Mapping of wheat stripe rust resistance gene Yr041133 by BSR-Seq analysis
Yhui Li, Ruiming Lin, Jinghung Hu, Xiohn Shi, Dn Qiu, Peipei Wu,Geremehin Hte Goitom, Siqi Wng, Hongjun Zhng, Li Yng, Hongwei Liu, Qiuhong Wu,Jingzhong Xie, Yng Zhou, Zhiyong Liu,*, Hongjie Li,*
a The National Engineering Laboratory of Crop Molecular Breeding, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
b Institute of Genetics and Developmental Biology, The Innovative Academy of Seed Design, Chinese Academy of Sciences, Beijing 100101, China
c State Key Laboratory for Biology of Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing 100193, China
d College of Humanities and Development Studies, China Agricultural University, Beijing 100083, China
Keywords:Bulked segregant analysis Genetic mapping Puccinia striiformis f. sp. tritici Triticum aestivum Yellow rust
ABSTRACT Puccinia striiformis Westend.f.sp.tritici(Pst)pathotype CYR34 is widely virulent and prevalent in China.Here,we report identification of a strpie rust resistance (Yr)gene, designated Yr041133,in winter wheat line 041133. This line produced a hypersensitive reaction to CYR34 and conferred resistance to 13 other pathotypes.Resistance to CYR34 in line 041133 was controlled by a single dominant gene.Bulked segregant RNA sequencing(BSR-Seq)was performed on a pair of RNA bulks generated by pooling resistant and susceptible recombinant inbred lines.Yr041133 was mapped to a 1.7 cM genetic interval on the chromosome arm 7BL that corresponded to a 0.8 Mb physical interval (608.9–609.7 Mb) in the Chinese Spring reference genome. Based on its unique physical location Yr041133 differred from the other Yr genes on this chromosome arm.
1. Introduction
Wheat stripe rust occurs in more than 60 countries, causing approximately 1% of global grain losses annually. It is well known for widespread distribution, rapid epidemic development and large-scale damage to wheat production [1,2]. The magnitude of yield losses depends on growth stage at initial infection, level of resistance, and environmental conditions favoring its spread [1].Stripe rust is considered the most important disease of wheat in China and major epidemics in 1950, 1956, 1958, 1960, 1962,1964,1990,2002,and 2009 caused losses estimated in the millions of tons[3,4].During 2006–2015,the average annual grain loss due to stripe rust was estimated at 159,000 t ranging from 66,500 t in 2011 to 257,400 t in 2009 [5].
The causal fungus of wheat stripe rust,Puccinia striiformis Westend. f. sp. tritici Erikss. (Pst), mainly infects leaves, but can also affect leaf sheaths, stalks, and spikes under favorable conditions,i.e., susceptible cultivars, high moisture, and favorable temperatures (7–12 °C at night and 20–25 °C during the day). Because of its economic importance stripe rust has to be managed countrywide by adoption of resistant cultivars (preferred) and/or use of fungicides [6]. Resistance to stripe rust at any level contributes reduced costs of production and protection of the environment[1].
Since Lupton and Macer[7]first proposed Yr to symbolize stripe rust resistance genes, 83 permanently designated genes or alleles and 71 tentatively designated genes are now identified[8,9].Most of these Yr genes confer race-specific or all-stage resistance (ASR).Experience has shown that resistance genes of that type fail to provide long-term protection as new or previously rare Pst variants emerge with the result that curently few such genes provide ongoing protection in widespread cultivation. For example, Pst isolate CYR34 has virulence for a number of previously effective genes and is therefore widespread throughout the country[10–14].There is thus an urgent need to identify more resistance genes to diversify sources of resistance to this virulent isolate.
In an evaluation of wheat cultivars or breeding lines against stripe rust, a breeding line, 041133,was highly resistant to isolate CYR34.A genetic analysis determined that a single dominant gene conferred its resistance.Here, bulked segregant analysis and RNASeq (BSR-Seq) carried out on a recombinant inbred line (RIL) population generated from a cross of line 041133 and a susceptible genotype enabled localization of the resistance gene to a unique position of the chromosome arm 7BL.
