Inheritance of marsh spot disease resistance in cranberry common bean(Phaseolus vulgaris L.)
Bosen Jia, Robert L. Conner, Nadeem Khan, Anfu Hou,*, Xuhua Xia, Frank M. You
a Ottawa Research and Development Centre, Agriculture and Agri-Food Canada, Ottawa, Ontario K1A 0C6, Canada
b Department of Biology, University of Ottawa, Ottawa, Ontario K1N 6N5, Canada
c Morden Research and Development Centre, Agriculture and Agri-Food Canada, Morden, Manitoba R6M 1Y5, Canada
Keywords:Common bean Cranberry bean Marsh spot Resistance Recombinant inbred line (RIL)Joint segregation analysis Major gene Polygene
ABSTRACT Common bean (Phaseolus vulgaris) is an annual legume crop that is grown worldwide for its edible dry seeds and tender pods.Marsh spot(MS)of the seeds is a physio-genic stress disease affecting seed quality in beans. Studies have suggested that this disease involves a nutritional disorder caused by manganese deficiency, but the inheritance of resistance to this disease has not been reported. A biparental genetic population composed of 138 recombinant inbred lines (RILs) was developed from a cross between an MS resistant cultivar ‘Cran09’ and an MS susceptible cultivar ‘Messina’. The 138 RILs and their two parents were evaluated for MS resistance during five consecutive years from 2015 to 2019 in sandy and heavy clay soils in Morden, Manitoba, Canada. The MS incidence (MSI) and the MS resistance index(MSRI) representing disease severity were shown to be both highly correlated heritable traits that had high broad-sense heritability values (H2) of 86.5% and 83.2%, respectively. No significant differences for MSI and MSRI were observed between the two soil types in all five- (MSI) or four-year (MSRI) data collection, but significant correlations among years were observed despite MS resistance was moderately affected by year. The MSIs and MSRIs displayed a right-skewed distribution, indicating a mixed genetic model involving a few major genes and polygenes.Using the joint segregation analysis method,the same four major genes with additive-epistasis effects showed the best fit for both traits, explaining 84.4% and 85.3%of the phenotypic variance for MSI and MSRI, respectively. For both traits,the M1,M2,M3 and m4 acted as the favorable (resistant) alleles for the four genes where M and m represent two alleles of each gene.However,due to epistatic effects,only the individuals of the M1M2M3M4 haplotype appeared to be highly resistant, whereas those of the m1m2m3M4 haplotype were the most susceptible. The m4 allele significantly suppressed the additive effects of M1M2M3 on resistance, but decreased susceptibility due to the additive effects of m1m2m3. Further quantitative trait locus (QTL) mapping is warranted to identify and validate individual genes and develop molecular markers for marker-assisted selection of resistant cultivars.
1. Introduction
Common bean(Phaseolus vulgaris L.,2n=10)is one of the most important grain legumes for human consumption. The wild common bean originates from two gene pools, namely the Mesoamerican and Andean gene pools,both derived from the same common ancestral population[1,2].These gene pools were further domesticated by farmers in Mexico and South America nearly 8000 years ago.Nowadays,common beans are an important source of protein,vitamins, and minerals. Common beans have been widely planted in both developed and developing countries, especially in Africa and Latin America[3].In Canada,the common bean is an economically important crop that also plays an important role in crop rotation.From 2015 to 2019,the area planted with common beans has increased from 105,200 to 160,000 ha, with dry bean production reaching 316,800 Mt in Canada by 2019 [4]. However, significant yield losses caused by abiotic and biotic stresses are common[5,6]. In tropical and subtropical regions, where acidic soil leads to mineral deficiency, low availability of soil minerals can be a major production constraint [7]. In addition, since climate change impacts crop production, genetic improvement for abiotic tolerance should be a promising approach to significantly increase common bean yield [4].
Marsh spot (MS) is a common disease affecting seed quality in market classes such as cranberry beans and pea. In susceptible varieties, a discolored brown spot often occurs at the center of the seeds,which reduces seed quality and affects consumption values [8]. To date, only a few studies on MS disease have been reported. MS was first observed in peas grown in Britain [9]. The brown spot symptoms were thought to be caused by a pathogen,and scientists initially attributed MS to bacterial infection,but this initial assumption was disregarded as they failed to isolate the putative causal pathogens from peas [10]. Later MS was observed to occur alongside oats (Avena sativa L.) affected with grey speck[8]. Because grey speck in oats is caused by manganese(Mn) deficiency,scientists speculated that it could also be the cause of MS in legumes.A hydroponics growing method was employed to further investigate the effects of Mn2+concentration on root length, flowering time, and seed formation in pea [11]. A refined pot-culture technique was later performed in Long Ashton, England, United Kingdom, to grow many pulse species, including peas (Pisum sativum L.), broad beans (Vicia faba L.), runner beans (Phaseolus coccineus L.), green and French (dwarf) beans (Phaseolus vulgaris L.),in Mn-deficient sand medium. Examination of the seeds revealed typical severe marsh spots in peas and mild to severe marsh spot symptoms in broad and runner beans[12].These results confirmed that MS is caused by Mn deficiencies.
