4)糖苷键还原端,获得多种寡糖产物,其中以-纤维三糖(G4G4G3G)和-纤维四糖(G4 G4G4G3G)为主,两者的比值DP3:DP4即为DP比值[11–12]。当DP比值为1~2.5时,-葡聚糖的溶解度较高[13]。燕麦-葡聚糖含量较高(6%~8%),具有较好的水溶性,是日常饮食中-葡聚糖的理想来源[14](表1)。
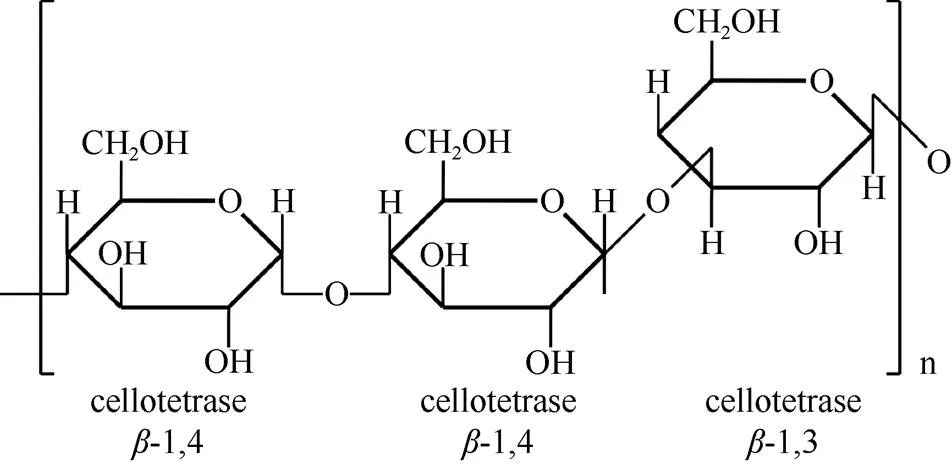
图1 (1,3;1,4)-β-d-葡聚糖化学结构(根据McFarlane等[10]重绘)

表1 作物中β-葡聚糖含量及溶解度
1.2 功能应用
Beta-葡聚糖在植物生长发育过程中发挥重要功能[15]。缺失-葡聚糖的突变体,植株生长速率较低,细胞壁变薄,花药和花丝变形导致雄性不育[16]。Beta-葡聚糖还与葡萄糖的贮藏利用有关[17],植物通过水解细胞壁中的-葡聚糖获得葡萄糖,由维管束运输到其他部位,提供代谢所需的能量[18–19]。
Beta-葡聚糖可有效降低血糖,改善血液中的胰岛素水平[20–21],参与人体胆汁酸的调节、胆固醇代谢、影响肠道微生物组成[22]。在小鼠试验中,-葡聚糖能增强免疫系统应答能力[23],降低呼吸道的易感性[24]。Chang等[25]报道稳定摄入-葡聚糖,可改善腹部脂肪堆积情况,降低人体肥胖度。在食品中添加-葡聚糖,显著降低血糖生成指数,在烘焙类食物及肉奶制品中,常用-葡聚糖替代脂肪[26]。Beta-葡聚糖有良好的皮肤渗透性,临床实验中表现出减少细纹生成[27],抑制微生物慢性感染,促进皮肤损伤后的愈合过程等功效[28],相关产品已投入生产。然而,-葡聚糖对麦芽啤酒酿造会产生不良影响, 发酵过程中残余的-葡聚糖增加过滤成本,并影响口感[29]。在单胃动物的饲料中,-葡聚糖会降低动物胃肠道的消化吸收效率[30]。
2 Beta-葡聚糖合成酶基因家族
2.1 Beta-合成酶基因家族成员
2.1.1亚基因家族
Beta-葡聚糖合成酶基因家族包括3个主要成员:、和亚基因家族,都属于类纤维素合酶(cellulose synthase-like)基因家族()。该基因家族与多种非纤维素多糖的合成有关。早期拟南芥()的研究表明,基因家族包括5个亚家族(~)[31]。2002年,Hazen等[5]在水稻基因组中鉴定到新的家族成员:和亚基因家族。由于双子叶植物中未鉴定到这两个亚基因家族存在,有学者推测和亚基因家族为单子叶植物特有的家族成员。
是首个被证实与-葡聚糖合成有关的亚基因家族。Burton等[6]将与-葡聚糖含量相关的数量性状位点(quantitative trait locus, QTL)比对到水稻基因组中,在7号染色体上鉴定到6个基因组成的基因簇(、、、、和),在8号和10号染色体上鉴定到和基因。通过农杆菌介导法,将3个基因、和转入拟南芥基因组中异源表达,在叶片表皮层细胞壁中检测到微量的-葡聚糖,证实基因参与-葡聚糖合成[6]。
物种间亚基因家族成员的数目存在差异,且部分基因只存在于特定物种中(表2)。Burton等[32]在大麦() 2号染色体上鉴定到与水稻相同的基因簇,除、、外,还包括新的基因, 而、和等基因分布在其他染色体上。Schreiber等[33]在重新组装的大麦基因组中,鉴定出3个在水稻中未发现的基因:、和。
基因的异源表达实验中,-葡聚糖的合成量较低,据此Doblin等[34]认为可能存在其他基因共同调控合成-葡聚糖,如大麦QTL位点附近的基因。通过将转入拟南芥中进行异源表达,Doblin等[34]报道叶片细胞壁中存在-葡聚糖沉积,证实家族参与-葡聚糖合成。亚基因家族在早熟禾亚科(Pooideae)的大麦和燕麦()中仅有单个成员,而在禾本科其他亚科的物种中则有多个成员[35]。
Little等[7]的研究指出,、和亚家族均可介导-葡聚糖的合成[7]。Farrokhi等[36]推测与和亚家族类似,单子叶植物中的亚家族可能参与-葡聚糖合成。其后在双子叶植物中,也发现基因存在,而该基因的功能一直未得到确认[37]。Little等[7]将双子叶植物中的亚家族命名为,目前亚家族仅分布在单子叶植物中。通过异源表达实验,Little等[7]证实亚基因家族成员基因参与-葡聚糖合成,表达活性类似亚家族。
燕麦-葡聚糖合成酶基因家族的研究不多。Newell等[38]比较了燕麦-葡聚糖含量相关的QTL位点,有1个QTL标记与水稻7号染色体上的基因簇具有序列同源性。Zhang等[39]采用同源序列搜索,在燕麦基因组中鉴定到5个基因家族成员(、、、和),与亚家族都仅具有单个成员,而基因簇在燕麦基因组中是否存在有待检测。
2.1.2 主效基因
基因是-葡聚糖合成中的主效基因。化学诱变实验表明,在大麦-葡聚糖缺失突变体中, 存在基因突变[40];敲除实验表明,、和基因敲除后,并不影响大麦籽粒-葡聚糖含量,而敲除基因后,大麦籽粒中检测不到-葡聚糖含量[41];基因过表达后,大麦籽粒-葡聚糖含量平均增长45%,而过表达基因后,大麦籽粒-葡聚糖含量无显著变化[18,42];在转录水平上,大麦籽粒基因的转录峰值时期与胚芽鞘中-葡聚糖含量达到峰值的时期相对应[32,43]。水稻和小麦()中的基因研究也得到了相似结果[44–46]。亚家族其他成员在-葡聚糖合成过程中的功能尚不明确。

