海马齿根系响应盐胁迫的转录组分析
李卫锦 钟才荣 张颖 袁长春 李仁茂
摘要:【目的】對盐胁迫下海马齿根系进行转录组测序分析,挖掘海马齿根系耐盐相关基因,为揭示海马齿耐盐的分子机制提供参考。【方法】利用Illumina测序技术对0 mmol/L NaCl(对照组)和400 mmol/L NaCl胁迫处理(盐胁迫处理组)下的海马齿根系进行转录组测序分析,从中筛选出差异表达基因,选取13个基因进行实时荧光定量PCR(qRT-PCR)检测,以验证转录组数据的可靠性。【结果】在海马齿根系转录组中共鉴定出305145个转录本,平均长度为622 bp,其中,对照组有146177个长度>300 bp的转录本,盐胁迫处理组有72173个长度>300 bp的转录本;共有65535条Unigenes在Nr、GO、Swiss-Prot、COG和KEGG五大数据库注释成功,占Unigenes总数的52.36%。对照组和盐胁迫处理组共有65535个差异Unigenes,其中,有182个热休克蛋白基因。对照组和盐胁迫处理组间共有24042个差异表达基因,从中选取13个基因进行qRT-PCR检测,结果显示,9个基因表达上调,其余4个基因表达下调,与转录组测序结果一致。24042个差异表达基因中,共有10106个显著差异基因富集到129条代谢通路,其中富集程度排名前10的代谢途径为核糖体、次级代谢生物合成、RNA转运、内吞作用、剪接体、甘油磷脂代谢、内质网加工、吞噬、醚脂类代谢和植物-病原体相互作用,参与盐胁迫相关的硫代谢、脯氨酸积累、活性氧(ROS)代谢、与盐胁迫相关的钙信号通路和过氧化氢代谢等途径的差异基因上调。【结论】在盐胁迫下海马齿差异表达基因如小分子量热激蛋白基因、抗氧化酶相关基因及与离子交换相关基因发挥了重要调控作用。
关键词: 海马齿;根;盐胁迫;转录组;耐盐基因
中图分类号: S156.4 文献标志码: A 文章编号:2095-1191(2022)03-0693-11
Analysis of the root transcriptomes in Sesuvium portulacastrum respond to salt stress
LI Wei-jin ZHONG Cai-rong ZHANG Ying YUAN Chang-chun LI Ren-mao
(1 School of Life Science and Technology, Lingnan Normal University, Zhanjiang,Guangdong 524048, China;
2 Hainan Academy of Forestry, Haikou, Hainan 571100, China)
Abstract:【Objective】To conduct transcriptome sequencing analysis on Sesuvium portulacastrum under salt tole-rance, and to find out genes related to salt tolerance in the root of S. portulacastrum, so as to provide reference for studying molecular mechanism of S. portulacastrum. 【Method】Illumina sequencing technology was applied to compare and analyze the transcriptomesand the differentially expressed genes (DEGs) related to salt tolerance inroots of S. portulacastrum under 0 mmol/L NaCl (control group) and 400 mmol/L NaCl salt stress (salt stress treatment group),respectively. Thirteen DEGs were selected to verify the reliability of transcriptome data by using quantitative real-time PCR(qRT-PCR) analysis. 