Manganese uptake may be a factor causing an Mn deficiency.Soil types can influence metal uptake by plants [13,14]. Soil characters such as metal concentration,soil pH,cation exchange capacity, organic matter and soil structure vary dramatically among different soil types and have proven to affect metal uptake [15].Sandy and heavy clay soils are two major mineral soil types in Canada. Sandy soil consists of larger particles compared to heavy clay, allowing water and nutrients to leach more easily in this coarse texture soil. Conversely, heavy clay soils have a greater water and nutrient retention capacity. Plant roots grow rapidly and smoothly in friable sandy soils,but water and metal can more easily become limiting. In heavy clay soil, though plenty of water and nutrients exist, clay soil may be too tight for plants to penetrate, so water and nutrients may be tightly absorbed and not available to the crop [16]. Moreover, pH is higher in heavy clay(6.5–7.0) than in sandy soil (5.5–6.5), which also affects metal uptake because metal solubility decreases in alkaline soil and increases in acidic soil [17]. Mn2+is the only metal form available to plants[18],and there is basically a proportional linear relationship between metal uptake and soil pH[17,19].Thus,Mn should be more soluble at equivalent Mn concentration, and therefore more available to plants in sandy than in heavy clay soils[20].Other factors, such as organic matter content, microbiome, quantity and reactivity of hydrous oxides, also influence metal uptake [20,21].
Genetic studies of MS resistance in beans have not been reported. Whether the MS is a quantitative or a qualitative trait and is controlled by few major genes, polygenes, or both is still uncertain. Joint segregation analysis (JSA) has been developed to jointly use phenotypic data from different generations of a biparental population to compare the frequency distribution in a real dataset with the theoretical distribution underlying the genetic model[22].Using JSA,major gene effects can be separated individually, while polygenes can be collectively detected. This method was initially used for a population comprising parents (P1and P2), F1, F2and F2:3generations [23,24], and was subsequently extended to the analysis of various biparental populations, such as doubled haploid (DH)/recombinant inbred line (RIL) [25],back-cross (BC) [26], back-cross inbred line (BIL) [27] and their combinations. The R package SEA, released in 2018 and recently updated, allows data analysis of 14 different population types and multiple genetic models with up to four major genes(https://cran.rproject.org/web/packages/SEA/index.html) [22]. JSA has been widely used since the 1990 s in genetic studies of several plant species such as soybean (Glycine max L.) [28,29], chickpea(Cicer arietinum L.)[30],wheat(Triticum aestivum L.)[31,32],maize(Zea mays L.) [33], rapeseed (Brassica napus L.) [34,35], melon(Cucumis melo L.) [36], pepper (Piper nigrum L.) [37], crape myrtle(Lagerstroemia indica L.) [38], chrysanthemum (Chrysanthemum × morifolium) [39], and iris (Iris germanica L.) [40].
The objective of this study was to determine the inheritance of MS in cranberry common beans using the developed biparental RIL population with the JSA method. This study constitutes the first report of MS inheritance in beans.
2. Materials and methods
2.1. Recombinant inbred line (RIL) population
A cross between the highly MS resistant cultivar ‘Cran09’ and the MS-susceptible cultivar ‘Messina’ was made to produce F1seeds. These F1seeds were planted in the greenhouse at the Morden Research and Development Centre,Morden,Manitoba,Canada to produce the F2generation. A total of 138 F2:8RILs were developed through single-seed descent from 2013 to 2015. The seeds of each individual plant in the last generation were bulkharvested to produce 138 RILs.
2.2. Evaluating marsh spot (MS) resistance and statistical analysis
The 138 RILs and their two parents were tested under two field soil types (sandy and heavy clay soil) for five years (2015–2019)using a partially-balanced lattice design with three replications,on the experimental fields of the Morden Research and Development Centre, Morden, Manitoba, Canada (49°11′N, 98°5′W). Five soil samples in each soil type of MS field sites were obtained each year.The soil samples were analyzed for pH values,Mn concentrations and other mineral material contents by the private firm FarmersEdge (https://www.farmersedge.ca/). According to the soil test results, textures of the soils at the experimental field sites were either heavy clay or sandy soil with low concentrations of manganese.Throughout the five-year field study,cereal crops were grown on each of the field sites in the year prior to the MS study.Each year,herbicide and fertilizer applications were made to maximize bean seed yield [41].
In a 5-m row with 75 cm row-spacing,95 seeds of each RIL were planted. All plants were harvested at maturity. Ten seeds of each RIL were randomly selected after harvest and graded for MS incidence (MSI) (percentage of seeds with symptoms) and for MS severity on a scale of 0–5, where 0 represents no symptom, and 5 represents severe symptoms (Fig. S1).
Marsh spot resistance index (MSRI), ranging from 0 to 5, was calculated to represent the disease severity or resistance level of each line:
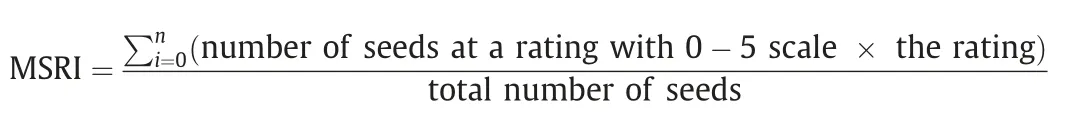
where, n is the total number of ratings and i = 0, 1, ..., 5,respectively.
2.3. Weather data collection
To determine whether the MS resistance is related to the weather conditions, the weather data, including temperature and precipitation during growth seasons of five years (2015–2019),were collected from the Morden Research and Development Centre, Agriculture and Agri-Food Canada.