表2 作物中β-葡聚糖合成酶基因家族成员
2.2 生物信息学分析
2.2.1 系统发育关系
目前的观点认为,-葡聚糖基因家族出现在约130~140百万年前,早于单双子叶植物的分化时间[7]。Yin等[37]认为-葡聚糖合成酶基因家族的祖先存在于陆地植物的共同祖先中,该家族的分化时间要早于陆地植物的分化。在陆地植物演化过程中,和亚家族在苔藓中丢失,仅残余序列片段。和亚家族的远缘关系说明,-葡聚糖合成酶基因家族来源于多次独立的亚家族分化事件。
Beta-葡聚糖合成酶基因家族成员位于系统发育树的不同分支:亚家族仅分布于部分鸭跖草类(Commelid)植物的分支,可能是由亚家族演化而来。而和亚家族在单子叶植物中广泛存在,其中亚家族是亚家族(真双子叶植物特有)的姊妹分支,亚家族是亚家族(广义双子叶植物特有)的姊妹分支,分别位于和分支[7]。
亚家族包括~共13个成员,成员间的演化关系较为清晰。与基因较早分化出来,其后通过复制事件产生基因簇,该基因簇包含除与外的大部分亚家族基因,并在禾本科物种中高度保守[47]。共线性分析的结果表明,基因簇来源于串联重复事件[48]。复制事件后的基因后代容易发生功能分化,如和基因,其在系统发育树上组成单独的分支,它们可能与木葡聚糖的合成有关[49]。和亚家族成员较少,缺少系统发育关系的专门研究。
Beta-葡聚糖合成酶基因家族成员普遍受到负选择压力,表明作为细胞壁组成成分,-葡聚糖在演化过程中具有保守性[47]。亚家族部分基因受到显著的正选择压力,如、、和基因,受到正选择压力的基因可能与植物的竞争优势有关。和亚家族受到的负选择压力稍弱,存在极高的核苷酸替换速率[47]。目前的研究多数是在大跨度进化框架下,探究-葡聚糖合成酶基因家族的系统发育关系,而着眼于作物及野生近缘种之间精细差别的研究较少。
2.2.2 表达模式
亚家族基因主要在生长旺盛的幼嫩组织中表达。基因簇内基因的表达模式并不相同, 例如基因在胚芽鞘组织中表达水平较高,而基因在叶基部组织中表达水平较高[32–33]。基因在作物的根尖、叶基部、籽粒等幼嫩组织中都具有较高的表达水平,而基因的表达部位尚不清楚[48]。
亚家族在叶尖等成熟组织中具有较高的表达水平,可能与细胞停止生长后,化合物的沉积过程有关[50–51]。二穗短柄草()籽粒萌发过程中,基因的表达水平与基因相似,而大麦和小麦萌发籽粒中并未检测到基因的表达[52]。亚家族表达模式的研究不多,仅在燕麦籽粒中检测到较高的表达水平[39]。对于-葡聚糖合成酶基因家族成员之间的差异表达模式,一种观点认为与组织间-葡聚糖溶解性差异有关[51],其他观点则认为可能是基因家族扩张后功能分化的结果[53]。
Beta-葡聚糖合成酶基因的表达水平受环境因素影响,尤其是光照。当光照增加时,玉米()胚轴中的表达水平增加,而的表达水平降低,推测与光照对生长素的调节有关[54–55]。Zhang等[39]在燕麦-葡聚糖合成酶基因家族的启动子区域(约2 000 bp)中检测到大量的光响应元件,并且叶片中基因的表达水平随光照强度上升而增加,-葡聚糖含量也随之增加[39]。除光照强度之外, 其他环境因素对作物-葡聚糖含量的影响尚未得到评估。
3 Beta-葡聚糖合成酶
Beta-葡聚糖合成酶氨基酸序列主要包含有: (1) 亲水催化功能域(PF00535);(2) 催化功能基序(D、D×D、D和Q××RW),其中Q是谷胺酰胺,R是精氨酸,W是色氨酸,×表示任意氨基酸;(3) 存在多个跨膜结构域(transmembrane helices, TMH)[56](图2)。

图2 Beta-葡聚糖合成酶蛋白二级结构的比较(根据Zhang等[39]重绘)
3.1 催化合成
Beta-葡聚糖合成酶存在两种催化活性,分别合成-(1,4)葡萄糖苷键和-(1,3)葡萄糖苷键。Fincher[57]认为,-葡聚糖合成酶仅具有添加-(1,3)葡萄糖苷键的活性,-(1,4)葡萄糖苷键来源于异源表达实验中,植物细胞自身存在的-(1,4)葡聚糖合酶。Kim等[56]选择无-(1,4)葡聚糖合酶的毕赤酵母作为材料,异源表达基因后成功检测到-葡聚糖的合成。据此认为,-葡聚糖合成酶具有催化两种糖苷键合成的活性。
Beta-葡聚糖合成酶催化-葡聚糖合成的具体过程已得到初步研究。Schwerdt等[47]提出大麦HvCSLF6酶蛋白的同源模型,认为在合成过程中存在明显的跨膜运输和催化中心。Kim等[56]进一步证明BdCSLF6酶蛋白的催化功能域、N-末端和C-末端都暴露在胞质中。并推测-葡聚糖的合成过程为:胞质内的合成底物尿苷二磷酸葡萄糖(UDP- glucose, UDP-Glc),通过暴露在胞质中的催化结构域,不断被添加到葡聚糖链上, 再穿过跨膜螺旋形成的膜孔,被运输到膜的另一侧。而-葡聚糖合成酶如何调控-(1,3)和-(1,4)糖苷键合成比例,以及克服合成过程中空间扭转的合成机制问题,还需更多的研究来解答。
3.2 亚细胞定位