2. Materials and methods
2.1. Plant materials
Line 041133,a winter wheat breeding line with unknown pedigree, originated from Qinghai province, China. It was crossed with stripe rust susceptible wheat landrace Qingxinmai, and the resultant F1,F2,and F2:5populations were used to identify a single stripe rust resistance gene. Mingxian 169 was used as the susceptible control in stripe rust tests. The 5 traditional differential wheat genotypes[15]were used to examine response patterns to an array of Pst pathotypes or isolates (Table 1).
2.2. Stripe rust seedling test
Twenty Pst isolates were used to examine the reactions of wheat parents 041133 and Qingxinmai to stripe rust. The original Pst isolates were collected from Tibet, Zhejiang, Sichuan, Hubei,and Gansu in 2017, 2018, and 2019, respectively, and maintained in the Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China (Table 1). The F1(15 plants), F2(460 plants),and F2:5(176 lines)populations were phenotyped with isolate CYR34 for genetic analysis of the disease resistance, and the genotyped F2:5RIL population was used to construct the genetic linkage map. Seeds of all genotypes were sown in plastic pots(6×6×6 cm).Fifteen seeds of each RIL were tested and tests were carried out twice. A previously published method for preparation of urediniospore suspensions, inoculation and infection type (IT)scoring was used [16]. Briefly, when primary leaves were fully unfolded, wheat seedlings were sprayed with a Pst urediniospore suspension in light mineral oil (Novec 7200) at 4 mg mL-1. Inoculated plants were incubated at 9–13 °C for 24 h in a dark dew chamber, and then placed on greenhouse benches for symptom development at 15–18 °C during the day and 10–14 °C at night with a supplemented photoperiod of 12–14 h at a light intensity of 5000–6000 lx. When disease symptoms on the susceptible control Mingxian 169 were fully developed, ITs of the primary leaves were rated on a 0–4 scale,where 0,no visible uredia;0;,hypersensitive flecks; 1, small uredia with necrosis; 2, small to mediumsized uredia with green islands and surrounded by necrosis or chlorosis; 3,large-sized uredia with chlorosis and with or without necrosis; and 4, large uredia without chlorosis and necrosis. A 2+represented infection type between 2 and 3, or a mixture of 2 and 3. Categorization of plant phenotypes was carried out based on the ITs: resistant, IT 0, 0; 1, and 2; and susceptible ITs 3 and 4.
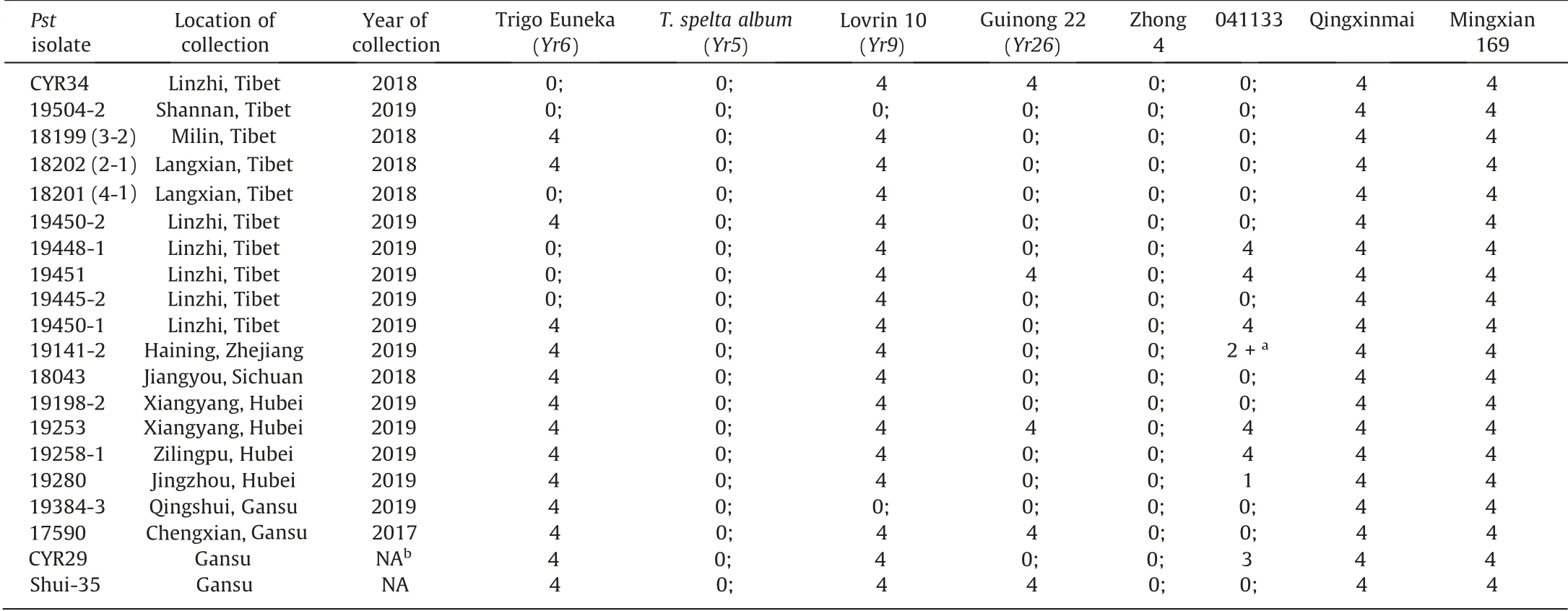
Table 1 Infection types of 041133 and Qingxinmai to 20 Puccinia striiformis Westend. f. sp. tritici Erikss. (Pst) isolates.
2.3. BSR-Seq analysis