【Result】 A total of 305145 transcripts with an average length of 622 bp were identified in the roots of S. portulacastrum transcriptome. In the control group, 146177 transcripts were greater than 300 bp; in the salt treatment group,72173 transcripts were greater than 300 bp. A total of 65535 unigenes were successfully annotated in the five databases of Nr, GO, Swiss-Prot, COG and KEGG, accounting for 52.36% of the total unigenes and including 182 heat shock protein genes. A number of 24042 DEGs between the control group and the salt stress treatment group were identified. Thirteen candidate genes were selected for qRT-PCR analysis, and the result showed that 9 genes were up-regulated and 4 genes were down-regulated, which was consistent with transcriptome sequencing analysis. Among the 24042 DEGs,10106 significant DEGs were enriched in 129 metabolic pathways. The top 10 enriched metabolic pathways were ribosome, biosynthesis of secondary metabolites, RNA transport, endocytosis, spliceosome, glycerophospholipid metabolism, protein processing in endoplasmic reticulum, phagocytosis, ether lipid metabolism and plant-pathogen interaction path. Differential genes in sulfur metabolism, proline accumulation, eactive oxygen species(ROS) metabolism, calcium signaling pathway and hydrogen peroxide metabolism related to salt stress were up-regulated. 【Conclusion】Under salt stress,DEGs in S. portulacastrum, such as small molecular heat shock protein genes, genes related to anti-oxidation and genes related to ion exchange, play an important role in regulation.1D9152E6-FF06-4DC8-B029-2C5CAEBFEE6C
Key words: Sesuvium portulacastrum; root; salt stress; transcriptome; genes related to salt tolerance
Foundation items:Special Basic Scientific Research Project of HainanTechnological Innovation in Scientific Research Institutes(KYYS-021-13); Zhanjiang Science and Technology Plan Project (2018A03024); Talent Project of Lingnan Normal University (ZL2003); University-level Project of Lingnan Normal University (1170918174)
0 引言
【研究的意义】海马齿(Sesuvium portulacastrum)为番杏科(Aizoaceae)海馬齿属(Sesuvium L.)多年生双子叶盐生植物,其作为红树林伴生植物,通常生长在世界各地沿海和内陆的沙滩上(Yi et al.,2014;Chang et al.,2016),具有耐盐雾、砂洗、贫瘠和耐高温的特性(Rabhi et al.,2010a)。海马齿属于两性生殖的盐生植物,具有独特的耐盐特性,不仅能产生大量的生物量,可通过细胞和组织积累大量的Na+,可达872 mg/株,且在浓度高达800 mmol/L的NaCl溶液中正常生长,利用该特性可实现盐渍土壤的改良(Rabhi et al.,2010b;Chang et al.,2016)。目前全球约20%的可耕地和7%的土地受到盐渍化危害(Rizwan et al.,2015)。因此,海马齿不仅对盐碱地有修复功能,还能固定沙丘、修复污染海岛和海岸带生态环境(范伟等,2010;Lokhande et al.,2013;丁国华等,2020)。