Beta-葡聚糖合成的亚细胞定位争议未决。纤维素[-(1,4)糖苷键链接]和胼胝质[-(1,3)糖苷键链接]在质膜合成[10,58],而果胶和其余非纤维素多糖在高尔基体合成[59]。早期学者根据体外合成实验,认为高尔基体是细胞内的-葡聚糖合成部位[60–61]。随着免疫细胞学方法应用[62],新的研究结果并不支持-葡聚糖在高尔基体上合成,小麦糊粉层细胞中的高尔基体仅有少量的免疫细胞学标记信号[63],大麦胚芽鞘细胞的高尔基体上也检测不到明显信号[64]。2010年,Carpita等[65]提出,-葡聚糖的合成运输是一个动态的过程,-葡聚糖在高尔基体中合成,经由囊泡运输到质膜,最终在细胞壁上沉积。
Beta-葡聚糖合成酶的亚细胞定位也存在不同的观点。Doblin等[35]将CSLH蛋白定位于高尔基体囊泡和内质网上。Kim等[56]将CSLF6蛋白定位于高尔基体,并观察到通过分泌途径的管腔转运。Wilson等[66]在内质网、高尔基体、分泌囊泡和质膜中都发现了CSLF6和CSLH蛋白信号,分布丰度不同。有学者认为,在-葡聚糖分泌和运输过程中,伴随着-葡聚糖合成酶蛋白从高尔基体到内质网的膜系统转移,这也是导致-葡聚糖合成酶蛋白亚细胞定位争议未决的根本原因[67]。
4 作物β-葡聚糖定向育种
Beta-葡聚糖合成酶蛋白上的特定氨基酸位点突变后,可显著影响-葡聚糖含量和精细结构,或可成为作物-葡聚糖定向育种的一种手段。通过构建CSLF6片段嵌合体蛋白,并分析其在本氏烟草()叶片瞬时表达的结果, Jobling等[68]报道HVCSLF6中的I757L氨基酸位点突变,可导致-葡聚糖DP比值明显下降,而在玉米ZMCSLF6中,同一位点突变则会导致DP比值明显上升,实验证明,氨基酸位点突变可能是造成不同作物-葡聚糖水溶性差异的主要原因。Dimitroff等[69]采用相似的方法,观察到SBCSLF6蛋白上G638D氨基酸位点突变后,会引起-葡聚糖产物DP比值变小, 且-葡聚糖含量随之下降。根据-葡聚糖酶蛋白的三维结构,解析特定氨基酸位点突变对-葡聚糖含量和精细结构的影响,是育种关注的研究方向。
染色体工程是一种常见的育种手段,通过附加、代换、消减和易位等染色体操作,改变研究对象的染色体组成,定向调整作物遗传特性[70]。通过染色体工程导入异源-葡聚糖合成酶基因后,如将大麦或山羊草属()物种中包含基因的染色体转移到小麦中,可观察到籽粒中-葡聚糖含量的增高[71–72]。
采用荧光原位杂交技术,实现基因在染色体上的精准定位,可应用于作物染色体工程育种。小麦D亚基因组上的基因,被认为在籽粒-葡聚糖合成中的贡献更大,通过导入异源基因,替换AB亚基因组上的基因后,小麦籽粒中的-葡聚糖含量明显增加[73]。Fogarty等[74]报道在-葡聚糖产量高的燕麦品种中,_C基因表达较低,而在-葡聚糖含量低的燕麦品种中,其表达水平较高,推测C基因组的_C基因在-葡聚糖合成中起负向调节作用,但对于燕麦中基因的分布,还缺乏染色体定位证据。
物种间的-葡聚糖含量的差异,是作物-葡聚糖定向育种研究的基础。燕麦属A、C基因组二倍体物种间存在明显的-葡聚糖含量差异,大西洋燕麦(, AA)的-葡聚糖含量,约为偏肥燕麦(, CC)的4倍[75]。小麦属近缘种中, 山羊草(, DD)籽粒的-葡聚糖含量, 约为乌拉图小麦(AA)的4倍[76]。Garcia-Gimenez等[77]报道,-葡聚糖在籽粒发育过程中,表现为明显的基因型依赖性积累。并且无论在何种生长条件下,高-葡聚糖品种籽粒的-葡聚糖含量, 均高于在同等条件下的低-葡聚糖品种[78]。因此,研究-葡聚糖含量形成的遗传机制,具有十分重要的育种价值。
5 展望
Beta-葡聚糖可以显著降低食品的血糖生成指数,改善人体脂肪堆积情况,对威胁人类健康的高血脂症和高血糖具有防治作用。燕麦中-葡聚糖含量较高,且具有良好的溶解度,是食物中理想的-葡聚糖来源。近年来在功能食品及化妆品领域,燕麦-葡聚糖相关的产品已形成一定的市场规模,而燕麦-葡聚糖育种仍然停留在杂交育种阶段,无法满足市场需求。通过染色体工程导入异源-葡聚糖合成酶基因,可显著改变作物籽粒中-葡聚糖含量。其中-葡聚糖合成酶基因在染色体上的定位, 是检测异源基因是否成功导入的关键。设计特异性探针,完成主效基因的染色体定位,是提高燕麦籽粒中-葡聚糖含量的突破口,可推动染色体工程在燕麦-葡聚糖定向育种中的应用。
随着燕麦属物种基因组数据积累,通过比较基因组学方法,探讨-葡聚糖合成酶基因家族的基因拷贝数目、染色体分布模式等特征与-葡聚糖含量的相关性,为在基因水平解释物种间-葡聚糖含量差异形成的原因提供可能性,也为燕麦属野生种质资源的开发利用提供科技支撑。结合作物不同发育时期的转录组数据,可筛选出与-葡聚糖合成酶基因家族成员共表达的基因,确定关键的调控基因或转录因子,来构建-葡聚糖合成的调控网络。
本文综述了-葡聚糖的化学特性、功能应用,-葡聚糖合成酶基因家族成员的系统发育关系、表达模式、-葡聚糖合成酶的亚细胞定位,及作物-葡聚糖定向育种中的研究进展。并提出开发燕麦属野生种质资源,促进染色体工程在作物-葡聚糖定向育种中的应用,是厘清-葡聚糖合成酶基因家族的调控网络的研究方向。