2.4. Statistical analysis
A significantly right-skewed distribution for both MSI and MSRI justified the use of data transformations of the phenotypic datasets such as a square root or logarithm,but these failed to improve the normality of the data distributions significantly. Thus, nonparametric statistical tests were adopted for statistical analyses.To test the statistical differences of MSI and MSRI between years and soil types, the paired samples Wilcoxon test was used with the R function ‘‘wilcox.test” (https://www.r-project.org/). The R function‘‘cor”was used to measure the Spearman rank correlation coefficients of MSI and MSRI between different years.All statistical figures were drawn using the R package ggplot2 (https://cran.rproject.org/web/packages/ggplot2/index.html).
To exclude effects due to years or soil types, the mean value of each line across years or soil types was calculated by removing year and/or soil type effects:
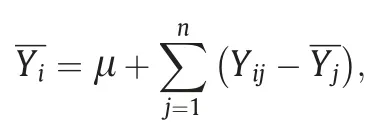
Furthermore, bidirectional cluster analysis with the Ward method was performed to group the RIL lines and years using the R package ‘pheatmap’ (https://cran.r-project.org/web/packages/pheatmap).
2.5. Broad-sense heritability estimation
The variances were estimated using the R package lme4 [42]according to a linear model as follows:
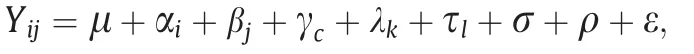
where Yijis the observed value of the ith row and jth column,μ is the population mean;αiand βjare the random effects of the ith row and jth column, respectively; γcis the fixed effect of the cth soil type; λkis the random effect of kth year, τlis the random genetic effect of the lth line, ρ and σ are the interaction effects between lines and years and between lines and soil types, respectively; and ε is the random error.
Broad-sense heritability (H2) was estimated using:
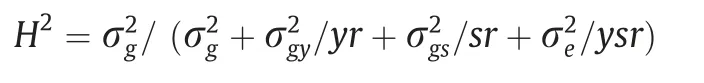
where σ2gis the genetic variance;σ2eis the error variance;σ2gyis the interaction variance between lines and years;σ2gsis the interaction variance between lines and soil types; r is the number of replications; y is the number of years; and s is the number of soil types.
2.6. Joint segregation analysis (JSA)
JSA was adopted to dissect the trait distribution of the RIL population to determine whether MS resistance was controlled by a few major genes and/or polygenes and to estimate the number of major genes and their additive dominant and epistatic effects. All the genetic analyses were performed using the R package SEA(https://cran.r-project.org/web/packages/SEA/index.html) [22]. A total of 35 genetic models available in SEA for hypothesizing zero to four major genes with additive, dominant and epistatic effects and/or polygenes were assessed to detect the most likely genetic models underlying MSI and MSRI (Table S1). JSA was conducted separately for a total of 18 phenotypic datasets of the 138 RILs and two parents, including ten for combinations of five individual years and two soil types, two mean datasets for two soil types across five years, five mean datasets for five years across two soil types, and one overall mean dataset for five years and two soil types.
Three criteria were used to select the best-fit genetic models:(1) the smallest Akaike Information Criterion (AIC) [22,43], (2)the highest heritability of the proposed gene model(the proportion of phenotypic variance explained by the model),and (3)no significant difference between the empirical data distribution and the theoretical distribution underlying the genetic model.AIC was calculated by the formula AIC = 2 k – 2lnL, where k is the number of estimated parameters in the model and L is the maximized value of the likelihood function for the model.Uniformity tests(U21,U22,and U23)for the mean,second moment and variance,as well as Smirnov(nW2) and Kolmogorov (Dn) statistical tests, were used to test the significant differences between the empirical data distribution and the theoretical distribution underlying the hypothesized models [22].
3. Results
3.1. Genetic variation of marsh spot (MS) resistance
The 138 RILs and parents (Cran09 and Messina) were rated for MS over five years in both soil types.Cran09 appeared to be highly resistant to MS in all years and soil types, having low average MSI(1.5% ± 2.5%) and MSRI ratings (0.04 ± 0.07). In contrast, Messina was highly susceptible with high average MSI (31.9% ± 6.5%) and MSRI ratings (0.8 ± 0.2) across all years and soil types, showing a substantial difference in MS rating between the two parents(Table 1).
The 138 RILs derived from Cran09 and Messina showed considerable variation in their MS rating, exceeding those of their two parents.The MSIs ranged from 0 to 43.67%,indicating transgressive segregation in the RILs compared to Can09(1.5)and Messina(31.9)(Table 1). Similar results were obtained for MSRI, with a range of 0–1.2 in the segregating population, and MSRIs of 0.04 and 0.8 for Cran09 and Messina, respectively (Table 1).
Both MSIs and MSRIs of the RIL population were right-skewed in all datasets (Fig. 1), indicating that the two traits were mostly controlled by a few major genes or a mixed genetic model involving major genes plus polygenes. Cluster analysis grouped the 138 RILs and the two parents into a resistant (R) and a susceptible (S)group for MSI (Fig. S2a) and MSRI (Fig. S2b). The parents Cran09 and Messina were classified separately into the R and S groups,respectively. Most of the RILs were included in the R group.