因此,开展在盐胁迫下海马齿根系的转录组测序分析,挖掘响应盐胁迫的功能基因,对探究海马齿对盐响应的分子机制、改良农作物耐盐性及解决土壤盐渍化具有重要的意义。【前人研究进展】盐胁迫对海马齿的影响研究主要集中在海马齿的形态结构基础(Yi et al.,2014;Chang et al.,2016)、生理特性(Rabhi et al.,2010a;Kannan et al.,2013)及分子调控机制等方面(Ghnaya et al.,2013)。研究发现,在盐胁迫下,海马齿叶片能保持足够的气体交换和色素组成(Rabhi et al.,2010b),且叶中参与离子结合、质子转运、光合作用和ATP合成的相关基因差异表达(Kannan et al.,2013;Yi et al.,2014)。目前耐盐相关基因已有较多报道,如海马齿的果糖-1,6-二磷酸醛缩酶基因(SpFBA)表达可提高海马齿对盐的耐受性(Fan et al.,2009)。甜菜的碱醛脱氢酶基因(SpBADH)表达产物可减少H2O2、增加脯氨酸和激活抗氧化酶,以改善活性氧(ROS)清除,提高植物对干旱或者渗透胁迫的耐受性(Yang et al.,2015)。水通道蛋白基因(SpAQP1)可通过增强植物的抗氧化性来提高其耐盐性(Chang et al.,2016)。Na+/H+逆向转运基因(SpNHX1)是响应盐胁迫的关键基因(Zhou et al.,2018)。盐超敏感基因1(SpSOS1)和H+-ATP基因(SpAHA1)共表达可提高拟南芥的耐盐性(Ji et al.,2013;Fan et al.,2019)。随着测序技术的快速发展,高通量转录组测序已成为一种快速、高效的基因表达研究方法(Bazakos et al.,2015)。至今,已对大量植物的盐敏感品种和耐盐品种进行转录组测序分析的研究报道,例如Taji等(2004)利用比较基因组学研究拟南芥和盐芥的耐盐基因,结果发现盐芥耐盐的原因可能是Fe-SOD、P5CS、PDF1.2、AtNCED、P-protein、β-葡萄糖苷酶基因和SOS1基因共表达的结果;Rabello等(2008)从旱稻中鉴定出22种可能与耐旱相关的蛋白;Qiu等(2011)从杨树中鉴定出与盐胁迫相关的脱落酸(ABA)合成基因;且Sun等(2010)研究发现,番茄耐盐品种的SOS途径更活跃,水杨酸结合蛋白2基因(SABP2)在其耐盐机制中可能发挥重要调控作用;Zahaf等(2012)研究发现,苜蓿bHLH转录因子可能在盐胁迫中发挥重要作用;Ma等(2013)从盐角草中鉴定出大量参与离子稳态和渗透调节相关基因;Zhang等(2014)研究发现,SnRK2、PYL、PP2C等差异表达基因与ABA的信号转导途径相关;Bazakos等(2015)研究发现,橄榄根系中有24个差异表达基因,其中9个上调基因,15个下调基因;在叶中有70个差异表达基因,其中14个下调基因,56个上调基因;Tian等(2018)研究发现,SnRK2、ABF、HST、GSTs和GSH1基因在盐胁迫中表现出高活性;Pan等(2019)研究发现,Unigenes有15321个微卫星标记基因,其中,有17个单核苷酸多肽(SNP)与6个盐胁迫相关的差异表达基因(DEGs);Wang等(2020a)研究发现,大穗结缕草中TaHSP23.9可能作为一种蛋白质伴侣来正向调节植株对盐胁迫的响应。【本研究切入点】目前鲜见有关海马齿根系响应盐胁迫转录组分析的研究报道。【拟解决的关键问题】利用Illumina测序技术对不同浓度NaCl胁迫处理的海马齿根系进行转录组测序分析,并挖掘响应盐胁迫的功能基因,为培育耐盐的农作物新品种及有效解决土壤盐渍化提供理论依据。1D9152E6-FF06-4DC8-B029-2C5CAEBFEE6C
1 材料与方法
1. 1 试验材料
供试材料海马齿采自海南省海口市东寨港口(东经110°33′59″,北纬19°57′12″)。选自同一植株的茎,每个分枝保留3个节和4片叶,并用1/2改良型Hoagland营养液进行培养。经过3周扦插生根后用400 mmol/L NaCl(盐胁迫组)和0 mmol/L NaCl(对照,CK)连续处理5周,每处理重复3次。RNAplant Plus试剂盒购自天根生化科技(北京)有限公司,SYBR Premix Ex Taq Kit购自宝日医生物技术(北京)有限公司。主要仪器设备:Agilent2100分析仪(Agilent,美国)、Nanodrop分光光度计(Thermo,美国)、ABI 7500荧光定量PCR仪(ABI,美国)等。
1. 2 RNA提取、文库制备及测序
按照RNAplant Plus试剂盒说明提取海马齿根系总RNA。用Agilent2100分析仪检测RNA的纯度。cDNA文库的制备参照Sharma(2015)的方法。对制备的cDNA文库进行PCR扩增以获得大量的连接片段,用NanoDrop分光光度計进行定量,并用Bioanalyzer检测其纯度。最后使用Illumina HiSeq 2000平台进行测序。
1. 3 组装、注释和差异表达基因分析