[1] SHEWRY P R, WAN Y F, HAWKESFORD M J, et al. Spatial distri- bution of functional components in the starchy endosperm of wheat grains [J]. J Cereal Sci, 2020, 91: 102869. doi: 10.1016/j.jcs.2019. 102869.
[2] STINARD P S, NEVINS D J. Distribution of noncellulosic-d- glucans in grasses and other monocots [J]. Phytochemistry, 1980, 19(7): 1467–1468. doi: 10.1016/0031-9422(80)80196-8.
[3] NIRMALA PRASADI P V, JOYE I J. Dietary fibre from whole grains and their benefits on metabolic health [J]. Nutrients, 2020, 12(10): 3045. doi: 10.3390/nu12103045.
[4] SHVACHKO N A, LOSKUTOV I G, SEMILET T V, et al. Bioactive components in oat and barley grain as a promising breeding trend for functional food production [J]. Molecules, 2021, 26(8): 2260. doi: 10. 3390/molecules26082260.
[5] HAZEN S P, SCOTT-CRAIG J S, WALTON J D. Cellulose synthase- like genes of rice [J]. Plant Physiol, 2002, 128(2): 336–340. doi: 10. 1104/pp.010875.
[6] BURTON R A, WILSON S M, HRMOVA M, et al. Cellulose synthase-likegenes mediate the synthesis of cell wall (1,3;1,4)-- d-glucans [J]. Science, 2006, 311(5769): 1940–1942. doi: 10.1126/ science.1122975.
[7] LITTLE A, SCHWERDT J G, SHIRLEY N J, et al. Revised phylogeny of thegene superfamily: Insights into cell wall evolution [J]. Plant Physiol, 2018, 177(3): 1124–1141. doi: 10.1104/ pp.17.01718.
[8] STAUDTE R G, WOODWARD J R, FINCHER G B, et al. Water- soluble (1→3),(1→4)--d-glucans from barley () endosperm: III. Distribution of cellotriosyl and cellotetraosyl residues [J]. Carbohydr Polym, 1983, 3(4): 299–312. doi: 10.1016/0144-8617 (83)90027-9.
[9] MIKKELSEN M S, JESPERSEN B M, LARSEN F H, et al. Molecular structure of large-scale extracted-glucan from barley and oat: Identification of a significantly changed block structure in a high-glucan barley mutant [J]. Food Chem, 2013, 136(1): 130–138. doi: 10.1016/j.foodchem.2012.07.097.
[10] MCFARLANE H E, DÖRING A, PERSSON S. The cell biology of cellulose synthesis [J]. Annu Rev Plant Biol, 2014, 65: 69–94. doi: 10. 1146/annurev-arplant-050213-040240.
[11] BULONE V, SCHWERDT J G, FINCHER G B. Co-evolution of enzymes involved in plant cell wall metabolism in the grasses [J]. Front Plant Sci, 2019, 10: 1009. doi: 10.3389/fpls.2019.01009.