MSI and MSRI showed broad-sense heritability (H2) of 86.51%and 83.29%, respectively, for the overall mean dataset across all five years and two soil types.In the heavy clay and sandy soil types,H2was estimated at 89.65% and 85.91% for MSI and 88.41% and 85.24% for MSRI, respectively. In the datasets of individual years and soil types, the highest H2estimates were 97.13% for MSI and 96.95%for MSRI in the S-2017 dataset.Overall,both traits had high H2regardless of years or soil types, indicating that MS resistance was highly heritable (Table 1). Significant high correlations were observed between the two traits in all datasets based on Spearman rank correlation coefficient of 0.96–0.99 (Table S2).

Table 1 Marsh spot incidence (MSI) and marsh spot resistance index (MSRI) of the 138 RILs and their two parents across 5 years and two soil types (sandy and heavy clay).
3.2. Marsh spot (MS) resistance affected by years and soil types
Due to the skewed distribution of MSI and MSRI in the RIL population,a non-parametric paired-sample Wilcoxon test was used to test the differences in MSRI and MSI between years or soil types.For MSI, no significant differences were detected between 2016 and 2017 or between 2017 and 2018, but the other comparisons were significantly different at 5% or 1% probability levels (Fig. 2a).Mean values across years were used for bi-directional cluster analysis. The latter grouped the 2016, 2017 and 2018 data together(Fig. S2a). For MSRI, no significant differences were observed among 2016, 2017 and 2018 or between 2018 and 2019 (Fig. 2b).Cluster analysis generated results similar to MSI in that the first three years (2016–2018) and the last two years (2019 and 2010)were clustered into two separate groups(Fig.S2b).Taken together,the disease ratings of the Cran09/Messina RIL population and its two parents were similar in 2016–2018 and in 2019–2020 but differed between the two groups(Fig. S2).
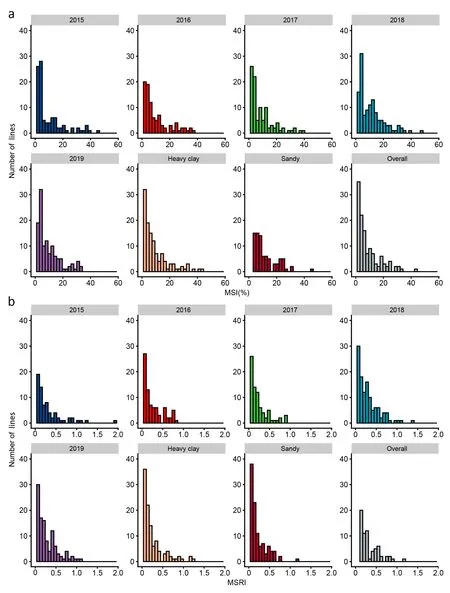
Fig. 1. Histograms of marsh spot incidence (MSI, %) (a) and marsh spot resistance index (MSRI) (b) in different years, soil types and overall mean values.
No significant differences were observed between the two soil types for MSI in all five years or in the overall mean dataset(Fig. 2c). Similar results were obtained for MSRI with the exceptions of 2017 and 2018, where a significant difference was observed between the two soil types (Fig. 2d). The overall results suggest that the MS ratings are not significantly affected by soil type.
To determine whether climate and additional soil conditions impact the MS resistance, the temperature and precipitation data during growth periods were collected,and pH and mineral concentration, including Mn concentration for both sandy soil and heavy clay fields, were tested. The patterns of temperature changes during growth periods were similar among the five years in Morden,Manitoba, Canada (Fig. S3a). The precipitation varied dramatically during five years from May to September, but no obvious patterns were observed (Fig. S3b).
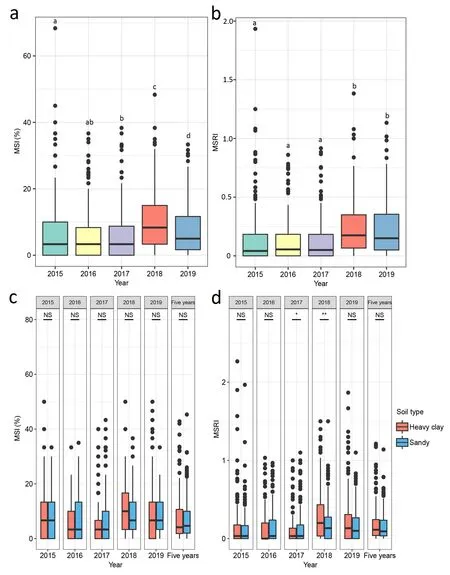
Fig.2. Boxplots of marsh spot incidence(MSI,%)(a and c)and marsh spot resistance index(MSRI)(b and d)across five years(a and b)and between the two soil types across five years and five-year means(c and d).The paired samples Wilcoxon test was used to test for significant differences between the two soil types.*,P <0.05;**,P <0.01;NS,not significant.
Soil pH varied from 7.2 to 7.6 and was similar in two soil types for five years(Fig.S4b).However,the soil Mn concentration varied largely in different years. During 2015–2017, the soil Mn concentrations were 19.5–48.5 mg L-1and 4.3–10.9 mg L-1for heavy clay and sandy soil fields, respectively. In contrast, in 2018 and 2019,the soil Mn concentrations decreased to 4.2–4.5 mg L-1and 3.1–3.4 mg L-1for two soil type fields, respectively (Fig. S4).
