A soybean NAC homolog contributes to resistance to Phytophthora sojae mediated by dirigent proteins
Guolong Yu, Jinn Zou, Jinhui Wng, Rongsheng Zhu, Zhoming Qi, Hongwei Jing,Zhenng Hu, Mingling Yng, Ying Zho, Xioxi Wu, Chunyn Liu, Cndong Li,e, Xue Yng,Zhendong Zhu, Qingshn Chen,*, Yongu Fu, Dwei Xin,*
a Key Laboratory of Soybean Biology of MOE, Key Laboratory of Soybean Biology and Breeding/Genetics of MOA, College of Agriculture, Northeast Agricultural University,Harbin 150030, Heilongjiang, China
b MOA Key Lab of Soybean Biology (Beijing), National Key Facility of Crop Gene Resource and Genetic Improvement, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
c Soybean Research Institute, Jilin Academy of Agricultural Science, Changchun 130033, Jilin, China
d School of Agriculture and Biology, Shanghai Jiao Tong University, Shanghai 200240, China
e Jiamusi Branch Academy of Heilongjiang Academy of Agricultural Sciences, Jiamusi 154007, Heilongjiang, China
f Collaboration Centre to Russia of Institute of Grass Industry, Heilongjiang Academy of Agricultural Sciences, Harbin 150000, Heilongjiang, China
Keywords:Phytophthora sojae Soybean GmNAC1 Dirigent Lignin
ABSTRACT Phytophthora sojae infection severely impairs soybean production. We previously identified a dirigent protein, GmDRR1 (Glycine max Disease Resistant Response 1), that increases soybean resistance to P.sojae. However, the molecular basis of GmDRR1 function remained largely uncharacterized. In the present study,analysis of GmDRR1-RNAi,GmDRR1-overexpressing,and CRISPR/Cas9-derived Gmdrr1 mutant lines revealed that GmDRR1 expression significantly restricted P. sojae growth. Combining coimmunoprecipitation with liquid chromatography–tandem mass spectrometry revealed a GmDRR1-interacting protein, GmDRR2, which is homologous to GmDRR1. An E-coniferyl alcohol coupling assay indicated that GmDRR1 promotes the synthesis of (+)-pinoresinol, which helps to protect plants from P. sojae. The GmNAC1 (Glyma.05G025500) transcription factor bound to the GmDRR1 promoter both in vitro and in vivo to upregulate GmDRR1 expression. Soybean resistance to P. sojae was increased by overexpression of GmNAC1.Our findings suggest a novel signaling pathway involving a NAC transcription factor that mediates soybean resistance to P. sojae. Specifically, GmNAC1 directly induces GmDRR1 expression to increase resistance of soybean plants to P. sojae.
1. Introduction
In plants,dirigent(DIR)proteins function in the correct patterning of lignin deposition and associated responses to biotic factors,including fungi and insects [1–3]. The formation of lignin oligomers and lignan is strictly regulated by DIRs[4].DIRs can mediate the polymerization of monolignols into correct three-dimensional structures [5–6]. Increases in lignan content can increase soybean resistance to biotic stresses [7]. Lignin serves mainly as a physical barrier that inhibits pathogen infections [8]. Secondary cell wall production is associated with the synthesis of lignin in the cell wall[9].(+)-Pinoresinol synthesis is a key step in lignin formation[10].It remains unclear how GmDIRs regulate soybean response to P.sojae.
The NAC (NAM, ATAF, and CUC) genes encode transcription factors (TFs) involved in controlling plant morphology, physiology,and stress responses [11]. This TF family was first identified in Petunia species [12]. Genome sequencing identified 151, 126, and 226 NAC genes in rice (Oryza sativa), Arabidopsis thaliana, and soybean (Glycine max), respectively [13–15]. As one of the largest TF families, NAC TFs have multiple regulatory roles that help plants avoid the adverse effects of biotic and abiotic stresses and maintain yield [16–18]. In Arabidopsis, NAC TFs are associated with various developmental processes, including secondary cell wall formation and leaf senescence [19,20]. They also positively regulate plant defense responses against diverse pathogens [21]. In Arabidopsis,AtNAC4 promotes pathogen-induced cell death [22], whereas in potato, two NACs (NTP1 and NTP2) interact with the RxLR (Nterminal domain with the sequence motif RXLR (Arg-Xaa-Leu-Arg) consensus sequence) effector of Phytophthora infestans, and silencing of NTP1 or NTP2 expression increases the susceptibility of plants to P. infestans [23]. However, the regulatory function of NAC TFs in the lignin synthesis associated with plant resistance to pathogens remains unexplored.
Phytophthora sojae is an oomycete that infects soybean plants,leading to severe yield losses. In an earlier study [2], we determined by instantaneous transformation of Nicotiana benthamiana that GmDRR1,a dirigent protein in soybean,contributes to soybean resistance to P.sojae.In the present study,to detect the function of GmDRR1 to P. sojae, GmDRR1-overexpressing and GmDRR1-RNAi transgenic soybean lines were used. Whether GmDRR1 can mediate lignin synthesis and the transcription factors might regulate GmDRR1 was also detected.
2. Materials and methods
2.1. Plant materials, bacteria, and oomycetes
The soybean cultivars Tianlong 1 [24] and Suinong 10 [2] were used in this study. P. sojae strain 6497 [25] was routinely maintained on 10% V8 juice medium at 15 °C in darkness before being used to inoculate soybean roots and cotyledons. Escherichia coli DH5α and DE3 cells were transformed with the recombinant plasmids pET-28a-GmNAC1 (with a C-terminal His tag) and pET-28a-GmDRR1 (with a C-terminal His tag) carrying GmNAC1 or GmDRR1 for the subsequent expression of fusion proteins. Agrobacterium tumefaciens EHA105 was used for the infiltration of Arabidopsis and N. benthamiana plants. Agrobacterium rhizogenes K599 was used to transform soybean hairy roots.