利用SolexaQA对Raw reads进行过滤处理后得到Clean reads。为得到高质量的测序数据方便后续分析,从中去除由于接头自连等原因导致没有插入片段的reads,以及舍弃adapter及质量修剪后长度小于20 bp的序列。利用Trinity(http://trinitynaseq.scourceforge.net)将Clean reads进行从头组接,设置参数K-mer graph(K=25),从而获得Unigenes。经组装后的转录本以差异倍数(Fold change)≥2,错误发现率(False dicovery rate)<0.05作为筛选标准筛选出差异表达基因。将差异表达基因在Nr(http://www.ncbi.nlm.nih.gov)、GO(http://www.geneontology.org)、Swiss-Prot(http://www.expasy.ch/sprot)、KEGG(http://www.genome.jp/kegg)和COG(http://www.ncbi.nlm.nih.gov/COG)五大数据库进行功能注释。采用KEGG数据库对差异表达基因进行功能分类和代谢途径富集分析。
1. 4 实时荧光定量PCR(qRT-PCR)检测
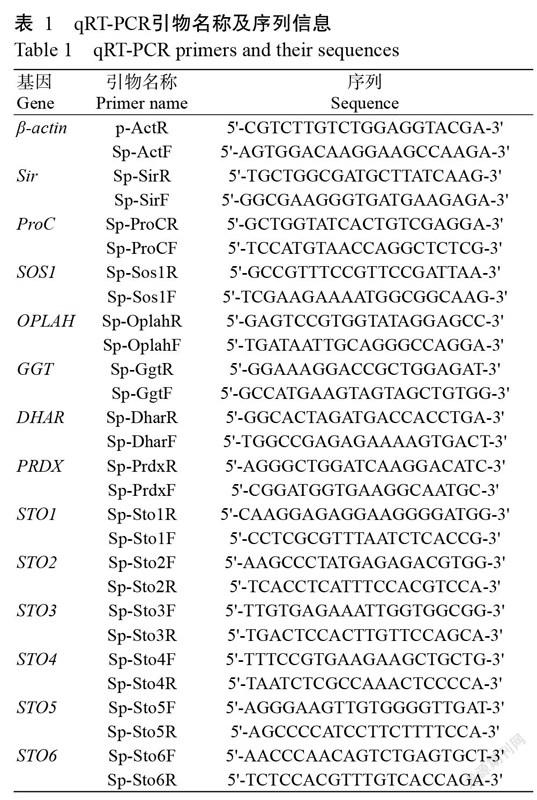
以第一链cDNA为模板,利用SYBR Premix Ex Taq Kit对随机选取13个差异表达基因进行qRT-PCR检测,以β-actin基因为内参。所有反应均设3次重复。qRT-PCR所用引物(表1)均使用Primer Express(Applied Bio systems)设计,并利用NCBI数据库的BLAST程序对所设计引物进行验证。最后用相对定量法(2-△△Ct)计算目的基因表达水平。
2 结果与分析
2. 1 海马齿转录组数据分析结果
利用Illumina配对末端测序法对海马齿根系的2个cDNA文库(CK和盐胁迫处理组)进行转录组测序,共获得138133008条Raw reads。经过对Clean reads进行拼接后,在海马齿根系转录组中共鉴定出305145个转录本,平均长度为622 bp,其中,对照组有146177个长度>300 bp的转录本,盐胁迫处理组有72173个长度>300 bp的转录本。差异表达的125173个转录本中,长度为300~500 bp的转录本占差异表达基因总数的64.4%,长度为501~1000 bp的转录本占18.9%,长度为1000~3000 bp的转录本占16.6%,不含长度>3000 bp的转录本。
2. 2 Unigenes功能注释结果
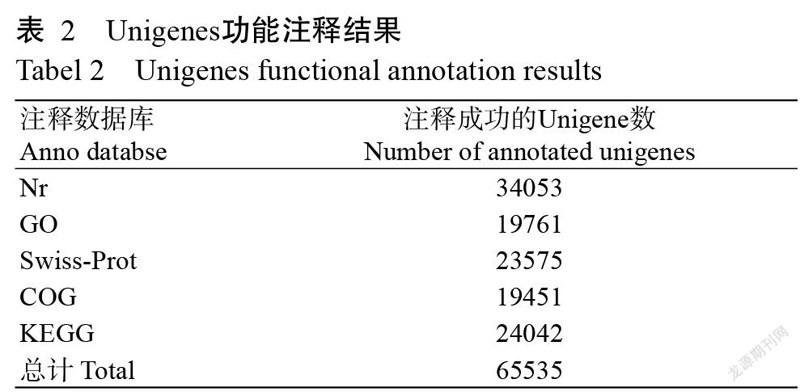
将Unigenes在Nr、GO、Swiss-Prot、COG和KEGG五大数据库中进行注释,结果(表2)发现,共有65535条Unigenes注释成功,占Unigenes总数的52.36%,剩下的59638条Unigenes均未获得注释占Unigenes总数的47.64%。在Nr数据库中被成功注释的Unigenes最多,比对上的同源物种有拟南芥、水稻、蒺藜苜蓿、大麦、海金藻等。在GO数据库中注释成功的Unigenes如图1所示。Unigenes被注释为生物学过程和细胞组分两大类别的数量较分子功能类别多。生物学过程类别中,富集程度最高的是细胞过程,其次是单一生物细胞过程和代谢过程;细胞组分类别中,富集程度较高的是细胞和细胞部分,其次是细胞器部分;分子功能类别中,富集程度最高的是催化活性和结合活性。
2. 3 Unigenes功能分析结果
对照组和盐胁迫处理组共有65535个差异Unigenes。与对照相比,盐胁迫处理组的上调Unigenes有14609个,下调Unigenes有50926个。在Unigenes中发现有182个热休克蛋白基因,如表3所示。这些蛋白质包括高分子量的Hsps(70 kD Hsp70和90 kD Hsp90)、低分子量的Hsps(18.2 kD class I、15.4 kD class V、22.7 kD class IV、26.5 kD Hsps,10 kD类伴侣蛋白,19 kD Hsps和23.5 kD ACD-sHsps)和分子伴侣(20 kD叶绿体分子伴侣、分子伴侣ClpB1及分子伴侣dnaJ 1,dnaj 2、dnaj 6、dnaj 10、dnaj 13、dnaj 16、含t-复合蛋白的分子伴侣、分子伴侣CPN60、分子伴侣GroEL、分子伴侣-60 kD和ch60),其中低分子量的HSPs表达上调。1D9152E6-FF06-4DC8-B029-2C5CAEBFEE6C
2. 4 差异表达基因的qRT-PCR检测结果
对照组和盐胁迫处理组共有24042个差异表达基因。为验证测序结果的可信度,从中随机选择13个差异表达基因进行qRT-PCR检测,結果(图2)发现,其中6个耐盐蛋白基因(STO1~STO6),吡咯啉-5-羧酸还原酶基因(ProC),亚硫酸还原酶基因(SIR)和盐超敏感基因(SOS1)上调表达,其余4个基因下调表达。差异表达基因的qRT-PCR检测结果与转录组测序结果表达趋势一致。说明测序文库较真实地反映盐胁迫下差异表达基因的表达情况。
