Effects of sgRNA length and number on gene editing efficiency and predicted mutations generated in rice
Xiaojing Liu, Jiangtao Yang, Yaya Song, Xiaochun Zhang, Xujing Wang, Zhixing Wang
Biotechnology Research Institute, Chinese Academy of Agricultural Sciences/MARA Key Laboratory on Safety Assessment (Molecular) of Agri-GMO, Beijing 100081, China
Keywords:CRISPR-Cas9 sgRNA number sgRNA length Editing efficiency
ABSTRACT CRISPR-Cas9 is a common tool for gene editing,and appropriate sgRNAs are the key factor for successful editing. In this study, the effect of sgRNA length and number on editing efficiency was analyzed in rice using CYP81A6 as the target gene. A series of CRISPR-Cas9 plant expression vectors containing single sgRNAs with different lengths (17, 18, 19, 20, 21, 22, 23 nt) or two sgRNAs were constructed and introduced into rice cultivar Zhonghua11 by Agrobacterium-mediated transformation.Analysis of the editing status of 1283 transgenic rice plants showed that 371 were successfully edited with base preference.Single A or T insertions were the most frequent among the six edited types. The editing efficiency of transgenic rice with two sgRNAs was higher than that with a single sgRNA. Editing efficiency and sgRNA length showed a normal distribution with 20 nt sgRNA(25%)being the most efficient.The editing efficiency decreased slightly with decreases of 1–2 bases (19 nt 20%, 18 nt 21%), but decreased significantly with a decrease of 3 bases (17 nt 4.5%). Editing efficiency was significantly reduced by adding 1 to 3 bases(21 nt 16.8%,22 nt 13%,23 nt 13%)to the sgRNA.These results provide data for successful gene editing or rice by CRISPR-Cas9.
1. Introduction
The CRISPR-Cas9 system that includes a CRISPR repeat-spacer array and a Cas protein is an RNA-guided DNA endonuclease system that targets specific genomic sequences [1]. It initiates DNA double strand breaks (DSBs) through the RuvC and HNH nuclease domains in the Cas9 enzyme and repairs occur through natural DNA repair pathways of cells,non-homologous end joining(NHEJ)and homologous recombination(HR)[2].As the most common current gene editing tool, the CRISPR-Cas9 system is low-cost, more precise and easy-to-use allowing targeted genetic manipulation and simultaneous editing at multiple sites across the genome. It is becoming widely used in gene functional analysis and crop breeding following its demonstration in rice, Arabidopsis and tobacco in 2013 [3–5].
Design of sgRNA is important for gene editing using the CRISPRCas system. There are numerous accessible online bioinformatics tools for designing sgRNA such as CRISPRlnc and sgRNA Scorer 2.0[6].The length and structure of the sgRNA determines the editing efficiency and specificity of CRISPR-Cas editing. Truncation of the 5′end or addition of two G nucleotides at 5′end of the gRNA improves the specificity of RNA-guided Cas9 and decreases editing efficiency [7,8]. Truncation of the length of the sgRNA improved specificity in 293T cells; 17 nt length sgRNA had the same target activity and lower off-target activity than 20 nt sgRNA [9]. However, in mesenchymal stem cells (MSCs) and induced pluripotent stem cells (iPSCs), both off-target activity and target activity of 17 nt sgRNA was lower than that of 20 nt sgRNA [10]. In rice, 20 nt esgRNAs showed higher conversion efficiency than 14- to 19-nt esgRNAs in a plant adenine base editing (ABE) system using OsEV and OsOD as the target genes [11]. To our knowledge, there are few reports on the effects of sgRNA lengths longer than 20 nt on editing efficiency.To evaluate the influence of sgRNA on editing efficiency, we designed sgRNAs of different lengths from 17 to 23 nt and also double sgRNAs to explore the relative effects of sgRNA length, sgRNA number, editing efficiency and off-target activity in rice.
Gene CYP81A6 (Bel) encodes cytochrome P450 monooxygenase and confers tolerance to bendazone and sulfonylurea herbicides in rice. The recessive mutant bel is sensitive to bendazone and is used as a lethal selective marker in hybrid breeding[12,13].In this study, we constructed seven vectors with sgRNA lengths varying from 17 to 23 nt and two double sgRNA vectors using the Bel gene as the target gene to determine their relative effects on editing efficiency in transgenic rice.The results showed that 20 nt sgRNA had the highest editing efficiency with sgRNA and editing efficiency being normally distributed. The majority of editing sites were single-nucleotide T or A insertions.