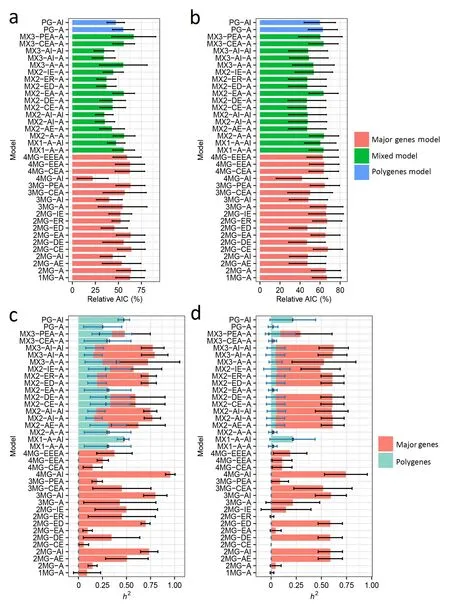
Fig. 3. Bar charts of the relative AIC values (a and b) and heritability (h2) estimates (c and d) of the 35 genetic models tested for the18 phenotypic datasets of marsh spot incidence(MSI,%)(a and c)and marsh spot resistance index(MSRI) (b and d)across five years and two soil types.For each phenotypic dataset,the highest AIC values in all genetic models were set as 100%.The relative AIC of each genetic model was calculated as a percentage of the AIC over the maximum AIC.The best model was the one with the lowest relative AIC.
3.3. Assessment of genetic models
A total of 35 genetic models(Table S1),including two with polygenes only(i.e., PG-AI and PG-A),17 with mixed models including 1–4 major genes in combination with polygenes, and 16 with 1–4 major genes only, were tested for both MSI and MSRI. For the RIL population, the SEA R package provides only major gene models for the proposed four-gene inheritance(i.e.,4MG-X,where X represents different genetic effects);thus,no polygenes were assessed in four-gene models (Table S1). For each of the proposed models, a total of 18 phenotypic datasets (Table 1) for MSI and MSRI were analyzed, and the results are listed in Tables S3 and S4. To choose the best-fit model, the average AIC values of 18 phenotypic datasets were calculated for each model. However, AIC values estimated from different phenotypic datasets were not comparable.For each phenotypic dataset, relative AIC values of genetic models were calculated by setting the highest AIC values in all genetic models to 100%, and then calculating a percentage of the AIC over the maximum AIC.The best-fit model was the one with the lowest average relative AIC value of the 18 phenotypic datasets.
Fig. 3a and b depict the average relative AIC values of all 35 genetic models for MSI and MSRI.The model 4MG-AI had the lowest AIC values for both traits with 22.10% and 42.93% for MSI and MSRI, respectively, and also explained the highest proportion of the phenotypic variance (h2) with 96% ± 40% and 74% ± 21% for MSI and MSRI,respectively(Fig.3c and d),indicating that the same model with four major genes with additive-epistasis was optimal for both traits.
Out of the 35 genetic models (Table S1), there were the other four-gene models (i.e., 4MG-EEEA, 4MG-EEA, and 4MG-CEA) that all included major genes with only additive effects. These models had much higher relative AIC values than 4MG-AI,and also showed fairly low h2estimates (15%–37%, for MSI and 11%–19% for MSRI)(Fig. 3c and d), thereby pointing toward the role of epistasis in MS resistance inheritance.
The 4MG-AI model was further statistically tested for uniformity using uniformity, Smirnov and Kolmogorov statistical tests to determine whether the theoretical distribution underlying the genetic models fits the empirical data distribution. Table S5 lists the statistical test results for the 4MG-AI model in eight mean datasets for MSI and MSRI. No significant differences were observed for most of the tests of goodness-to-fit (U21, U22, U23, nW2,and Dn) in 4MG-AI (Table S5) while most of the tests for the other models showed significant differences(Tables S3,S4).Therefore,it was reasonable to deduce that MSI and MSRI were controlled predominately by four major genes with both additive and epistatic effects (4MG-AI).
3.4. Genetic analysis of the best-fit genetic model
Table 2 lists the first- and second-order genetic parameters for the best-fit model 4MG-AI,estimated in the eight mean phenotypic datasets. These parameters included additive effects of the four major genes (da, db, dc, and dd), epistatic effects of paired genes(iab-icd),genetic variances of major genes(σ2M)and polygenes plus residual (σ2R+P), and heritability (h2, a proportion of phenotypic variance explained by major genes).The additive effects of the first three major genes(a, b, and c) appeared to be negative in contrast to the fourth gene(d),which appeared to be positive for both traits in most datasets(Table 2). The four major genes explained 84.37%to 98.17%of the phenotypic variance for MSIs and 85.30%to 98.54%of phenotypic variance for MSRI in the phenotypic datasets.Although the additive effects of the four genes were all negative in some genetic models estimated from different datasets(Table 2),the genetic architecture of the four genes in these models performed similarly. Hereafter only the best-fitted models obtained from the overall mean phenotypic dataset are exhibited (Table 2).