[12] CHANG S C, SALDIVAR R K, LIANG P H, et al. Structures, biosynthesis, and physiological functions of (1,3;1,4)--d-glucans [J]. Cells, 2021, 10(3): 510. doi: 10.3390/cells10030510.
[13] LAZARIDOU A, BILIADERIS C G. Molecular aspects of cereal- glucan functionality: Physical properties, technological applications and physiological effects [J]. J Cereal Sci, 2007, 46(2): 101–118. doi: 10.1016/j.jcs.2007.05.003.
[14] KIM H J, WHITE P J. Impact of the molecular weight, viscosity, and solubility of-glucan onoat starch digestibility [J]. J Agric Food Chem, 2013, 61(13): 3270–3277. doi: 10.1021/jf305348j.
[15] KIEMLE S N, ZHANG X, ESKER A R, et al. Role of (1,3)(1,4)-- glucan in cell walls: Interaction with cellulose [J]. Biomacromolecules, 2014, 15(5): 1727–1736. doi: 10.1021/bm500 1247.
[16] BAIN M, VAN DE MEENE A, COSTA R, et al. Characterisation of cellulose synthase like F6 () mutants shows altered carbon metabolism in-d-(1,3;1,4)-glucan deficient grain in[J]. Front Plant Sci, 2021, 11: 602850. doi: 10.3389/fpls. 2020.602850.
[17] MORRALL P, BRIGGS D E. Changes in cell wall polysaccharides of germinating barley grains [J]. Phytochemistry, 1978, 17(9): 1495–1502. doi: 10.1016/S0031-9422(00)94628-4.
[18] BURTON R A, COLLINS H M, KIBBLE N A J, et al. Over-expression of specificcellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)--d-glucans and alters their fine structure [J]. Plant Biotechnol J, 2011, 9(2): 117–135. doi: 10. 1111/j.1467-7652.2010.00532.x.
[19] BURTON R A, FINCHER G B. Current challenges in cell wall biology in the cereals and grasses [J]. Front Plant Sci, 2012, 3: 130. doi: 10. 3389/fpls.2012.00130.
[20] SCHUSTER J, BENINCÁ G, VITORAZZI R, et al. Effects of oats on lipid profile, insulin resistance and weight loss [J]. Nutr Hosp, 2015, 32(5): 2111–2116. doi: 10.3305/nh.2015.32.5.9590.
[21] YANG C J, CHEN M S, DAI T T, et al. Research advances in func- tional properties and application of oat-glucan [J]. J Chin Inst Food Sci Technol, 2021, 21(6): 301–311. [杨成峻, 陈明舜, 戴涛涛, 等. 燕麦-葡聚糖功能与应用研究进展 [J]. 中国食品学报, 2021, 21(6): 301–311. doi: 10.16429/j.1009-7848.2021.06.036.]
[22] JOYCE S A, KAMIL A, FLEIGE L, et al. The cholesterol-lowering effect of oats and oat beta glucan: Modes of action and potential role of bile acids and the microbiome [J]. Front Nutr, 2019, 6: 171. doi: 10. 3389/fnut.2019.00171.
[23] PAN W, HAO S S, ZHENG M X, et al. Oat-derived-glucans induced trained immunity through metabolic reprogramming [J]. Inflammation, 2020, 43(4): 1323–1336. doi: 10.1007/s10753-020-01211-2.
[24] CHEN O, MAH E, DIOUM E, et al. The role of oat nutrients in the immune system: A narrative review [J]. Nutrients, 2021, 13(4): 1048. doi: 10.3390/nu13041048.
[25] CHANG H C, HUANG C N, YEH D M, et al. Oat prevents obesity and abdominal fat distribution, and improves liver function in humans [J]. Plant Foods Hum Nutr, 2013, 68(1): 18–23. doi: 10.1007/s11130-013- 0336-2.
[26] ZHU F M, DU B, XU B J. A critical review on production and industrial applications of beta-glucans [J]. Food Hydrocoll, 2016, 52: 275–288. doi: 10.1016/j.foodhyd.2015.07.003.
[27] FU H, WU D, BING X, et al. Study on the skin care effect of oat- glucan extracted by bidirectional fermentation [J]. China Surfactant Deterg Cosmet, 2021, 51(4): 324–330. [付豪, 吴迪, 邴雪, 等. 双向发酵提取燕麦-葡聚糖的护肤功效研究 [J]. 日用化学工业, 2021, 51(4): 324–330. doi: 10.3969/j.issn.1001-1803.2021.04.010.]
[28] RIBEIRO D M L, CARVALHO JÚNIOR A R, VALE DE MACEDO G H R, et al. Polysaccharide-based formulations for healing of skin- related wound infections: Lessons from animal models and clinical trials [J]. Biomolecules, 2019, 10(1): 63. doi: 10.3390/biom 10010063.
[29] GUPTA M, ABU-GHANNAM N, GALLAGHAR E. Barley for brewing: Characteristic changes during malting, brewing and applications of its by-products [J]. Compr Rev Food Sci Food Saf, 2010, 9(3): 318–328. doi: 10.1111/j.1541-4337.2010.00112.x.
[30] BRENNAN C S, CLEARY L J. The potential use of cereal (1→3;1→4)--d-glucans as functional food ingredients [J]. J Cereal Sci, 2005, 42(1): 1–13. doi: 10.1016/j.jcs.2005.01.002.
[31] RICHMOND T A, SOMERVILLE C R. The cellulose synthase super- family [J]. Plant Physiol, 2000, 124(2): 495–498. doi: 10.1104/pp.124. 2.495.