Phenotypically contrasting RILs tested with CYR34 were used to construct homozygous resistant and susceptible RNA pools for RNA-sequencing analysis. Fifty homozygous resistant (Bulk-R)and homozygous susceptible (Bulk-S) lines, representing by equal sized segments of primary leaves from single plants of each line,were separately pooled. BSR-Seq was performed following a pipeline described by Xie et al. [17]. Briefly, RNA was extracted from the two bulks with an Illumina TruSeq RNA sample preparation kit (Illumina, Inc., San Diego, CA, USA) and subjected to RNAsequencing on an Illumina HiSeq 4000 platform at Beijing Northern Genome Research Technology.Adapter and low-quality sequences were trmmed using Trimmomatic v0.36 [18]. High-quality reads were aligned against the Chinese Spring reference genome sequence v1.0 (http://wheat-urgi.versailles.infa.fr) with the aid of software STARv2.5.1b [19,20]. Single nucleotide polymorphism(SNP)variants were identified from the unique and confident read alignments using the ‘‘HaplotypeCaller” module in software GATK v3.6 [21], with the following criteria: P-value of the Fisher’s Exact Test (FET) <1e-8 and the allele frequency difference (AFD) >0.6.
2.4. Development and analysis of SSR and InDel markers
The SSR loci that fell in the target genomic region identified by the BSR-Seq analysis were extracted by JBrowse from the TriticeaeMulti-omics Center and SSR primer pairs were designed using Primerserver tools (http://202.194.139.32). Sequences (3 kb) that flanked the candidate InDels were also used to search the Chinese Spring reference genome and the resulting homologous scaffold sequences were used to design InDel primer pairs on the GSP website (http://probes.pw.usda.gov/GSP/) [18].
Leaves of the parents and RILs after phenotyping were sampled for DNA extraction using a CTAB method. The phenotypically contrasting DNA bulks consisted of separate pools of equal quantifies of DNA (50 ng mL-1) from 8 resistant or 8 susceptible lines for identifying the polymorphic markers. DNA amplification was performed in a Biometra T3000 Thermocycler (Applied Biosystems,New York, NY, USA). Each 10 μL reaction mixture comprised 5 μL of PCR mixture (including Taq polymerase, dNTPs, and 10× PCR buffer with Mg2+), 2 μL ddH2O, 1 μL DNA (50 ng μL-1), and 1 μL of 10 μmol L-1each of the forward and reverse primers. The PCR profile included initial denaturation at 98 °C for 3 min; 98 °C for 10 s, annealing at 54–62 °C (depending on the primers used) for 10 s, and 72 °C for 25 s, 35 cycles; and extension at 72 °C for 10 min. Polymorphism of SSR and InDel markers was determined by comparing amplification patterns between the two parents and phenotypically contrasting DNA bulks on 2% agarose gels or 8% non-denaturing polyacrylamide gel (Acr:Bis = 39:1). The resultant polymorphic markers were used to genotype the RIL population for constructing a genetic linkage map.