2. 5 差异表达基因的KEGG代谢通路富集分析结果
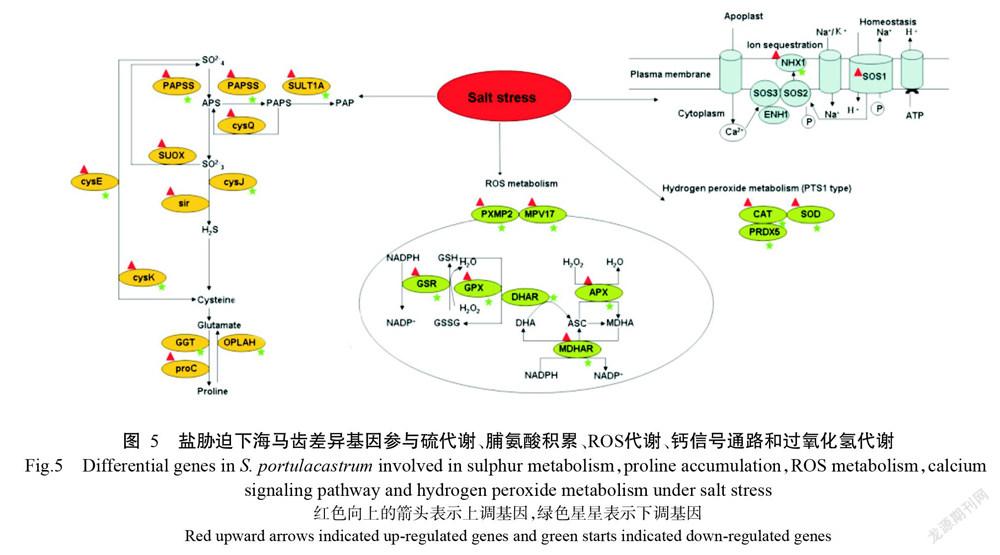
将获得的24042个差异表达基因与KEGG数据库进行比对,结果发现有10106个显著差异基因富集到129条代谢通路,其中富集程度排名前10的代谢途径为核糖体、次级代谢生物合成、RNA转运、内吞作用、剪接体、甘油磷脂代谢、内质网加工、吞噬、醚脂类代谢和植物—病原体相互作用(图3)。由图4可知,富集差异表达基因最多的通路为内质网加工蛋白通路,为948个基因,其中上调基因145个,下调基因803个,其次是吞噬体通路,为946个,其中上调基因120个,下调基因826个。此外,差异表达基因参与盐胁迫密切相关的途径包括硫代谢、脯氨酸积累、活性氧(ROS)代谢、与盐胁迫相关的钙信号通路和过氧化氢代谢(图5)。在硫代谢中上调基因为SOS1和Na+/H+转运蛋白基因(NHX1);脯氨酸累积中,proC基因上调,而谷胱甘肽水解酶基因(GGT)和5-氧脯氨酸酶基因(OPLAH)下调;ROS代谢中,脱氢抗坏血酸还原酶基因(DHAR)表达下调,而过氧化物酶体膜蛋白基因2(Pxmp2)和线粒体内膜蛋白基因17(Mpv17)基因表达上调。
3 讨论
高度保守的热激蛋白是一种组成型表达蛋白,并具有分子伴侣的功能,参与多种生物学过程,如转录、翻译和翻译后修饰、蛋白质折叠及蛋白质的聚集和解聚(Tiwari et al.,2015)。5个保守的Hsps家族(Hsp100、Hsp90、Hsp70、Hsp60和sHsp)和小热休克蛋白(sHsp)在植物中普遍存在,在生物或非生物胁迫下其基因表达上调,可作为分子伴侣保护其他蛋白免受非生物胁迫的破坏(Elizabeth et al.,2020)。本研究发现,有7种以上的sHsps基因表达上调,推测其参与耐盐机制。前人研究发现,TaHsp23.9、PfHsp17.2等sHsp基因的表达均提高了转基因拟南芥的耐盐性,推测sHsp保护了某些酶和蛋白质在盐胁迫下免于破坏和降解(Zhang et al.,2018;Wang et al.,2020b)。但Sun(2016)研究表明,转基因拟南芥中AsHsp17基因表达降低植株对盐的耐受性,其原因可能是不同种类sHsp对盐胁迫的响应机制不同。本研究还发现,与ROS代谢相关的2个膜蛋白基因Pxmp2和Mpv17表达上调,表明盐胁迫下海马齿根系的ROS合成代谢加强,其原因可能是盐胁迫刺激下产生大量ROS(Mittler,2017),导致与ROS代谢相关基因表达上调。此外,ROS作为信号分子,将盐胁迫信号传递给sHsp基因,从而导致sHsp基因表达上调(Wrzaczek et al.,2013)。盐胁迫会引起蛋白质错误折叠或未折叠蛋白质的累积,使内质网中编码分子伴侣基因及其他提高蛋白质折叠能力基因的表达,有助于内质网恢复其稳态(Walter and Ron,2011)。海马齿在盐胁迫下,差异表达基因中多种分子伴侣基因互作参与海马齿的盐胁迫响应,该结论在大穗结缕草(Zhang et al.,2018)亦得到证实。
在盐胁迫下,海马齿中参与到抗坏血酸-谷胱甘肽循环(AsA-GSH)代谢途径中的脱氢抗坏血酸还原酶基因(DHAR)显著下调,与燕麦在盐胁迫下的研究结果(刘建新等,2021)一致,但水稻DHAR在拟南芥中过表达可提高植株耐盐能力(Ushimaru et al.,2006)。海马齿的单脱氢抗坏血酸还原酶基因(MDHAR)和抗坏血酸过氧化物酶基因(APX)基因均显著上调。该结论与盐胁迫下大豆MDHAR基因表达受到抑制,但APX基因表达量升高的结论存在差异(Rahman et al.,2021)。此外,脯氨酸作为应激反应的衔接分子,在自然界中作用广泛,在植物逆境胁迫的抗氧化反应中发挥重要作用。脯氨酸处理过的烟草幼苗中APX和谷胱甘肽过氧化物酶(GPX)活性增强,说明脯氨酸参与提高烟草幼苗的抗氧化能力(Boudmyxay et al.,2019),推测脯氨酸在海马齿盐胁迫响应中间接发挥作用。今后将通过转基因技术进一步深入研究海马齿中盐胁迫响应基因的分子调控机制。
4 结论
盐胁迫下海马齿差异表达基因如小分子量热激蛋白基因、抗氧化酶相关基因及与离子交换相关基因发挥了重要调控作用。
参考文献:
丁国华,阮雪玉,张雪妍,王旭初. 2020. 滨海盐生植物海马齿耐盐机制解析及其在生态环保中应用研究[J]. 中国科技成果,21(7):18-23. [Ding G H,Ruan X Y,Zhang X Y,Wang X C. 2020. Study on salt-tolerance mechanism of marine halophytes Sesuvium portulacastrum L. and its application in ecological and environmental protection[J]. China Science and Technology Achievements,21(7):18-23.] doi:10.3772/j.issn.1009-5659.2020.07.006.