Soybean seeds were surface-sterilized as previously described[26], and then germinated in vermiculite. Soybean plants were incubated in a growth chamber at 25 °C and 70% humidity with a 8-h light/16-h dark cycle for phenotypic analysis.The plants were then inoculated with P.sojae 6497 according to a published procedure [2]. The P. sojae 6497 biomass was quantified at 48 h after inoculation. Nicotiana benthamiana plants were grown in vermiculite with B&D medium in a phytotron (Imtech, Gouda, Netherlands) set at 22 °C with a 16-h light/8-h dark cycle for 3 weeks before the leaves were infiltrated with an A. tumefaciens suspension[27].Arabidopsis plants were grown under long-day conditions(16-h light/8-h dark) with a light intensity of 100 μmol m-2s-1.
To evaluate the effects of abiotic stresses, Tianlong 1 seedlings were treated with 100 μmol L-1MeJA (Methyl Jasmonate), ABA(Abscisic acid), and SA (Salicylic acid), as well as 200 mmol L-1NaCl at 2 weeks after germination. Seedlings were collected at 0,1, 3, 6, 9, 12, and 24 h after treatment and immediately frozen in liquid nitrogen for subsequent RNA extraction and qRT-PCR analysis. Untreated soybean seedlings served as controls [28].
To test the effects of biotic stress, 2-week-old Tianlong 1 seedlings were inoculated with P. sojae 6497 as previously described[2].Cotyledons were collected at 0,6,12,24,and 48 h after inoculation and immediately frozen in liquid nitrogen for subsequent RNA extraction and qRT-PCR analysis. Untreated seedlings were used as controls.
2.2. DNA constructs
Details of cloning strategies and primers used are listed in Table S2. Recombinant plasmids were generated based mostly on the Golden Gate cloning system described by Liu et al. [24].
PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) was used to identify potential functional motifs in the GmDRR1pro sequence. A DNA segment from the start codon to 3000 bp upstream was examined to identify promoter elements.
Primers were designed based on the GmDRR1 genomic sequence, including the promoter, CDS, and terminator. The PCR product was ligated to the entry vector Fu28 [29] to generate the Fu28-GmDRR1pro:CDS::GFP::GmDRR1ter recombinant plasmid.The GmDRR1pro:CDS::GFP::GmDRR1ter construct was transferred to the binary vector pSoy7 via the LR reaction to produce the expression vector pSoy7-GmDRR1pro:CDS:GFP:GmDRR1ter, which was inserted into A. tumefaciens EHA105 cells. The transformed A.tumefaciens cells were then injected into the leaves of threeweek-old tobacco plants.Three days later,the subcellular localization of the GFP-tagged GmDRR1 was identified with a confocal microscope.
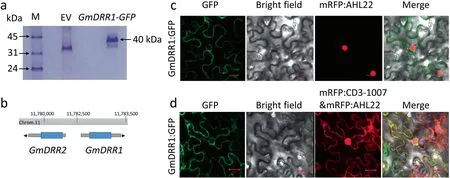
Fig.1. Identification of the location and candidate interaction protein of GmDRR1.(a)SDS-PAGE of candidate proteins after Co-IP.Total proteins from hairy roots expressing the GFP-tagged GmDRR1 were immunoprecipitated with anti-GFP antibody and the protein gel was stained with Coomassie Brilliant Blue.EV,empty vector.(b)The candidate gene GmDRR2 location in the soybean genome.(c,d)Subcellular localization of the GmDRR1 in N.benthamiana.The pSoy7:GmDRR1pro:CDS::GFP::GmDRR1ter was constructed and transformed into A.tumefaciens EHA105.GmDRR1-GFP signal was detected on the membrane by comparison with the nuclear marker AtAHL22 and cytoplasmic marker pm-rk-CD3-1007.
The pSoy7-GmDRR1pro:GUS::GmDRR1ter recombinant plasmid was inserted into wild-type Arabidopsis (Col-0) plants. Of the resulting 11 T1transgenic lines, three were selected to produce homozygous T3lines for further analyses. The seeds of the homozygous transgenic plants were germinated in plates containing Murashige and Skoog medium(Sigma,St.Louis,MO,USA).The plants were grown under long-day conditions(16-h light/8-h dark)with a light intensity of 100 μmol m-2s-1. GUS activity was detected as previously described [24]. Briefly, samples were fixed in 90% acetone for 5 min, placed on ice for 20 min, washed in distilled water for 5 min, and then stained with X-Gluc (5-bromo-4-chloro-3-indolyl-β-D-glucuronide) solution at 37 °C overnight.Samples were destained in 70% ethanol and then examined with a microscope and photographed. At least 10 plants per transgenic line were examined,with similar results.The protocol used to stain soybean roots was similar to that used to stain Arabidopsis plants.
2.3. Biochemical and physical assay
EMSA (Electrophoresis mobility shift assay) was completed with the LightShift Chemiluminescent EMSA Kit [Thermo Fisher Scientific (China) Co., Ltd.]. The GmNAC1 binding site on the GmDRR1 promoter sequence was predicted with an online tool(http://neomorph.salk.edu/dap_web/pages/browse_table_aj.php).The EMSA probes were designed based on the predicted binding sites and the results of a ChIP (Chromatin Immunoprecipitation Sequencing) experiment. Two pairs of biotin-labeled probes and the corresponding competitive probes were designed.