2. Materials and methods
2.1.sgRNA design and in vitro determination of DNA cleavage activity
The design of six sgRNA of 20 bp length was based on a sequence located in the first exon of CYP81A6 gene using the CRISPR-P web tool (http://cbi.hzau.edu.cn/cgi-bin/CRISPR) [14].The sgRNA (named Guide 1 to Guide 6) were synthesized using a T7 in vitro transcription kit. The CYP81A6 gene was amplified by PCR from the rice genome using primer pair 3-200F/3-200R. We performed in vitro DNA cleavage assays and cleavage site sequencing according to method reported by Wang et al. [15] and Shan et al. [16].
Based on the detection results Guide 1 was selected as the target sequence to design sgRNA truncated lengths of 19, 18, and 17 nt and augmented lengths of 21, 22, and 23 nt. The DNA cleavage of these sgRNAs was detected in vitro.
2.2. Construction of CRISPR-Cas9 plant expression vector
All sgRNA were synthesized as oligonucleotide pairs. The synthesized sgRNA carrying an extra DNA sequence was inserted between the OsU3 promoter and sgRNA scaffold of the pp1c.3 vector through homologous recombination.The double sgRNAs(Guide 1-Guide 3 and Guide 3-Guide 4)were cloned into the plant binary vector pp1c.7 using the same method.The derived expression vectors were named 17 nt, 18 nt, 19 nt,20 nt, 21 nt, 22 nt,23 nt,1–3 and 4–3, respectively.
2.3. Rice transformation and growth
The plant expression vectors were moved into Agrobacterium tumefaciens strain EHA105 by the freeze–thaw method and then transformed into japonica cultivar Zhonghua 11 using the published method [17]. Transgenic rice was grown in the net house of the Chinese Academy of Agricultural Sciences.
2.4. Detection and analysis of gene edited mutant plants
Genomic DNA was extracted from leaves of transgenic rice by the CTAB method. Cas9 was amplified from transgenic rice DNA using primer pair RTCas9-F/RTCas9-R with the following reaction conditions: 30 s at 95 °C, followed by 34 cycles of 30 s at 95 °C,30 s at 60 °C, 30 s at 72 °C and finally 72 °C for 10 min.
Primer pair Guide1-20jianF/Guide1-20jianR and Guide1-3-4F/Guide1-4R were used to amplify the single sgRNA and double sgRNA target site sequences with the following reaction conditions: 95 °C for 30 s, followed by 34 cycles of 95 °C for 30 s, 55–58 °C for 30 s, 70 °C for 1 min, and finally 72 °C for 10 min. PCR products were sequenced through Sanger sequencing method.Sequencing results were compared with wild-type to analyze gene editing mutations.
2.5. Off-target site detection
Primers were designed according to the five most likely offtarget gene sequences of Guide 1, Guide 3 and Guide 4 predicted by CRISPR-P. High-fidelity DNA polymerase PCR was performed using two groups of transgenic rice genomic DNA as template.One group contained Cas9 protein for editing the target gene.The other group contained Cas9 protein, but the target gene was not edited. Ten plants from each group were selected to extract genomic DNA. PCR products were sequenced and aligned with the corresponding gene sequence of Zhonghua 11. All predicted off-target sequences are shown in Table S1 and all primers sequences are shown in Table S2.
3. Results
3.1. Effect of sgRNA length on DNA cleavage efficiency
Cas9 protein cleaves the linear DNA of the CYP81A6 gene into two fragments at a predicted site under guidance of the designed sgRNA Guide 1, Guide 2, Guide 3, Guide 4, and Guide 6 (Fig. S1).The length of sgRNA had a significant effect on DNA cleavage efficiency.100%of target DNA was cleaved within 5 min by Cas9 protein under guidance of 18, 19 and 20 nt Guide 1; 62.23%, 62.8%,62.98%and 27%of target DNA was cleaved with 5 min under guidance of 17, 21, 22, and 23 nt Guide 1, respectively, and 72.92%,82.3%, 73.86% and 50.49% of target DNA was cleaved at 30 min,respectively (Fig. 1A, B).