范伟,李文静,付桂,张治礼. 2010. 一种兼具研究与应用开发价值的盐生植物-海马齿[J]. 热带亚热带植物学报,18(6):689-695. [Fan W,Li W J,Fu G,Zhang Z L. 2010. Sesuvium portulacastrum L.,a promising halophyte in research and application[J]. Journal of Tropical and Subtro-pical Botany,18(6):689-695.] doi:10.3969/j.issn.1005-3395.2010.06.017.1D9152E6-FF06-4DC8-B029-2C5CAEBFEE6C
劉建新,刘瑞瑞,贾海燕,刘秀丽,卜婷,李娜. 2021. 硫化氢对盐碱胁迫下裸燕麦叶片抗坏血酸—胱甘肽循环的调控效应[J]. 应用生态学报,32(12):3988-3996. [Liu J X,Liu R R,Jia H Y,Liu X L,Bu T,Li N. 2021. Regulation effects of hydrogen sulfide on ascorbate-glutathione cycle in naked oat leaves under saline-alkali stress[J]. Chinese Journal of Applied Ecology,32(11):3988-3996.] doi:10.13287/j.1001-9332.202111.023.
Bazakos C,Manioudaki M E,Sarropoulou E,Spano T,Kalaitzis P. 2015. 454 pyrosequencing of olive (Olea europaea L.) transcriptome in response to salinity[J]. PLoS One,10(11):e0143000. doi:10.1371/journal.pone.0143 000.
Boudmyxay K,沈镭,钟帅,孙艳芝,杨慧芹. 2019. 脯氨酸引发提高烟草种子和幼苗抗逆性及其与抗氧化系统的关系[J]. 山西农业科学,47(1):39-48. [Boudmyxay K,Shen L,Zhong S,Sun Y Z,Yang HQ. 2019. Improving the antioxidant system and its stress resistance to tobacco seeds and seedling by proline priming[J]. Journal of Shanxi Agri-cultural Sciences,47(1):39-48.] doi:10.3969/j.issn.1002- 2481.2019.01.11.
Chang W J,Liu X W,Zhu J H,Fan W,Zhang Z L. 2016. An aquaporin gene from halophyte Sesuvium portulacastrum,SpAQP1,increases salt tolerance in transgenic tobacco[J]. Plant Cell Reports,35(2):385-395. doi:10.1007/s00299-015-1891-9.
Elizabeth R,Waters,Elizabeth V. 2020. Plant small heat shock proteins–evolutionary and functional diversity[J]. New Phytologist Trust,227(1):24-37. doi:10.1111/nph.16536.
Fan W,Chang W J,Liu X W,Xiao C,Yang J L,Zhang Z L. 2017. Identification of up-regulated genes provides integrated insight into salt-induced tolerance mechanisms in Sesuvium portulacastrum roots[J]. Acta Physiologiae Plan-tarum,39(3):1-11. doi:10.1007/s11738-017-2383-z.
Fan W,Zhang Z L,Zhang Y L. 2009. Cloning and molecular characterization of fructose-1,6-bisphosphate aldolase gene regulated by high-salinity and drought in Sesuvium portulacastrum[J]. Plant Cell Reports,28(6):975-984. doi:10.1007/s00299-009-0702-6.
Fan Y F,Yin X C,Xie Q,Xia Y Q,Wang Z Y,Song J,Zhou Y,Jiang X Y. 2019. Co-expression of SpSOS1 and SpAHA1 intransgenic Arabidopsis plants improves salinity tolerance[J]. BMC Plant Biology,19(1):74. doi:10.1186/s12870-019-1680-7.
Ghnaya T,Zaier H,Baioui R,Sghaier S,Lucchini G,Sacchi GA,Lutts S,Abdelly C. 2013. Implication of organic acids in the long-distance transport and the accumulation of lead in Sesuvium portulacastrum and Brassica juncea[J]. Chemosphere,90(4):1449-1454. doi:10.1016/j.chemosphere.2012.08.061.