The recombinant plasmid pEarleygate201-GmDRR1 was constructed from pGWC-GmDRR1 and pEarleygate201 (with the nYFP tag)via the LR reaction.The recombinant plasmid pEarleygate202-GmDRR2 was constructed from pGWC-GmDRR2 and pEarleygate202 (with the cYFP tag) via the LR reaction. The two recombinant plasmids were inserted into separate A. tumefaciens EHA105 cells. The transformed A. tumefaciens cells were injected into the same tobacco leaves. After a three-day incubation under long-day conditions, fluorescence was detected with a confocal microscope[30].
The recombinant plasmid pGWB20-GmDRR2 constructed from pGWC-GmDRR2 and pGWB20 (with the Myc tag) via the LR reaction was inserted into A. tumefaciens EHA105 cells. A CoIP (Coimmunoprecipitation) experiment was performed with proteins extracted from the leaves of tobacco plants co-injected with A.tumefaciens cells carrying pSoy7-GmDRR1pro:CDS::GFP::GmDRR1ter and pGWB20-GmDRR2 (with a C-terminal Myc tag).The proteins were immunoprecipitated with the anti-GFP antibody, after which a Western blot assay was performed with the anti-Myc antibody.
The E-coniferyl alcohol coupling assay was performed using a slightly modified published procedure [10]. The His-tagged GmDRR1 produced in E.coli cells was purified for the assay,which was completed with a reaction mixture comprising 10 μmol mL-1E-coniferyl alcohol, 5 nmol mL-1GmDRR1 protein, and 10 μmol mL-1ammonium peroxydisulfate in buffer (0.1 mol L-1MES-HEPES-sodium acetate, pH 5.0) in a final volume of 250 μL.Control reaction mixtures consisted of only standard E-coniferyl alcohol or (+)-pinoresinol in buffer. The reaction mixtures were incubated at 30 °C for 3 h, after which the reaction product was extracted three times with 500 μL ethyl acetate.The extracts were combined and evaporated in a rotary evaporator. The residue was resuspended in a 50%methanol-water solution.The products were separated by high-performance liquid chromatography (HPLC).
2.4. Soybean transformation
The pB7GWIWG2(I)-GmDRR1-RNAi recombinant plasmids were constructed and inserted into A. tumefaciens EHA105 cells, which were used for the cotyledonary node transformation of Tianlong 1 soybean to produce GmDRR1-RNAi transgenic lines. The transgenic lines were screened based on Basta herbicide resistance,after which transgene expression was assessed by qRT-PCR. The GmDRR1-overexpressing lines were generated by insertion of pFGC5941-GmDRR1::GFP into Tianlong 1 soybean via A. tumefaciens-mediated cotyledonary node transformation. A Western blot and a qRT-PCR assay were used to identify GmDRR1-overexpressing lines. pHairyRed-GmNAC1::GFP and pSoy10-GmDRR1-Cas9 were constructed and inserted into soybean hairy roots via A. rhizogenes K599 [31].
The two target sites on the GmDRR1 gene selected with CRISPRGE(http://skl.scau.edu.cn/)were separated by 350 bp(Fig.5a).The sgRNA (single guide RNA) was synthesized and cloned into the pSoy10-Cas9 vector(for red fluorescence),which was inserted into A. rhizogenes K599 cells for the transformation of soybean hairy roots. Soybean hairy roots transformed with K599 cells carrying the pSoy10-Cas9 empty vector were used as controls.
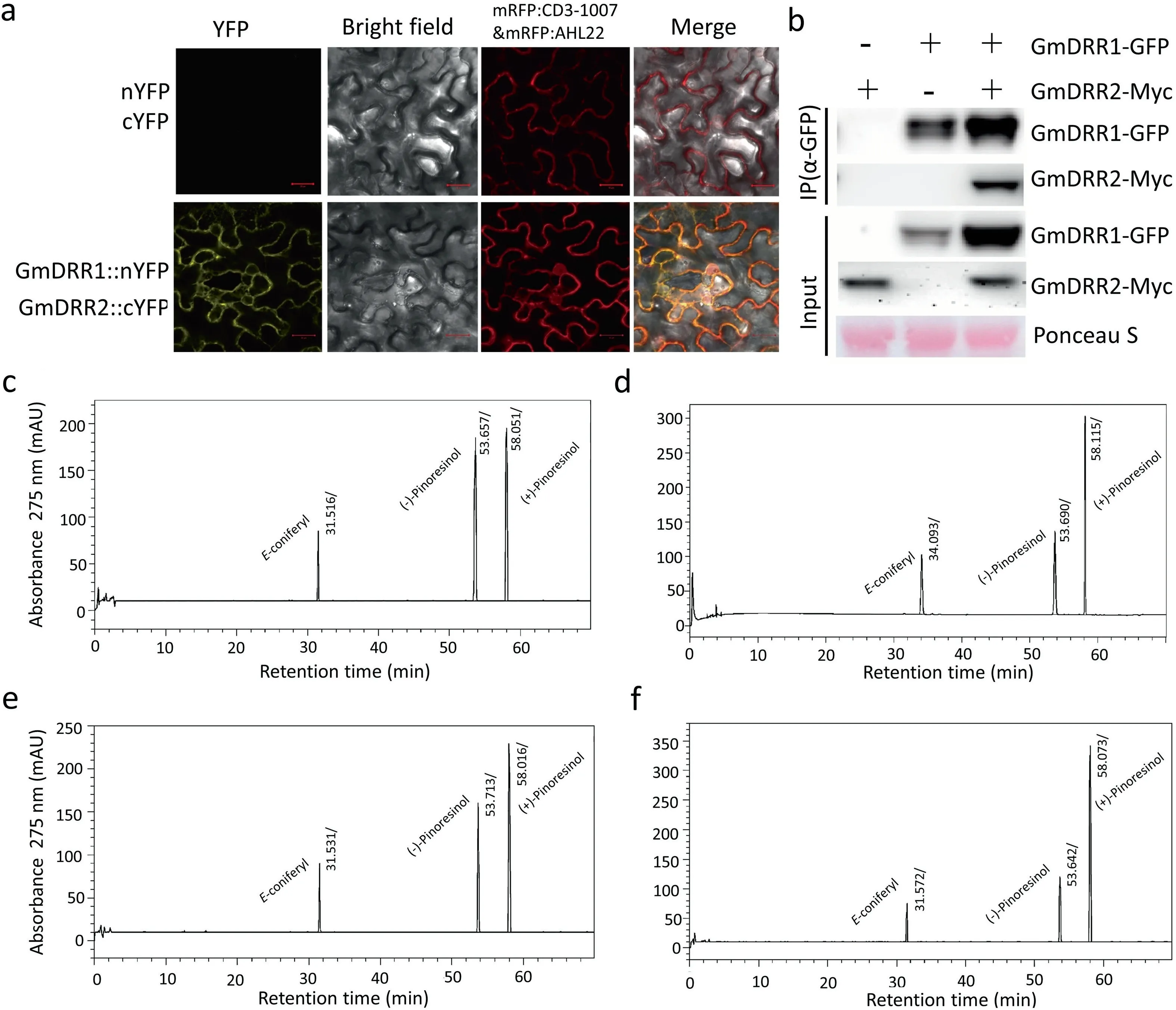