3.2. Gene-editing efficiency of different sgRNA lengths in transgenic rice
When A. tumefaciens strain EHA105 containing different vector plasmids was used, 371 of 1283 transgenic rice plants were correctly edited through PCR detection (Fig. S2). Sanger sequencing showed that all Cas9 enzyme activity led to edits at the third base upstream the PAM sequence and there were six edited types,including single nucleotide A,T,C and G insertions,and single base or multiple base deletions (Fig. S3). Single base insertion was the majority form of mutation,and the proportion of single A or T base insertions was highest in transgenic rice plants with different single sgRNAs. For transgenic rice with double sgRNA, the most common editing type was large fragment deletion, followed by single-base T insertion. These results suggest that Cas9-mediated mutation in plants involves a base preference with single base A or T insertions (Fig. 2A–C). Allele editing analysis indicated that heterozygous and biallelic mutations were most common in single sgRNA-targeted editing and double sgRNA-targeted editing,respectively (Fig. 2D, E).
CRISPR vectors containing sgRNAs with different lengths had different editing efficiencies. In the case of sgRNA with the same length the editing efficiency of double sgRNA was significantly higher than that of single sgRNA. The editing efficiency was 25%,66%and 80% in transgenic rice containing 20 nt,1–3 and 3–4 vectors,respectively(Table 1).sgRNA length also had an obvious effect on editing efficiency. Canonical 20 nt sgRNA length showed the highest editing efficiency (25%). sgRNA length ranging from 17 to 19 nt and 21 to 23 nt showed decreasing editing efficiencies.Truncations of one or two bases (19 or 18 nt) did not significantly decrease editing efficiency, truncation to 17 nt greatly reduced editing activity. Editing efficiency was significantly reduced by adding bases (21, 22, and 23 nt) to the 20 nt sgRNA length. There was a normal distribution of sgRNA length and editing efficiency(Fig. 2F). As for the effect of sgRNA length on editing efficiency,the trend of change in transgenic rice was basically consistent with that in vitro. It is noted that the cleavage efficiency of 17 nt was higher than that of 23 nt in vitro, but the editing efficiency of 17 nt was the lowest in transgenic rice.
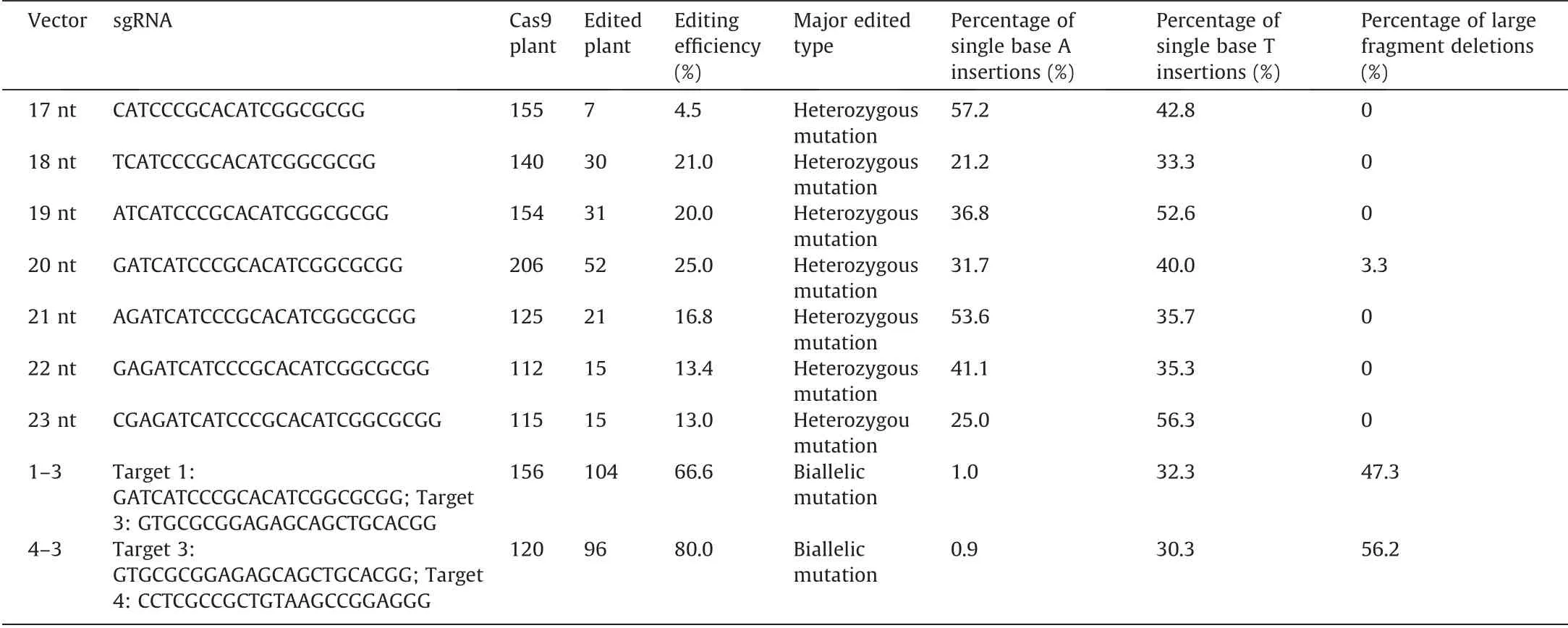
Table 1 Comparison of editing efficiency and editing types between single and double sgRNAs.