The additive effects of the four major genes were-3.53,-1.99,-3.74 and +1.94 for MSI, and -0.05, -0.05, -0.10 and +0.01 for MSRI, respectively. Large digenic epistatic effects, even greater than additive effects, were also observed, ranging from -3.67 to 3.40 for MSI and from -0.09 to 0.08 for MSRI. Since MSI and MSR are two non-independent traits and they were highly correlated (Table S2), we hypothesized the two traits shared the same four genes. Thus, for simplicity, gene symbols M1, M2, M3 and M4 corresponding to four genes (a–d) were assigned to both traits with alleles M and m. If M1, M2, and M3 are considered favorable alleles for the first three genes due to their negative effects on symptom development, then m4 would be a favorable allele for the M4 gene due to its positive effect on symptom development.
Based on the 4MG-AI model, the posterior probability of all 16 genotypes of four genes for 138 RILs was estimated (Tables S6,S7), which allows us to predict the possible genotype of each RIL.The most likely genotype of an RIL was the one that had the highest posterior probability among all 16 genotypes.If two or more genotypes shared the same highest probability, all of them wereassigned to the RIL. In addition, to clarify the possible genotypes graphically, a value of zero was assigned to the remaining genotypes.As such,the heat maps of the highest posterior probabilities with the possible genotypes are depicted in Fig. 4a for MSI and Fig. 4b for MSRI. All RILs were assigned to 14 genotypes for MSI(Fig. 4a) and to all 16 genotypes for MSRI (Fig. 4b).

Table 2 First and second order genetic parameters of the 4MG-AI model based on eight phenotypic datasets of marsh spot incidence (MSI, %) and marsh spot resistance index (MSRI).
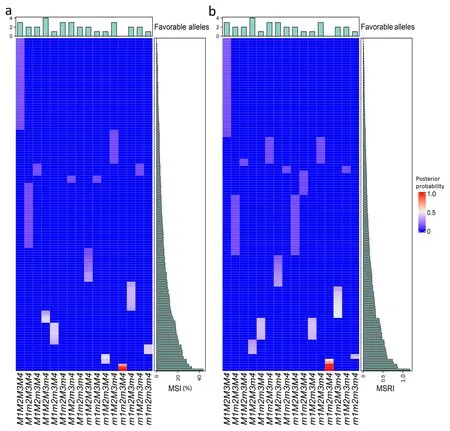
Fig.4. Posterior probability of each of 16 genotypes of four major genes in the 4MG-AI model for marsh spot incidence(MSI,%)(a)and marsh spot resistance index(MSRI)(b)in the RIL population of 138 lines. The mean dataset over five years and two soil types were used.
For MSI(Fig.4a),the three most susceptible lines(30.3%–43.7%MSI)had the haplotype m1m2m3M4,i.e.,the four unfavorable alleles.In contrast,the 39 top resistant lines(0–2.3%MSI)had the haplotype M1M2M3M4 that had three of the four favorable alleles(M1–M3). However, if the RILs had all four favorable alleles(M1M2M3m4), they turned out to be relatively susceptible(14.0%–17.7%of MSI).Similarly,for MSRI,the four most susceptible lines(0.75–1.2 MSRI)had a haplotype m1m2m3M4,all unfavorable alleles.The top 41 resistant lines(0–0.05 MSRI)had the haplotype M1M2M3M4 containing three of the four favorable alleles (M1–M3).The outcome was the same as for MSI in which the lines with all four favorable alleles (M1M2M3m4) were susceptible (0.5–0.6).
Further analysis indicated that MS ratings decreased with favorable alleles from zero to three, confirming that three of the four genes were primarily additive, but the fourth may suppress the expression of the other three genes (Fig. 5).
Therefore,the epistatic interactions among the four genes were further analyzed (Fig. S5). Although complex digenetic interaction effects were observed among the four genes, the first three genes(M1,M2,and M3)behaved mostly in an additive manner(Fig.S5b–d, f–h), while M4 significantly promoted or impeded expression of additive resistance conferred by the first three genes(Fig.S5a,e).
The M4 gene had significant epistatic effects on the M1,M2,and M3 genes. The M4 allele with the M1M2M3 haplotype resulted in the highest resistance,whereas the m4 allele suppressed the additive effects of M1M2M3. Conversely, the M4 allele assisted the expression of the additive effects of m1m2m3,resulting in the most susceptible plants.
The resistant parent Cran09 had an average MSI and MSRI of 1.5%and 0.04, respectively, while MSI and MSRI values in the susceptible parent Messina were 31.9%and 0.80,respectively.According to the ratings of the parents,it seemed reasonable to infer that Cran09 had the haplotype of M1M2M3M4, and that Messina may have the haplotype m1m2m3M4, m1m2m3m4 or m1M2m3M4.However, because 14–16 genotypes were inferred in the RILs(Fig. 4), m1m2m3m4 was the most likely haplotype that could account for the segregation of 16 genotypes.