Fig. 2. Interaction protein confirmation and functional analysis of GmDRR1. (a) BiFC (Bimolecular Fluorescence Complementation) assay of protein interaction between GmDRR1 and GmDRR2 proteins in N. benthamiana cells. (b) Co-immunoprecipitation assay of GmDRR1 interaction with GmDRR2 proteins. (c, d, e, f) Chromatogram of Econiferyl alcohol free-radical coupling reaction. (c) HPLC chromatogram of E-coniferyl alcohol free-radical coupling-reaction products without GmDRR1 proteins. (d) HPLC chromatogram of E-coniferyl alcohol free-radical coupling-reaction products with GmDRR1. (e) HPLC chromatogram of E-coniferyl alcohol free-radical coupling-reaction products with DRR2. (f) HPLC chromatogram of E-coniferyl alcohol free-radical coupling-reaction products with GmDRR1 and GmDRR2.

Fig. 3. GUS expression levels of GmDRR1 promoters. (a–j) GUS staining of T3 generation of GmDRR1pro:GmDRR1::GUS transgenic Arabidopsis plants. (a–c) GUS staining of GmDRR1pro: GmDRR1::GUS in Arabidopsis lines (T3 generation). (d–f) Enlarged image of roots in (a–c). (g–i) GUS staining of flowers and seeds in GmDRR1pro:GmDRR1::GUS Arabidopsis plants. (j) GUS staining of GmDRR1pro:GmDRR1::GUS Arabidopsis two days after flowering. (k–r) GUS staining of GmDRR1pro:GmDRR1::GUS transgenic soybean roots. (o–r) Root tips of roots in the figures of the (k–n).
The transgenic soybean hairy roots were examined with a fluorescence microscope to detect red fluorescence(Fig.S1a).DNA was extracted from the transgenic soybean hairy roots for PCR to identify mutants(Fig.5b,c).The PCR product was ligated to the pGWC vector and then sequenced.
Total RNA was extracted from the soybean cultivar Tianlong 1 with the Magen RNA kit (TIANGEN Biotech, Beijing, China). A qRT-PCR assay was conducted as described by Wang et al. [26].The qRT-PCR primers were designed with Beacon Designer 7.0 software (http://www.premierbiosoft.com/qOligo/Oligo.jsp?PID = 1)(Table S2). The GmUNK1 gene was selected as the reference gene for normalizing data. Relative gene expression levels were calculated by the 2-ΔΔCTmethod [26].
2.5. Identification of candidate proteins interacting with GmDRR1
To identify proteins that interact with GmDRR1, soybean hairy roots overexpressing GFP-tagged GmDRR1 (Fig. S1b) were used for a CoIP–MS analysis.A.rhizogenes K599 cells were used to transform soybean with pHairyRed-GmDRR1::GFP. Two weeks later,transgenic soybean hairy roots overexpressing GmDRR1 were identified based on DsRed fluorescence(Fig.S1a).Specifically,GmDRR1 was detected in transgenic soybean hairy roots transformed with pHairyRed-GmDRR1::GFP (Fig. S1c). Proteins were extracted from transgenic hairy roots in lysis buffer compatible with the buffer used for protein isolation in CoIP experiment. Anti-GFP antibody was conjugated to the surface of magnetic beads with the cross linker disuccinimidyl suberate. The beads were then incubated with the transgenic hairy root protein extract to isolate GmDRR1 and its interacting proteins.The empty vector protein extract was used as a negative control. After proteins were eluted from the beads,the IP samples were analysed by liquid chromatography-tandem mass spectrometry (Shanghai Applied Protein Technology, Shanghai, China) (Fig. 1a).