Fig. 2. Editing efficiency of sgRNAs with different lengths in rice and preference analysis of Cas9-mediated mutations. (A) Statistics of editing types of Cas9-mediated mutations by different sgRNA lengths. IA,single-nucleotide A insertion;IT, single-nucleotide T insertion;IC,single-nucleotide C insertion;IG,single-nucleotide G insertion;ITT, TT nucleotide insertion; DG, single-nucleotide G deletion; D >2, polybase deletion. (B) Statistics of editing types of Cas9-mediated mutations with double sgRNAs. DD,large fragment deletion; SD, single-nucleotide deletion; SI, single-nucleotide insertion. (C) Statistics of main insertion types for double sgRNAs. IA, single-nucleotide A insertion; IT, single-nucleotide T insertion; IG, single-nucleotide G insertion. (D) Allele gene editing types of Cas9-mediated mutations in different sgRNA lengths. (E) Allele gene editing types of Cas9-mediated mutations by double sgRNAs. (F) Comparison of editing efficiencies by sgRNAs of different length.
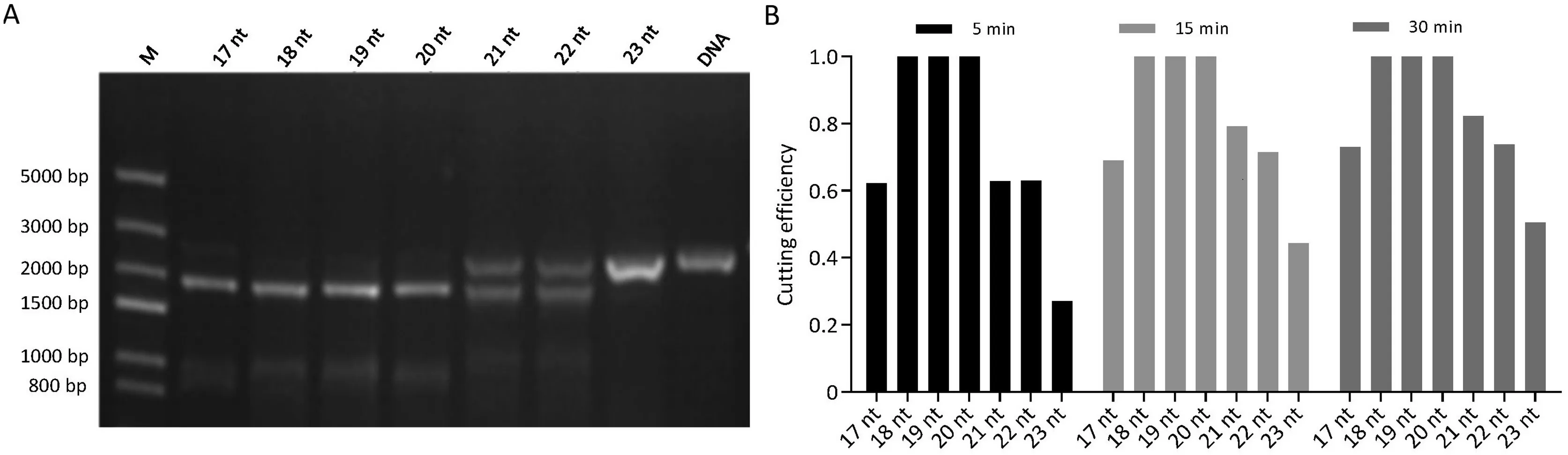
Fig.1. DNA cutting efficiency by different lengths of sgRNA in vitro.(A)Effects of different lengths of sgRNA in cutting DNA in agarose gels at 5 min.(B)Efficiency of cutting DNA by different lengths of sgRNA at 5, 15, and 30 min.