2.5. Statistical analysis and genetic map construction
The goodness of fit for the observed segregation data from the expected segregation ratio of the F2(3:1)and F2:5(1:1)populations was examined by the Chi-squared (χ2) test in the SAS statistical software package (version 8.01; SAS Institute Inc., Cary, NC, USA).Polymorphic markers identified, together with previously published molecular markers closely linked to the Yr genes on the chromosome arm 7BL, were used to genotype the RIL population for constructing the genetic linkage map using software Mapdraw V2.1[22].Genetic distance between the polymorphic markers and the target resistance gene was estimated using software Mapmaker 3.0, with a logarithm of the odd ratio (LOD) of 3.0 and the maximum genetic distance allowed of 50 cM [23].
2.6. Syntenic analysis of the stripe rust resistance genomic region
Physical locations of the target gene and the other Yr genes were determined by blasting the Chinese Spring reference genome(http://wheat-urgi.versailles.infa.fr) [19], using the sequences of flanking gene markers. Annotated genes in the target genomic region were obtained using the online JBrowse program (https://urgi.versailles.inra.fr/jbrowseiwgsc/gmod_jbrowse/). Orthologous genes were identified from the genome databases of durum wheat[T. turgidum subsp. durum (Desf.) Husnot] accession Svevo [24],wild emmer [T. turgidum ssp. dicoccoides (Körnicke ex Asch. &Graebn.) Thell.] accession Zavitan [25], and 10+ wheat genome sequences [26].
2.7. qRT-PCR analysis
Expression of candidate gene(TraesCS7B01G352400.1)was analyzed using the forward primer 5′-TCCATCTGGAGGCCAGTTTG-3′and the reverse primer 5′-ACGCGTTGAACCAATATGTGAG-3′. Total RNA was extracted from the primary leaves of line 041133 and Qingxinmai at 12 and 24 h after inoculation (hai) with Pst isoalte CYR34 with two biological replicates, using a Plant RNA Kit(Vazyme Biotech Co.,Ltd.,Nanjing).First-strand cDNA was synthesized using a Hifi-MMLV cDNA Kit (CWBIO, Beijing). qRT-PCR was performed with Taq Pro Universal SYBR qPCR Master Mix(Vazyme Biotech Co., Ltd., Nanjing) in an ABI 7500 Real Time PCR system(Applied Biosystems, Foster City, CA, USA). Three repeats for each RNA sample were run as technical replicates.The wheat Actin gene was amplified as the reference gene. Relative expression was determined using the 2-ΔΔCTmethod [27].
3. Results
3.1. Inheritance of stripe rust resistance in line 041133
All Pst isolates tested were virulent on Mingxian 169 and Qingxinmai,but avirulent on Zhong 4 and T.spelta album(Yr5)(Table 1).Fourteen isolates were avirulent on 041133, twelve of which,including CYR34, produced a hypersensitive reaction (IT 0;), one isolate IT 1, and one isolate IT 2+each (Table 1). The remaining six isolates were virulent on 041133 (IT 3 or 4).
Pst isolate CYR34 was used in the genetic analysis of stripe rust resistance. Line 041133 displayed a hypersensitive reaction (IT 0;)and Qingxinmai produced a highly susceptible reaction(IT 4)when inoculated with CYR34 (Fig. 1). The phenotypes of F1plants from cross Qingxinmai × 041133 (IT 0;) and F2population segregated in a 3 resistant:1 susceptible ratio.The RIL population(176 lines)segregated 1 resistant:1 susceptible(Table 2).Thus the stripe rust resistance of line 041133 was controlled by a single dominant allele, which was temporarily designated Yr041133.