3. Results
3.1. GmDRR1 interacts with GmDRR2 and promotes lignin synthesis

Fig.4. Phenotype analysis of GmDRR1 resistant to P.sojae. (a,b,c)Response of GmDRR1-overexpressing transgenic soybean cotyledons to P. sojae. (a) qRT-PCR analysis and Western blot of a positive transgenic line overexpressing GmDRR1.(b)Phenotype of GmDRR1-overexpressing-soybean cotyledon after infection by P.sojae.(c)Relative P.sojae biomass of GmDRR1-overexpressing transgenic cotyledons of soybean plants infected with P. sojae 48 h after inoculation. (d, e, f) Response of GmDRR1-RNAi transgenic soybean cotyledons to P.sojae.(d)qRT-PCR analysis of positive transgenic lines of GmDRR1-RNAi.(e)Phenotype of GmDRR1-RNAi cotyledons of soybean plants after infection by P.sojae.(f)Relative P.sojae biomass of GmDRR1-RNAi transgenic soybean cotyledons infected with P.sojae 48 h after inoculation.qRT-PCR analysis of P.sojae 6497 relative biomass based on the transcript level of the P.sojae Ps Actin gene in infected cotyledons after 48 h of inoculation.Soybean gene GmUKN1 was used as an internal reference for data normalization. Experiments were performed for three biological replicates, and means were compared using Student’s t-test (*, P <0.05; **, P <0.01). Bars indicate standard error of the mean.
A total of 22 candidate proteins that interact with GmDRR1 were identified following a (CoIP-MS (mass spectrometry)) analysis and functionally characterized via a BLAST search of the UniProt database (https://sparql.uniprot.org/) (Fig. 1a; Table S1). Two DIR homologous proteins, GmDRR2 (Glyma.11G150300) and GmDRR3(Glyma.02G169800), were among the identified proteins. Six of the proteins, which were not associated with disease response,were identified as heat shock proteins, zinc-finger proteins, signal transduction proteins, L-arginine biosynthesis I proteins, and structural constituents of ribosome-related proteins. GmDRR1 and GmDRR2 were detected at the same chromosome 11 locus,sharing a bidirectional promoter(Fig.1b).In an analysis involving AtAHL22 and pm-rk-CD3-1007 as nuclear and membrane markers, respectively [32–33], GmDRR1 was localized to the cell membrane(Fig. 1c, d).
On the basis of our CoIP-MS results, we hypothesized that GmDRR1 interacts with GmDRR2. We evaluated this potential interaction in a bimolecular fluorescence complementation (BiFC)assay. In the tobacco sample expressing GmDRR1::nYFP and GmDRR2::cYFP,yellow fluorescence was detected in the cell membrane,in agreement with the cellular location of GmDRR1(Fig.2a).
This interaction was further verified in a CoIP assay after transient expression in tobacco leaves (Fig. 2b. These analyses indicated direct interaction between GmDRR1 and GmDRR2. In the presence of GmDRR1 protein, the peak value of (+)-pinoresinol in the product was significantly higher than that of (–)-pinoresinol by the analysis of E-coniferyl alcohol free free-radical coupling reaction (Fig. 2c–f) and the product is mainly promoted by GmDRR1 but not GmDRR2(Fig. 2d–f), showing that GmDRR1 promotes the synthesis of (+)-pinoresinol.
3.2.GmDRR1 promoter activity in Arabidopsis and soybean hairy roots
To further investigate GmDRR1 function in plants,we evaluated GmDRR1 promoter activity in Arabidopsis. GmDRR1-GUS signals were observed predominantly at the leaf edge as well as in the hypocotyl, leaf veins, and lateral roots (Fig. 3a–f). Very weak signals were detected in the cotyledons and mature root. Signal was undetectable in flowers and seeds (Fig. 3g–j). We also inserted the pSoy7-GmDRR1pro::GUS recombinant plasmid into soybean hairy roots and assessed GmDRR1 promoter activity. Strong GUS signal was detected in soybean root primordia and root meristematic zones (Fig. 3k–r). A subsequent examination of the roots of transgenic Arabidopsis expressing GFP-tagged GmDRR1 indicated that GmDRR1 was present in the root tip and middle column sheath cells(Fig.S2).The GUS signal was restricted to the vascular bundle, root tips, and lateral roots. These results suggested that GmDRR1 is functional in the root, including at the root tip.
3.3. Effects of GmDRR1 on P. sojae resistance in soybean
We next investigated whether GmDRR1 is physiologically important for P.sojae resistance,using the cotyledons of transgenic soybean plants overexpressing GmDRR1. After a 48 h incubation with P. sojae 6497 zoospores, the disease symptoms on GmDRR1-overexpressing plants (Fig. 4a–c were less severe than those on plants in which GmDRR1 was silenced by RNAi (GmDRR1-RNAi)(Fig.4d–f).This phenotypic analysis was confirmed by an examination of the soybean hairy roots in which GmDRR1 was edited by CRISPR/Cas9. The Gmdrr1 knockout mutants were susceptible to the P. sojae 6497 infection (Fig. 5d, e), suggesting that GmDRR1 expression increases soybean resistance to P. sojae 6497. Thus,overexpressing GmDRR1 in soybean plants increased resistance to P. sojae and Gmdrr1 mutants as well as GmDRR1-RNAi transgenic soybean plants showed increased susceptibility to P. sojae.