[32] BURTON R A, JOBLING S A, HARVEY A J, et al. The genetics and transcriptional profiles of the cellulose synthase-likegene family in barley [J]. Plant Physiol, 2008, 146(4): 1821–1833. doi: 10.1104/pp.107.114694.
[33] SCHREIBER M, WRIGHT F, MACKENZIE K, et al. The barley genome sequence assembly reveals three additional members of the(1,3;1,4)--glucan synthase gene family [J]. PLoS One, 2014, 9 (3): e90888. doi: 10.1371/journal.pone.0090888.
[34] DOBLIN M S, PETTOLINO F A, WILSON S M, et al. A barley cellulose synthase-likegene mediates (1,3;1,4)-beta-d-glucan synthesis in transgenic[J]. Proc Natl Acad Sci USA, 2009, 106(14): 5996–6001. doi: 10.1073/pnas.0902019106.
[35] WU B, ZHANG Z W. Cloning and analysis of β-glucan synthase geneinL. [J]. Acta Agron Sin, 2011, 37(4): 723–728. [吴斌, 张宗文. 燕麦葡聚糖合酶基因的克隆及特征分析 [J]. 作物学报, 2011, 37(4): 723–728. doi: 10.3724/SP.J.1006.2011. 00723.]
[36] FARROKHI N, BURTON R A, BROWNFIELD L, et al. Plant cell wall biosynthesis: Genetic, biochemical and functional genomics approaches to the identification of key genes [J]. Plant Biotechnol J, 2006, 4(2): 145–167. doi: 10.1111/j.1467-7652.2005.00169.x.
[37] YIN Y B, HUANG J L, XU Y. The cellulose synthase superfamily in fully sequenced plants and algae [J]. BMC Plant Biol, 2009, 9(1): 99. doi: 10.1186/1471-2229-9-99.
[38] NEWELL M A, ASORO F G, SCOTT M P, et al. Genome-wide association study for oat (L.) beta-glucan concentration using germplasm of worldwide origin [J]. Theor Appl Genet, 2012, 125(8): 1687–1696. doi: 10.1007/s00122-012-1945-0.
[39] ZHANG J, YAN L, LIU M X, et al. Analysis of-d-glucan bio- synthetic genes in oat reveals glucan synthesis regulation by light [J]. Ann Bot, 2021, 127(3): 371–380. doi: 10.1093/aob/mcaa185.
[40] TAKETA S, YUO T, TONOOKA T, et al. Functional characterization of barley betaglucanless mutants demonstrates a unique role forin (1,3;1,4)--d-glucan biosynthesis [J]. J Exp Bot, 2012, 63(1): 381– 392. doi: 10.1093/jxb/err285.
[41] GARCIA-GIMENEZ G, BARAKATE A, SMITH P, et al. Targeted mutation of barley (1,3;1,4)--glucan synthases reveals complex relationships between the storage and cell wall polysaccharide content [J]. Plant J, 2020, 104(4): 1009–1022. doi: 10.1111/tpj.14977.
[42] LIM W L, COLLINS H M, SINGH R R, et al. Method for hull-less barley transformation and manipulation of grain mixed-linkage beta- glucan [J]. J Integr Plant Biol, 2018, 60(5): 382–396. doi: 10.1111/ jipb.12625.
[43] GIBEAUT D M, PAULY M, BACIC A, et al. Changes in cell wall polysaccharides in developing barley () coleoptiles [J]. Planta, 2005, 221(5): 729–738. doi: 10.1007/s00425-005-1481-0.
[44] NEMETH C, FREEMAN J, JONES H D, et al. Down-regulation of thegene results in decreased (1,3;1,4)--d-glucan in endosperm of wheat [J]. Plant Physiol, 2010, 152(3): 1209–1218. doi: 10.1104/pp. 109.151712.
[45] VEGA-SÁNCHEZ M E, VERHERTBRUGGEN Y, CHRISTENSEN U, et al. Loss of cellulose synthase-like F6 function affects mixed-linkage glucan deposition, cell wall mechanical properties, and defense responses in vegetative tissues of rice [J]. Plant Physiol, 2012, 159(1): 56–69. doi: 10.1104/pp.112.195495.
[46] MARCOTULI I, COLASUONNO P, BLANCO A, et al. Expression analysis of cellulose synthase-like genes in durum wheat [J]. Sci Rep, 2018, 8(1): 15675. doi: 10.1038/s41598-018-34013-6.
[47] SCHWERDT J G, MACKENZIE K, WRIGHT F, et al. Evolutionary dynamics of the cellulose synthase gene superfamily in grasses [J]. Plant Physiol, 2015, 168(3): 968–983. doi: 10.1104/pp.15.00140.
[48] WANG L Q, GUO K, LI Y, et al. Expression profiling and integrative analysis of thesuperfamily in rice [J]. BMC Plant Biol, 2010, 10(1): 282. doi: 10.1186/1471-2229-10-282.
[49] LITTLE A, LAHNSTEIN J, JEFFERY D W, et al. A novel (1,4)-- linked glucoxylan is synthesized by members of the cellulose synthase- like F gene family in land plants [J]. ACS Cent Sci, 2019, 5(1): 73–84. doi: 10.1021/acscentsci.8b00568.
[50] ERMAWAR R A, COLLINS H M, BYRT C S, et al. Distribution, structure and biosynthetic gene families of (1,3;1,4)--glucan in[J]. J Integr Plant Biol, 2015, 57(4): 429–445. doi: 10. 1111/jipb..