3.3. Detection of off-target mutations
We examined five predicted off-target sites for each of guide sgRNA Guide 1,Guide 3 and Guide 4.Ten plants were selected from each vector for off-target detection. A potential off-target site was detected in mutant plant 4–3–97 with a 5-bp deletion at the target site (Fig. S4). The overall results indicated that the off-target frequency was very low.
Everyone has a family. We live in it and feel very warm. There are three persons2 in my family, my mother, father and I. We live together very happily3 and there are many interesting stories about my family.
4. Discussion
Previous studies showed that sgRNA lengths less than 20 nt reduce editing efficiency in cells and rice [12]. In this study, we found a normal distribution among sgRNA length and editing efficiency with a maximum at 20 nt. This was similar previously reported experience. When the length of sgRNA was more than 20 nt, the editing efficiency decreased with the increasing sgRNA length(20 nt 25%,21 nt 16.8%,22 nt 13.39%,23 nt 13%).The reason could be that the added bases affect the formation of the R-loop created by the Cas9-sgRNA complex and thereby reduce the cutting activity of Cas9. For example, stem loop 1 of sgRNA plays a very important role in the function of the Cas9-sgRNA-DNA complex, whereas stem loops 2 and 3 stabilize the complex. This indicates that all three stem loop structures can affect the editing efficiency of sgRNA [18]. A later study found that the formation of the R loop regulates spCas9 conformation changes in key processes by connecting active nucleic acids. A specifically designed hairpin in the RNA secondary structure can be added at the 5′end of the spacer of the sgRNA,and the resulting hairpin structure can be used as a space and energy barrier similar to the R loop,increasing resistance to off-target nuclease activity and improving the specificity and efficiency of sgRNA editing [19].
Here,we counted the gene editing types of all 371 edited plants.The edits had a discernible bias with single-base A or T insertionsas the main editing type. Similar results were obtained with K562 cells[20].A possible explanation is that Cas9 binds to the proximal end of the PAM sequence, and the mismatched 1 nt at the distal end of the PAM sequence binds to the DNA polymerase and reconnects through the NHEJ pathway causing many single-nucleotide insertions to occur [21,22]. This effect could increase canonical end-joining activity during NHEJ repair.The prevalence of thymine insertions could indicate that the DNA repair enzymes (especially polymerases) have a specific preference leading to a difference in their demand for triphosphate nucleotides for incorporation.Another explanation is that when thymine is present, Cas9 tends to miscut rather than blunt cut. The most frequent deletion was removal of one or two repeated nucleotides, which depends on the two nucleotides near the cleavage site[14,16,23].These results indicate that we can design precise single-nucleotide insertions and deletions in gene sequences to study gene function. This implies that suitable sgRNAs can eventually be selected to obtain the predicted beneficial mutations affecting key traits in crop species.Here,we designed only three sgRNAs for one gene;many further trials will be needed to verify whether similar results will be achieved with other sgRNAs.
CRediT authorship contribution statement
Xiaojing Liu:Writing - original draft.Jiangtao Yang:Writing -review&editing.Yaya Song:Writing-review&editing.Xiaochun Zhang:Writing-review&editing.Xujing Wang:Project administration, Writing- review& editing.Zhixing Wang:Project administration, Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the Central Public-interest Scientific Institution Basal Research Fund.
Appendix A. Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2021.05.015.
- The Crop Journal的其它文章
- Origin, evolution, and molecular function of DELLA proteins in plants
- Far-red light: A regulator of plant morphology and photosynthetic capacity
- A rice XANTHINE DEHYDROGENASE gene regulates leaf senescence and response to abiotic stresses
- Dissection of heterotic loci for grain yield using interconnected chromosome segment substitution lines in rice
- A soybean NAC homolog contributes to resistance to Phytophthora sojae mediated by dirigent proteins
- The boron transporter SiBOR1 functions in cell wall integrity, cellular homeostasis, and panicle development in foxtail millet