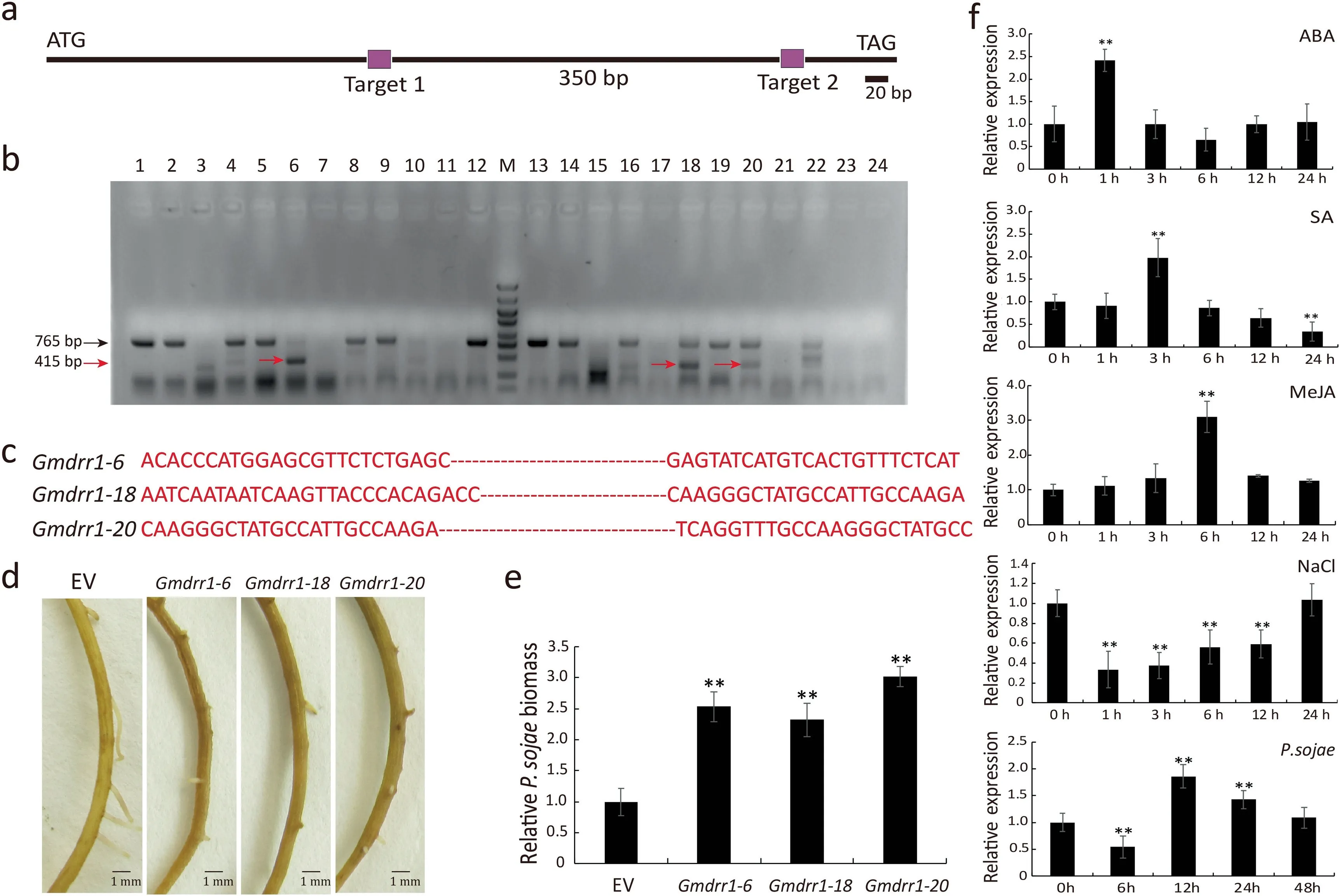
Fig. 5. Phenotype of Gmdrr1 CRISPR/Cas9 mutant resistant to P. sojae. (a) Schematic of CRISPR/Cas9 target regions. (b) PCR identification of candidate positive transgenic soybean roots.(c)Deleted sequences of positive CRISPR/Cas9-edited roots.(d)Phenotype analysis of Gmdrr1 transgenic roots after infection by P.sojae for 48 h.(e)Relative P.sojae biomass of CRISPR/Cas9-Gmdrr1 transgenic soybean hairy roots infected with P.sojae 48 h after inoculation.(f) GmDRR1 expression detection by qRT-PCR under NaCl,MeJA,ABA,SA,and P.sojae stresses.EV,empty vector.Soybean GmUKN1 gene was used as an internal reference for data normalization.Experiments were performed with at least three biological replicates, and means were compared using Student’s t-test (**, P <0.01). Bars indicate standard error of the mean.
Quantitative real-time (qRT)-PCR was used to measure GmDRR1 expression in response to P. sojae. A transcriptional peak was detected at 12 h after an inoculation with P. sojae 6497(Fig. 5f). We also evaluated whether GmDRR1 expression is influenced by abiotic stresses due to SA, MeJA, ABA, and NaCl.GmDRR1 expression was significantly changed following MeJA,ABA, and NaCl treatments (Fig. 5f). These results suggest a close relationship between GmDRR1 with MeJA, ABA signalling and salt stress.
3.4.GmNAC1 promoted expression of GmDRR1 and increased soybean resistance to P. sojae
GmDRR1 expression increased soybean resistance to P.sojae and expression of GmDRR1 was induced by inoculation with P. sojae(Figs. 4, 5). An earlier analysis [2] of the gene sequence diversity in soybean germplasm revealed a lack of significant differences between P. sojae-resistant and -sensitive cultivars [2]. However, a significant difference was identified in the GmDRR1 promoter region sequence. We accordingly hypothesized that the GmDRR1 function is influenced by the regulation of its promoter.
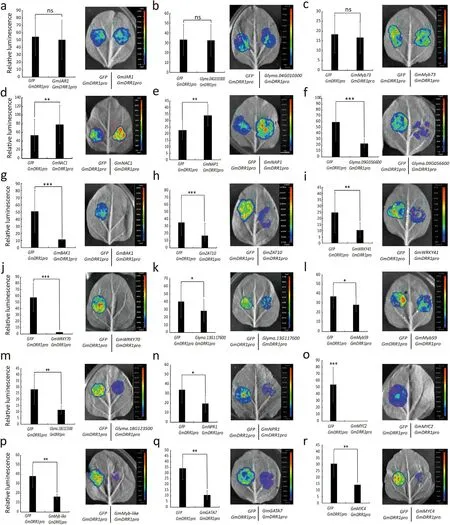
Fig.6. Screening for potential transcriptional regulators of GmDRR1.Twenty-five pieces of tobacco leaves were injected in each transcriptional activation experiment.Images are representative. The figures were drawn from the mean of the relative fluorescence of the experimental group with the control group. Experiments were performed for three biological replicates, and means were compared using Student’s t-test (ns, P >0.05; *, P <0.05; **, P <0.01; ***, P <0.001). Bars indicate standard error of the mean.