[51] ERMAWAR R A, COLLINS H M, BYRT C S, et al. Genetics and physiology of cell wall polysaccharides in the model C4grass,spp [J]. BMC Plant Biol, 2015, 15(1): 236. doi: 10.1186/s 12870-015-0624-0.
[52] CHRISTENSEN U, ALONSO-SIMON A, SCHELLER H V, et al. Characterization of the primary cell walls of seedlings of: A potential model plant for temperate grasses [J]. Phytochemistry, 2010, 71(1): 62–69. doi: 10.1016/j.phytochem.2009. 09.019.
[53] ZHANG G B, GE C X, XU P P, et al. The reference genome ofilluminates the evolution of Saccharinae [J]. Nat Plants, 2021, 7(5): 608–618. doi: 10.1038/s41477-021-00908-y.
[54] VAN ERP H, WALTON J D. Regulation of the cellulose synthase-like gene family by light in the maize mesocotyl [J]. Planta, 2009, 229(4): 885–897. doi: 10.1007/s00425-008-0881-3.
[55] DARAS G, TEMPLALEXIS D, AVGERI F, et al. Updating insights into the catalytic domain properties of plant cellulose synthase (CesA) and cellulose synthase-like (Csl) proteins [J]. Molecules, 2021, 26(14): 4335. doi: 10.3390/molecules26144335.
[56] KIM S J, ZEMELIS S, KEEGSTRA K, et al. The cytoplasmic locali- zation of the catalytic site of CSLF6 supports a channeling model for the biosynthesis of mixed-linkage glucan [J]. Plant J, 2015, 81(4): 537– 547. doi: 10.1111/tpj.12748.
[57] FINCHER G B. Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses [J]. Plant Physiol, 2009, 149 (1): 27–37. doi: 10.1104/pp.108.130096.
[58] SCHELLER H V, ULVSKOV P. Hemicelluloses [J]. Annu Rev Plant Biol, 2010, 61: 263–289. doi: 10.1146/annurev-arplant-042809-112315.
[59] ATMODJO M A, HAO Z Y, MOHNEN D. Evolving views of pectin biosynthesis [J]. Annu Rev Plant Biol, 2013, 64: 747–779. doi: 10. 1146/annurev-arplant-042811-105534.
[60] GIBEAUT D M, CARPITA N C. Synthesis of (1→3),(1→4)-beta-d- glucan in the golgi apparatus of maize coleoptiles [J]. Proc Natl Acad Sci USA, 1993, 90(9): 3850–3854. doi: 10.1073/pnas.90.9.3850.
[61] URBANOWICZ B R, RAYON C, CARPITA N C. Topology of the maize mixed linkage (1→3),(1→4)--d-glucan synthase at the golgi membrane [J]. Plant Physiol, 2004, 134(2): 758–768. doi: 10.1104/pp. 103.032011.
[62] MEIKLE P J, HOOGENRAAD N J, BONIG I, et al. A (1→3,1→4)--glucan-specific monoclonal antibody and its use in the quantitation and immunocytochemical location of (1→3,1→4)--glucans [J]. Plant J, 1994, 5(1): 1–9. doi: 10.1046/j.1365-313x. 1994.5010001.x.
[63] PHILIPPE S, SAULNIER L, GUILLON F. Arabinoxylan and (1→3), (1→4)--glucan deposition in cell walls during wheat endosperm development [J]. Planta, 2006, 224(2): 449–461. doi: 10.1007/s00425- 005-0209-5.
[64] WILSON S M, BURTON R A, DOBLIN M S, et al. Temporal and spatial appearance of wall polysaccharides during cellularization of barley () endosperm [J]. Planta, 2006, 224(3): 655– 667. doi: 10.1007/s00425-006-0244-x.
[65] CARPITA N C, MCCANN M C. The maize mixed-linkage (1→3), (1→4)--d-glucan polysaccharide is synthesized at the golgi membrane [J]. Plant Physiol, 2010, 153(3): 1362–1371. doi: 10.1104/pp.110. 156158.
[66] WILSON S M, HO Y Y, LAMPUGNANI E R, et al. Determining the subcellular location of synthesis and assembly of the cell wall polysaccharide (1,3;1,4)--d-glucan in grasses [J]. Plant Cell, 2015, 27(3): 754–771. doi: 10.1105/tpc.114.135970.
[67] KIM S J, ZEMELIS-DURFEE S, JENSEN J K, et al. In the grass species, the production of mixed-linkage (1,3;1,4)--glucan (MLG) occurs in the golgi apparatus [J]. Plant J, 2018, 93(6): 1062–1075. doi: 10.1111/tpj.13830.
[68] JOBLING S A. Membrane pore architecture of the CslF6 protein controls (1–3,1–4)--glucan structure [J]. Sci Adv, 2015, 1(5): e1500069. doi: 10.1126/sciadv.1500069.
[69] DIMITROFF G, LITTLE A, LAHNSTEIN J, et al. (1,3;1,4)--glucan biosynthesis by the CSLF6 enzyme: Position and flexibility of catalytic residues influence product fine structure [J]. Biochemistry, 2016, 55(13): 2054–2061. doi: 10.1021/acs.biochem.5b01384.
[70] YANG X Y, SU H D, ZHANG M Z, et al. Polyploidization and domestication [J]. Sci Sin Vitae, 2021, 51(10): 1457–1466. [杨学勇, 苏汉东, 张梦卓, 等. 多倍化和驯化研究进展与展望 [J]. 中国科学: 生命科学, 2021, 51(10): 1457–1466. doi: 10.1360/SSV-2021-0220.]