Ji H T,Pardo J M,Batelli G,Van O M J,Bressan R A,Li X. 2013. The salt overly sensitive(SOS) pathway:Established and emerging roles[J]. Molecular Plant,6(2):275-286. doi:10.1093/mp/sst017.1D9152E6-FF06-4DC8-B029-2C5CAEBFEE6C
Kannan P R,Deepa S,Kanth S V,Rengasamy R. 2013. Growth,osmolyte concentration and antioxidant enzymes in the leaves of Sesuvium portulacastrum L. under salinity stress[J]. Applied Biochemistry and Biotechnology,171(8):1925-1932. doi:10.1007/s12010-013-0475-9.
Lokhande V H,Gor B K,Desai N S,Nikam T D,Suprasanna P. 2013. Sesuvium portulacastrum,a plant for drought,salt stress,and fixation,food andphytoremediation. A review[J]. Agronomy for Sustainable Development,33(2):329-348. doi:10.1007/s13593-012-0113-x.
Ma J B,Zhang M R,Xiao X L,You J J,Wang J R,Wang T,Yao Y N,Tian C Y. 2013. Global transcriptome profiling of Salicornia europaea L. shoots under NaCl treatment[J]. PLoS One,8(6):e65877. doi:10.1371/journal.pone. 0065877.
Mittler R. 2017. ROS are good[J]. Trends in Plant Science,22(1):11-19. doi:10.1016/j.tplants.2016.08.002.
Pan L,Yu X L,Shao J J,Liu Z C,Gao T,Zheng Y,Zeng C,Liang C Z,Chen C Y. 2019. Transcriptomic profiling and analysis of differentially expressed genes in asparagus bean(Vigna unguiculata ssp. sesquipedalis)under salt stress[J]. PLoS One,14(7):e0219799. doi:10.1371/journal.pone.0219799.
Qiu Q,Ma T,Hu Q J,Liu B B,Wu Y X,Zhou H H,Wang Q,Wang J,Liu J Q. 2011. Genome-scale transcriptome ana-lysis of the desert poplar,Populus euphratica[J]. Tree Physiology,31(4):452-461. doi:10.1093/treephys/tpr015.
Rabello A R,Guimar?es C M,Rangel P H N,da Silva F R,Seixas D,de Souza E,Brasileiro A C M,Spehar C R,Ferreira M E,Mehta A. 2008. Identification of drought-responsive genes in roots of upland rice(Oryza sativa L.)[J]. BMC Genomics,9(1):485. doi:10.1186/1471-2164-9-485.
Rabhi M,Ferchichi S,Jouini J,Hamrouni M H,Koyro H W,Ranieri A,Abdelly C,Smaoui A. 2010a. Phytodesalination of a salt-affected soil with the halophyte Sesuvium portulacastrum L. to arrange in advance the requirements for the successful growth of a glycophytic crop[J]. Bioresource Technology,101(17):1336-1341. doi:10.1016/j.biortech.2010.03.097.
Rabhi M,Giuntini D,Castagna A,Renorini D,Baldan B. Smaoui A,Abdelly C,Ranieri A. 2010b. Sesuvium portulacastrum maintains adequate gas exchange,pigment composition,and thylakoid proteins under moderate and high salinity[J]. Journal of Plant Physiology,167(16):1336-1341. doi:10.1016/j.jplph.2010.05.009.
Rahman M,Rahman K,Sathi K S,Alam M M,Nahar K,Fujita M,Hasanuzzaman M. 2021. Supplemental selenium and boron mitigate salt-induced oxidative damages in Glycine max L.[J]. Plants(Basel),10(10):2224. doi:10.3390/plants10102224.1D9152E6-FF06-4DC8-B029-2C5CAEBFEE6C
Rizwan M,Ali S,Ibrahim M,Farid M,Adrees M,Bharwana S A,Zia-Ur-Rehman M,Qayyum M F,Abbas F. 2015. Mechanisms of silicon-mediated alleviation of drought and saltstress in plants:A review[J]. Environmental Science and Pollution Research,22(20):15416-15431. doi:10.1007/ s11356-015-5305-x.
Sharma R,Mishra M,Gupta B,Parsania C,SinglaPareek SL,Pareek A. 2015. De novo assembly and characterization of stress transcriptome in a salinity-tolerant variety CS52 of Brassica juncea[J]. PLoS One,10(5):e0126783. doi:10.1371/journal.pone.0126783.
Sun W,Xu X N,Zhu H S,Liu A H,Liu L,Li J M,Hua X J. 2010. Comparative transcriptomic profiling of a salt-tole-rant wild tomato species and a salt-sensitive tomato cultivar[J]. Plant & Cell Physiology,51(6):997-1006. doi:10.1093/pcp/pcq056.
Sun X B,Sun C Y,Li Z G,Hu Q,Han L B,Luo H. 2016. AsHSP17,a creeping bentgrass small heat shock protein modulates plant photosynthesis and ABA-dependent and independent signalling to attenuate plant response to abio-tic stress[J]. Plant,Cell & Environment,39(6):1320-1337. doi:10.1111/pce.12683.