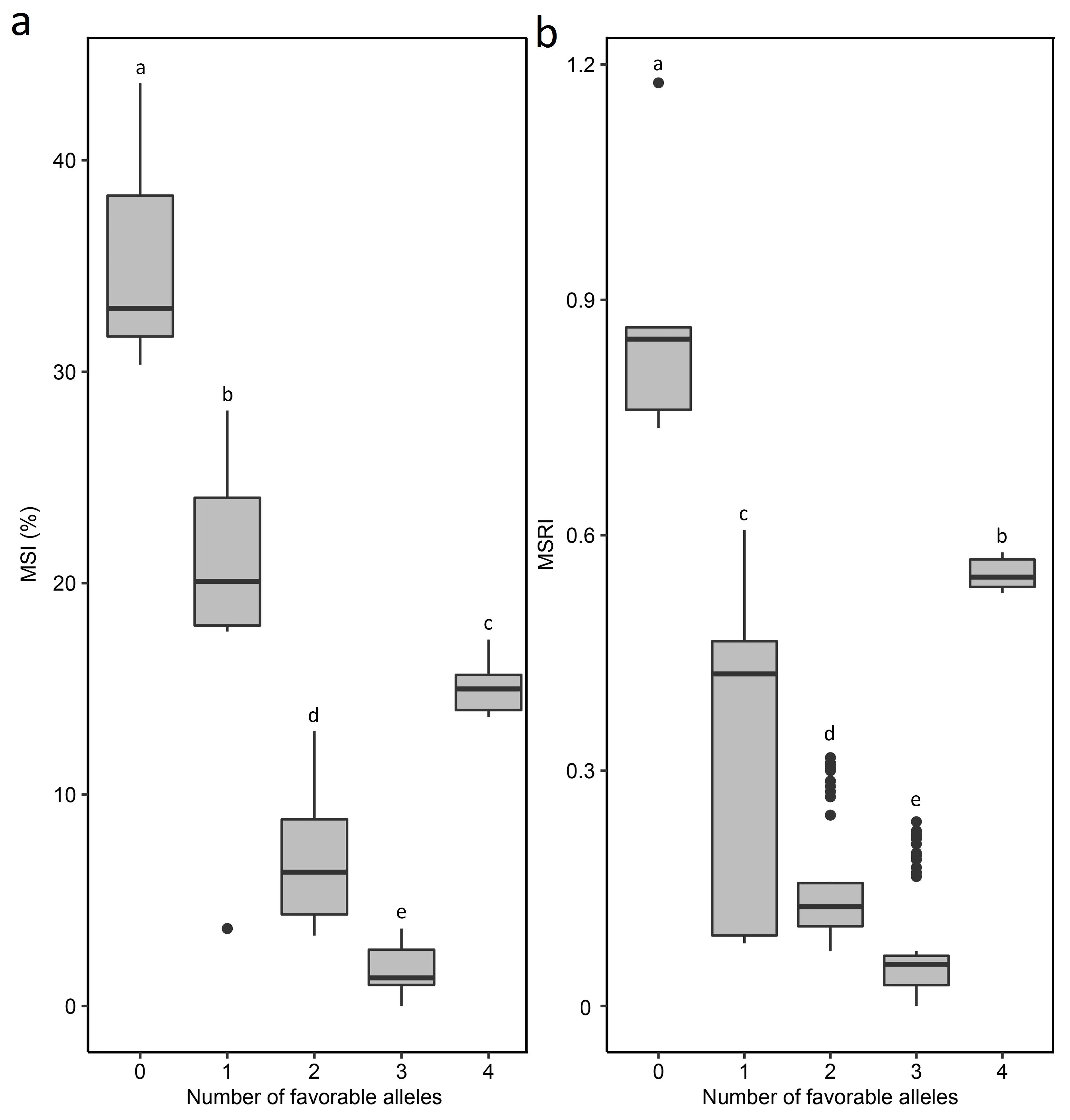
Fig.5. Relationship between marsh spot incidence(MSI,%)(a)and marsh spot resistance index(MSRI)(b)with the number of favorable alleles in the RIL population of 138 lines underlying the 4MG-AI model. The letters on the top of boxes represent statistical significance at the 5% probability level.
4. Discussion
MS is a common disease leading to seed quality and yield losses in cranberry beans and other common beans.Although MS disease was reported in beans in the 1930s[11,44],the genetics of its resistance remained unknown. This study is the first report on the inheritance of MS resistance in cranberry or other common beans.The study will facilitate discovering the genetic mechanisms controlling MS resistance, identifying associated genes, and developing molecular markers for resistance breeding.
This study developed a new genetic population consisting of 138 RILs derived from a cross between the resistant cultivar Cran09 and the susceptible cultivar Messina. MS ratings of the 138 RILs were evaluated over five years under two soil types,demonstrating that the genes associated with MS resistance in the two parents segregated and recombined to produce a genetic variation exceeding the performance of the two parents, thereby rendering this population useful for genetic studies and quantitative trait locus (QTL) mapping.
To study MS resistance,MSI and MSRI,which represent the incidence and severity of MS disease, respectively, were evaluated.Spearman rank correlation and genetic analysis of the phenotypes suggested that these two traits were highly correlated (r >0.96)and most likely had similar or the same genetic architecture.
Previous studies have confirmed that MS disease is most likely caused by Mn deficiency caused by low soil Mn or poor Mn uptake[9–11,45]; hence, the potential link between MS and soil type[13,14]. Thus, in this study, MS resistance was evaluated under the two soil types, sandy and heavy clay soil. The two soil types had similar Mn levels but different soil structures. No significant difference in MSI or MSRI between soil types was found. In addition, similar MS ratings among 2015, 2016 and 2017 and between 2018 and 2019 were observed, but significant differences existed between the first three years and the last two years. We analyzed the changes in temperature and precipitation during growth periods of five years, but no patterns of either temperature or rainfall were found to relate to the differences of MS resistance between years. However, the soil Mn concentrations in 2015–2017 were significantly higher than those in 2018–2019 (Fig. S5a), which may result in the differences of MS resistance between the years of 2015–2017 and the years of 2018–2019. This result also indirectly confirms that MS disease is associated with Mn deficiency.However, although the soil Mn concentration is much lower in sandy soil than in heavy clay, the Mn nutrition is more easily absorbed by plants in sandy soil [20], resulting in no significant differences in MS resistance between the two soil types. Overall,the evaluation of the MS resistance of the population across five years and two soil types strongly indicates that MS rating is less affected by environment than by genes. High broad-sense heritability estimates of both traits also support this conclusion. Thus,MS resistance is a relatively stable and heritable character.