The GmDRR1 promoter contains some regulatory elements associated with response to biotic and abiotic stresses, including the W-box, MBS, CGTCA-motif, TAC-element, NACRS, ABRE, HSE,ARE,and TC-rich repeats(Fig.S3)(PlantCARE).To identify the regulator of GmDRR1 expression, 18 candidate TFs associated with soybean resistance to P.sojae were selected for an analysis of their effect on the GmDRR1 promoter (Table S3) [34]. Promoter activity analysis indicated that the GmDRR1 promoter was unaffected by TF1, TF2, and TF3 (Fig. 6a–c). In contrast, TF6–18 suppressed the GmDRR1 promoter activity (Fig. 6f–r), whereas TF4 and TF5 showed the opposite effect (Fig. 6d, e). Given that GmDRR1 increased soybean resistance to P. sojae, TFs upregulating GmDRR1 expression might contribute to this resistance.
Both TF4 and TF5 belong to the NAC TF family. TF4(GmNAC1) was selected for further analyses. Three regions (P3,P4, and P9) within the GmDRR1 promoter and terminator were identified as potential binding sites for GmNAC1. An additional six regions were selected as control sites (Fig. 7a). The pHairyRed-GmNAC1::GFP recombinant plasmid and the empty pHairyRed vector were inserted into soybean hairy roots via A.rhizogenes K599. The results of ChIP coupled with qRT-PCR(quantitative real-time PCR) analysis indicated that GmNAC1 had a relatively high affinity for P4 and P5 (Fig. 7b). An EMSA was conducted to confirm the ChIP–qRT-PCR results. Two probes were designed based on the P3 and P4 sequences (Fig. 7c). After purifying the His-tagged GmNAC1, we conducted the EMSA,which revealed that GmNAC1 had a higher affinity for probe B than for probe A (Fig. 7d). Compared with the non-transgenic control, the GmNAC1 and GmDRR1 expression levels were 4-fold and 2-fold higher, respectively, in GmNAC1-overexpressing roots (Fig. 7e). In response to P. sojae 6497 infection, GmNAC1 transcription peaked at 12 h after inoculation (Fig. 7f). P. sojae showed significantly lower accumulation in GmNAC1-overexpressing lines than in empty vector control lines(Fig. 7g–i). Thus, GmNAC1 likely promoted GmDRR1 expression,thereby increasing soybean resistance to P. sojae.
4. Discussion
GmDRR1 expression peaked in the stems of a P. sojae-resistant cultivar (Suinong 10) 12 h after inoculation with P. sojae race 1.Genomic sequencing indicated no significant differences in the transcribed region of GmDRR1 among soybean cultivars, implying that GmDRR1 is highly conserved [2]. This support that the different expression level of GmDRR1 might the main factor underlying soybean resistance to P.sojae.Differences in the GmDRR1 promoter region sequence between P.sojae-resistant and-sensitive cultivars suggested that transcriptional regulation is an important factor influencing GmDRR1 functions,including those involved in P. sojae resistance.
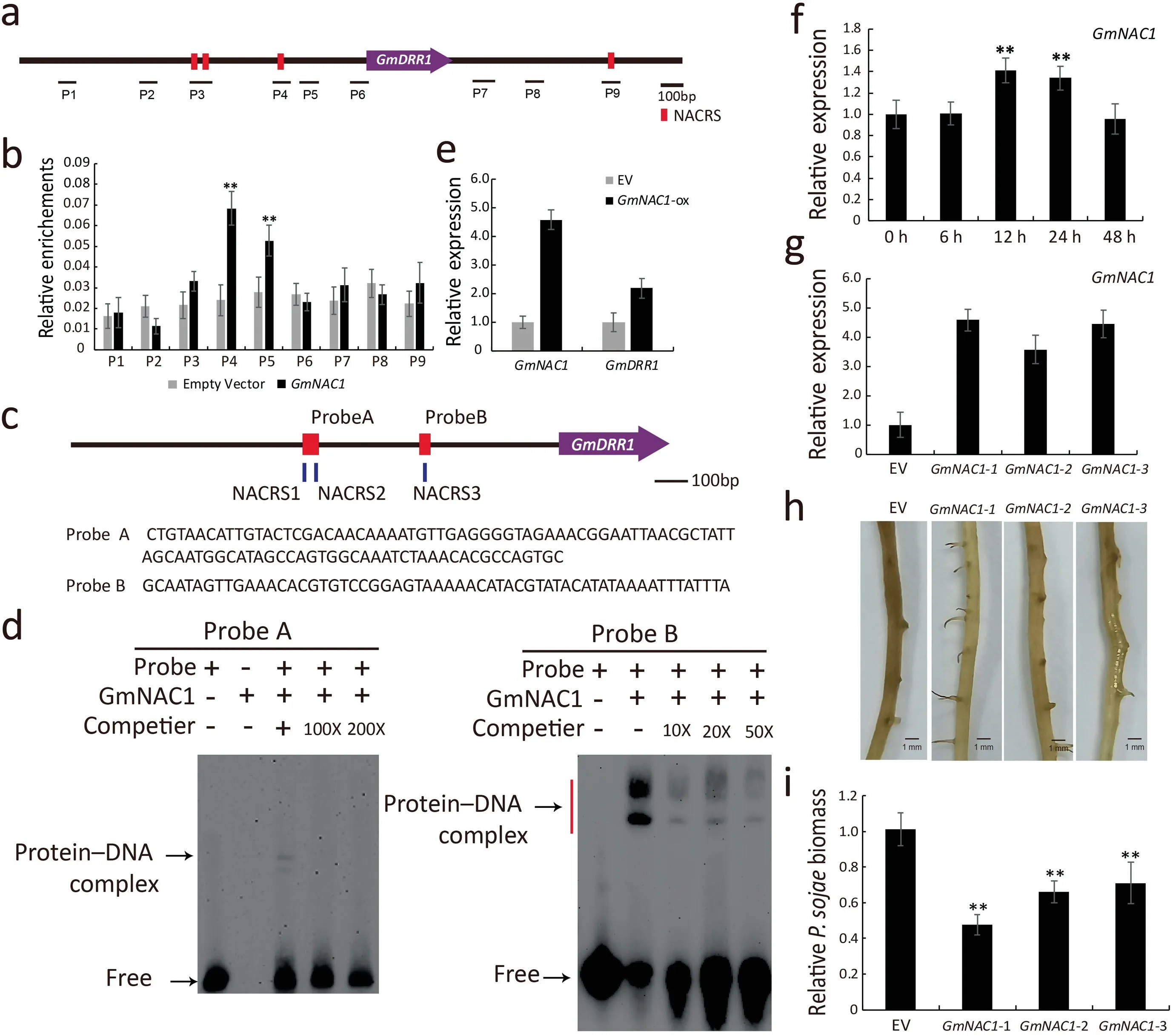
Fig.7. Detection of binding sites of GmNAC1 on the promoter region of GmDRR1.(a)Schematic diagram of GmDRR1 genomic sequence,indicating nucleotide sites of ChIP-PCR detection to which GmNAC1 binds.Red squares represent NACRS(NAC recognition sites).(b)ChIP-PCR results for GmNAC1 binding to GmDRR1 promoter,relative enrichment calculation used as IP (Immunoprecipitation)/Input. Experiments were performed for three biological replicates, and means were compared using Student’s t-test (**,P <0.01). Bars indicate standard error of the mean. (c, d) EMSA of GmNAC1 binding to the promoter region of GmDRR1 gene. Red squares represent probes. (e) qRT-PCR analysis of GmNAC1 and GmDRR1 transcripts in GmNAC1-overexpressing transgenic roots. (f) GmNAC1 expression identification after infection by P. sojae. (g) qRT-PCR detection of candidates overexpressing GmNAC1.