Taji T,Seki M,Satou M,Sakurai T,Kobayashi M,Ishiyama K,Narusaka Y,Narusaka M,Zhu J K,Shinozaki K. 2004. Comparative genomics in salt tolerance between Arabidopsis and Arabidopsis-related halophyte salt cress using Arabidopsis microarray[J]. Plant Physiology,135(3):1697-1709. doi:10.1104/pp.104.039909.
Tian X M,Wang Z Y,Zhang Q,Ci H C,Wang P S,Yu L,Jia G X. 2018. Genome-wide transcriptome analysis of thesalt stress tolerance mechanism in Rosa chinensis[J]. PLoS One,13(7):e0200938. doi:10.1371/journal.pone.0200938.
Tiwari S,Thakur R,Shankar J. 2015. Role of heat-shock proteins in cellular function and in the biology of fungi[J]. Biotechnology Research International,(2):1-11. doi:10.1155/2015/132635.
Ushimaru T,Nakagawa T,Fujioka Y,Daicho K,Naito M,Yamauchi Y,Nonaka H,Amako K,Yamawaki K,Murata N. 2006. Transgenic Arabidopsis plants expressing the rice dehydroascorbate reductase gene are resistant to salt stress[J]. Journal Plant Physiology,163(11):1179-84. doi:10.1016/j.jplph.2005.10.002.
Walter P,Ron D. 2011. The unfolded protein response:From stress pathway to homeostatic regulation[J]. Science,334:1081-1086. doi:10.1126/science.1209038.
Wang J,Gao X ,Dong J,Tian X Y,Wang J Z,Palta JA,Xu S B,Fang Y,Wang Z H. 2020a. Over-expression of the heat-responsive wheat geneTaHSP23.9 in transgenic arabidopsis conferred tolerance to heat and saltstress[J]. Frontiers in Plant Science,11:243. doi:10.3389/fpls.2020.00243.1D9152E6-FF06-4DC8-B029-2C5CAEBFEE6C
Wang R,Wang X,Liu K,Zhang X J,Zhang L Y,Fan S J. 2020b. Comparative transcriptome analysis of halophyte Zoysia macrostachya in response to salinity stress[J]. Plants,9(4):458. doi:10.3390/plants9040458.
Wrzaczek M,Brosché M,Kangasj?rvi J. 2013. ROS signaling loops—Production,perception,regulation[J]. Current Opi-nion in Plant Biology,16(5):575-582. doi:10.1016/j.pbi. 2013.07.002.
Yang C L,Zhou Y,Fan J,Fu Y H,Shen L B,Yao Y,Li R M,Fu S P,Duan R J,Hu X W,Guo J C. 2015. SpBADH of the halophyte Sesuvium portulacastrum strongly confers drought tolerance through ROS scavenging in transgenic Arabidopsis[J]. Plant Physiology and Biochemistry,96:377-387. doi:10.1016/j.plaphy.2015.08.010.
Yi X P,Sun Y,Yang Q,Guo A P,Chang L L,Wang D,Tong Z,Jin X,Wang L M,Yu J L,Jin W H,Xie Y M,Wang X C. 2014. Quantitative proteomics of Sesuvium portulacastrum leaves revealed that ion transportation by V-ATPase and sugar accumulation in chloroplast played crucial roles in halophyte salt tolerance[J]. Journal of Proteomics,99:84-100. doi:10.1016/j.jprot.2014.01.017.
Zahaf O,Blanchet S,de Zélicourt A,Alunni B,Plet J,Laffont Carole,de Lorenzo L,Imbeaud S,Ichanté JL,Diet A,Badri M,Zabalza A,González E M,Delacroix H,Gruber V,Frugier F,Crespi M. 2012. Comparative transcriptomic analysis of salt adaptation in roots of contrasting Medicago truncatula genotypes[J]. Molecular Plant,5(5):1068-1081. doi:10.1093/mp/sss009.
Zhang L,Hu W J,Gao Y K,Pan H T,Zhang Q X. 2018. A cytosolic class II small heat shock protein,PfHSP17.2,confers resistanceto heat,cold,and salt stresses in transgenic Arabidopsis[J]. Genetics and Molecular Biology,41(3):649-660. doi:10.1590/1678-4685-GMB-2017-0206.
Zhang X,Liao M S,Chang D,Zhang F C. 2014. Comparative transcriptome analysis of the Asteraceae halophyte Kareli-nia caspica under salt stress[J]. BMC Research Notes,7:927. doi:10.1186/1756-0500-7-927.
Zhou Y,Yang C L,Hu Y P,Yin X C,Li R M,Fu S P,Zhu B B,Guo J C,Jiang X Y. 2018. The novel Na+/H+ antipor-ter gene SpNHX1 from Sesuvium portulacastrum confers enhanced salt tolerance to transgenic yeast[J]. Acta Phy-siologiae Plantarum,40(3):61. doi:10.1007/s11738-018-2631-x.
(責任编辑 陈 燕)1D9152E6-FF06-4DC8-B029-2C5CAEBFEE6C