[71] RAKSZEGI M, MOLNÁR I, LOVEGROVE A, et al. Addition ofU and M chromosomes affects protein and dietary fiber content of wholemeal wheat flour [J]. Front Plant Sci, 2017, 8: 1529. doi: 10.3389/fpls.2017.01529.
[72] DANILOVA T V, FRIEBE B, GILL B S, et al. Development of a complete set of wheat-barley group-7 robertsonian translocation chro- mosomes conferring an increased content of-glucan [J]. Theor Appl Genet, 2018, 131(2): 377–388. doi: 10.1007/s00122-017-3008-z.
[73] DANILOVA T V, POLAND J, FRIEBE B. Production of a complete set of wheat-barley group-7 chromosome recombinants with increased grain-glucan content [J]. Theor Appl Genet, 2019, 132(11): 3129– 3141. doi: 10.1007/s00122-019-03411-3.
[74] FOGARTY M C, SMITH S M, SHERIDAN J L, et al. Identification of mixed linkage-glucan quantitative trait loci and evaluation ofhomoeologs in hexaploid oat [J]. Crop Sci, 2020, 60(2): 914– 933. doi: 10.1002/csc2.20015.
[75] TANG X Q. Determination of-glucan content andgene cloning amongspecies [D]. Ya’an: Sichuan Agricultural University, 2014. [唐雪琴. 燕麦属物种-葡聚糖含量测定及基因克隆 [D]. 雅安: 四川农业大学, 2014.]
[76] MARCOTULI I, COLASUONNO P, CUTILLO S, et al.-glucan content in a panel ofandgenotypes [J]. Genet Resour Crop Evol, 2019, 66(4): 897–907. doi: 10.1007/s10722-019- 00753-1.
[77] GARCIA-GIMENEZ G, RUSSELL J, AUBERT M K, et al. Barley grain (1,3;1,4)--glucan content: Effects of transcript and sequence variation in genes encoding the corresponding synthase and endohy- drolase enzymes [J]. Sci Rep, 2019, 9(1): 17250. doi: 10.1038/s41598- 019-53798-8.
[78] DOEHLERT D C, MCMULLEN M S, HAMMOND J J. Genotypic and environmental effects on grain yield and quality of oat grown in North Dakota [J]. Crop Sci, 2001, 41(4): 1066–1072. doi: 10.2135/ cropsci2001.4141066x.
Research Progress of-Glucan Synthase Gene Families in Cereal Crops
YUAN Hongyu1,2,3, CUI Dongli1,2,3, John Seymour Heslop HARRISON1,2,4, LIU Qing1,2,5*
(1. Key Laboratory of Plant Resources Conservation and Sustainable Utilization, South China Botanical Garden, Chinese Academy of Sciences,Guangzhou 510650, China; 2. South China National Botanical Garden,Guangzhou 510650, China; 3. University of Chinese Academy of Sciences,Beijing100049, China; 4. Department of Genetics and Genome Biology, University of Leicester, Leicester, LE1 7RH, UK; 5. Center for Conservation Biology, Core Botanical Gardens, Chinese Academy of Sciences,Guangzhou510650, China)
Beta-glucan is a noncellulosic polysaccharide linked by-(1,3) and-(1,4) glycosidic bonds, mainly distributed in the endosperm and aleurone layer of cereal crop grains with the synthesis in the Golgi apparatus, transportation to the plasma membrane by vesicles, and deposition in the cell wall. Beta-glucan is effective in reducing cholesterol and blood sugar levels by increasing bile acid excretion and delaying glucose absorption. Members of the-glucan synthase gene families were firstly identified in rice (), and subsequently discovered in other cereal crops. There are three main subfamilies (,and) in-glucan synthase with the formation via convergent evolution. These three subfamilies originated from different clades and evolved their respective functions independently.During evolution, the purifying selection pressure resulted in the high conservation of sequences for members of-glucan synthase gene families.subfamily members are relatively large and often form gene clusters on chromosomes, andis the key gene mediating-glucan synthesis.subfamily members showed the relatively high expression levels in young tissues such as leaf bases, and they were affected by light intensity obviously. There are relatively few members inandsubfamilies,genes presented the relatively high expression levels in mature tissues such as leaf tips, whilegenes showed the relatively high expression levels in young one like grains. The research progress on the phylogenetic relationships for members of-glucan synthase gene families, the subcellular localization of-glucan synthase, and the directional breeding in cereal crops were summarized. The accurate localization of-glucan synthase genes on chromosomes is prospected for the future research. The review aims to promote the directionally breeding of cereal crops with high-glucan content by the chromosome engineering.
Beta-glucan; Cereal crop directional breeding; Gene family evolution; Chromosome engineering
10.11926/jtsb.4645
2022-03-30
2022-05-08
国家自然科学基金项目(32070359);广东省自然科学基金面上项目(2021A1515012410);科学技术部高端外国专家引进计划(G2021030013);中国科学院华南植物园海外知名学者项目(No. Y861041001)资助
This work was supported by the National Natural Science Foundation of China (Grant No. 32070359), the project for Natural Science in Guangdong (Grant No. 2021A1515012410), the Project for High-end Foreign Expert of Ministry of Science and Technology of the People’s Republic of China (Grant No. G2021030013), and the Project for Overseas Distinguished Scholar of SCBG (Grant No. Y861041001).
袁泓宇(1998年生),男,在读硕士生,主要从事作物野生种基因组资源和分子细胞遗传学研究。E-mail: yuanhongyu@scbg.ac.cn
通讯作者Corresponding author.E-mail: liuqing@scbg.ac.cn