The significantly right-skewed distribution of MSI and MSRI observed suggested the reasonable hypothesis of MSI and MSRI being most likely controlled by few major genes plus some minor-effect polygenes.Joint segregation analysis provided an efficient approach to dissect this type of skewed distribution to determine whether the trait is controlled by major genes and/or polygenes in genetic populations such as double haploid [46] and RIL [22,24,25,28,29]. It has been successfully applied to dissect some complex quantitative traits,especially those involving major genes plus polygenes,such as disease resistance[29],male sterility[37],and flowering date[25].In the present study,JSA showed that for both MSI and MSRI, 4MG-AI (four major genes with additive and epistatic effects) was the best genetic model among the 35 tested (Table S1). This is the first empirical dataset of plant quantitative traits that fits a four major gene system using JSA.Theoretically,only the genetic model supporting a maximum of four pairs of major genes without polygenes has been developed and evaluated using simulation data of a RIL population [28], and implemented in the R package SEA V1.0 (https://cran.r-project.org/web/packages/SEA/index.html). Though 4MG-AI was considered the best-fit model based on the AIC criterion, various statistical tests,and heritability of the model(proportion of phenotypic variance explained by the model),additional major genes and/or polygene should not be excluded because of the theoretical limit in the development of genetic models with more major genes and polygenes. In addition, we tested the models with one to three major genes plus polygenes, from which polygenes explained 0–47.19%of the phenotypic variance. Therefore, we deduce that polygenes could also be a portion that confers MS resistance.
The SEA package provides a function that estimates the posterior probability of all genotypes underlying genetic models, which can be used to predict the most probable genotype of individuals of a population by selecting the genotype with the highest probability as the candidate genotype. Based on the predicted genotypes of population individuals, we can further analyze additive and epistatic effects of all genes in the best-fit genetic models in detail.Using this method, we found that digenetic epistatic interaction significantly suppressed additive effects of genes in MS resistance.
5. Conclusions
The evaluation of MS resistance in a RIL population of 138 lines suggested that MSRI and MSI were stable and heritable traits. The JSA of the RIL population and its parents revealed that both traits were controlled by four pairs of major genes (M1, M2, M3, and M4) with additive and epistatic effects. Among them, M4 performed as a key suppressor that affected additive effects of the other three genes on MS resistance. The individuals with the M1M2M3M4 haplotype were the most resistant, but those with the M1M2M3m4 haplotype were susceptible.Further QTL mapping would be useful to identify the individual genes or QTL on chromosomes,explore their genetic mechanisms,and develop corresponding markers for molecular breeding of resistant cultivars.
CRediT authorship contribution statement
Bosen Jia:Methodology,Data analysis,Manuscript preparation,Writing - original draft.Robert Conner:Field experiment design and implementation, Phenotypic data generation, Writing -reviewing and editing.Nadeem Khan:Data analysis, Writing -reviewing and editing.Anfu Hou:Conceptualization, Genetic population generation, Writing - reviewing and editing.Xuhua Xia:Conceptualization, Writing - reviewing and editing.Frank You:Conceptualization, Methodology, Data analysis, Manuscript preparation, Writing - original draft, Review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors thank Dr. Kenneth B. McRae (formerly of Kentville Research and Development Centre) for preparing the partiallybalanced lattice designs for the field studies and Waldo C. Penner,Dennis B.Stoesz,Dena Young,Janet Gruenke,and Nadine Dionne of the Morden Research and Development Centre, Agriculture and Agri-Food Canada (AAFC) for their technical support. We thank Steve Sager of the Morden Research and Development Centre,AAFC for providing the related weather data. We gratefully acknowledge the financial support provided by the Manitoba Pulse and Soybean Growers,AAFC,the Canadian Agricultural Partnership Pulse Science Cluster, and NSERC (RGPIN/2018-03878). We thank Dr. Yuanming Zhang for providing additional information of the R package SEA. We also thank Drs. Sylvie Cloutier, Tara Edwards,and Madeleine Lévesque-Lemay for their constructive comments and editing of the manuscript.
Appendix A. Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2021.05.013.
- The Crop Journal的其它文章
- Origin, evolution, and molecular function of DELLA proteins in plants
- Far-red light: A regulator of plant morphology and photosynthetic capacity
- A rice XANTHINE DEHYDROGENASE gene regulates leaf senescence and response to abiotic stresses
- Dissection of heterotic loci for grain yield using interconnected chromosome segment substitution lines in rice
- A soybean NAC homolog contributes to resistance to Phytophthora sojae mediated by dirigent proteins
- The boron transporter SiBOR1 functions in cell wall integrity, cellular homeostasis, and panicle development in foxtail millet