(h,i)Phenotype and biomass analysis of GmNAC1-overexpressing-lines.EV,empty vector.Soybean gene GmUKN1 was used as an internal reference gene for data normalization, experiments were performed with three biological replicates, and means were compared using Student’s t-test (*,P <0.05; **P <0.01). Bars indicate standard error of the mean.

Fig.8. Model of GmNAC1 regulation of GmDRR1 expression in soybean in response to P. sojae. NACRS, NAC recognition sites. Red GmDRR1 represents the GmDRR1 protein after Phytophthora induction.
Our finding that GmNAC1 regulates GmDRR1 expression is consistent with that of two earlier studies of NAC-regulated lignin biosynthesis showing that expression of GmDRR1 was induced by P. sojae [2,35–37]. Besides the transcription factor GmNAC1,GmNAP1 was also found to activate the promoter function of GmDRR1 (Fig. 6e). Recently, GmNAP1 was identified to affect trichome morphology and pavement cell ploidy by regulating actin filament assembly [38]. This result also suggested that GmDRR1 is involved in pavement cell development.As the plant surface cell is a natural barrier to environmental stress.
On the basis of our results,we propose a working model(Fig.8)in which GmNAC1 regulates GmDRR1 expression, leading to increased lignin synthesis and greater resistance to P. sojae. When soybean plants are infected by P. sojae, GmNAC1 expression is upregulated, after which the encoded TF increases GmDRR1 transcription. The resulting GmDRR1 induces lignin synthesis in soybean, inhibiting the ability of P. sojae to infect plants. The functionality of GmDRR1 may depend on the formation of the GmDRR1–GmDRR2 dimer. Thus, we may have identified a novel signaling pathway mediating resistance of soybean plants to P.sojae. Because the DRR1 and NAC1 sequences are highly conserved in the plant kingdom,our findings suggest roles of NAC TFs during P. sojae infections of other plant species.
CRediT authorship contribution statement
Qingshan Chen, Yongfu Fu, Guolongyu, Zhendong Zhu and Dawei Xin:drafted and revised the manuscript and contributed to data and gene analysis.Jianan Zou, Jinhui Wang, Zhaoming Qi,Rongsheng Zhu,Xiaoxia Wu,Zhenbang Hu,Mingliang Yang,Xue Yang and Chunyan Liu:performed the experiments and analysed the data. All authors read and approved the final manuscript for publication.
Declaration of competing interest
Authors declare that there are no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (U20A2027, 32070274, 32072014) and the Heilongjiang Postdoctoral Science Foundation (LBH-Q16014).
Appendix A. Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2021.08.009.
- The Crop Journal的其它文章
- Origin, evolution, and molecular function of DELLA proteins in plants
- Far-red light: A regulator of plant morphology and photosynthetic capacity
- A rice XANTHINE DEHYDROGENASE gene regulates leaf senescence and response to abiotic stresses
- Dissection of heterotic loci for grain yield using interconnected chromosome segment substitution lines in rice
- The boron transporter SiBOR1 functions in cell wall integrity, cellular homeostasis, and panicle development in foxtail millet
- RNAi-mediated suppression of the abscisic acid catabolism gene OsABA8ox1 increases abscisic acid content and tolerance to saline–alkaline stress in rice (Oryza sativa L.)