Fig. 1. Phenotypic reactions of resistant parent 041133, susceptible parent Qingxinmai, and F1 progeny to Pst isolate CYR34.
3.2. BSR-Seq analysis of the RNA bulks from plants with contrasting responses to Pst isolate CYR34
RNA-Seq analysis generated 2,620,296 and 2,720,867 raw reads for the R and S bulks, respectively. Quality control resulted in 2,514,928 and 2,640,430 high-quality reads, and 96% and 97% of them were mapped on the Chinese Spring reference genome,from which 1092 candidate SNPs and InDels associated with the disease resistance were identified.These SNPs and InDels were anchored to all chromosomes except 1D and nine were not assigned on any chromosome (Fig. 2A). The most abundant enrichment of the disease resistance-associated SNPs and InDels were observed on chromosomes 2B (432), 7B (361), and 5B (26). The majority of SNPs and InDels on chromosome 2B were scattered on the whole length of the chromosome with significant enrichments in the 17.0–18.5 Mb and 162–176 Mb physical intervals(Fig.2B).Despite only 26 SNPs being detected on chromosome 5B, six of them were significantly associated with disease resistance (Fig. 2A). Among the SNPs anchored on chromosome 7B, 62 were enriched in a 20 Mb interval (600–620 Mb) on the long arm (Fig. 2C).
3.3. Linkage analysis of SSR and InDels on chromosomes 2B and 5B
Twenty-two and 53 SSR primer pairs were designed from the clustered SNPs in the physical intervals 17.36–18.26 Mb and 162.8–176.2 Mb,respectively,on chromosome 2B.Another 17 primer pairs were developed based on chromosome 2B InDels between the two RNA bulks (Table S1). Six SSR primer pairs detected polymorphism between the two parents but not between the DNA bulks from the RIL population (Table S1). Similarly, the nine SSR primers pairs designed from the SNPs on chromosome 5B (Table S1) also failed to show an association with stripe rust response. We concluded that the InDel and SSR markers on chromosomes 2B and 5B were unlikely to be linked to resistance gene Yr041133 and they were not considered in further analysis.
3.4. Development of SSR markers on the chromosome arm 7BL and linkage map construction
SSR motifs in the genomic interval of 600–620 Mb on the chromosome arm 7BL were used to design 364 SSR primer pairs.Twenty-five primer pairs detected polymorphisms between 041133 and Qingxinmai.Nine(Xicst42,Xicst63,Xicst69,Xicst79,Xicst133, Xicst162, Xicst180, Xicst238, and Xicst338) of these markers were also polymorphic between the resistant and susceptible DNA bulks and showed linkage to Yr041133 after genotyping the entire RIL population (Fig. 3; Table S2). A genetic linkage map was constructed and Yr041133 was positioned between markers Xicst133 and Xicst338 with genetic distances of 0.6 and 1.1 cM,respectively (Fig. 4B).
3.5.Analysis of molecular markers linked to the known Yr genes on the chromosome arm 7BL
Twenty molecular markers linked to the known Yr genes, i.e.,
Yr39, Yr52, Yr59, Yr67, Yr79, YrZH84, YrTp2, YrSuj, and YrMY37 on the chromosome arm 7BL, were used to amplify line 041133 and Qingxinmai to identify markers that linked to Yr041133. Polymorphism was detected in markers Xcfa2040 and Xwmc557 linked to YrZH84/Yr59 and Xwmc526 and Xwmc476 lined to YrSuj/YrZH84 and YrMY37/Yr79. However, none was polymorphic between the present contrasting DNA bulks (Table S3). Markers Xwgp45,Xgpw1144, Xbarc32, and Xbarc182 linked to genes Yr39, Yr52, and Yr67 were not informative in the current population since they amplified only monomorphic DNA products. Consequently, these molecular markers could not be used to genotype the RIL population.The physical positions of the known Yr genes on chromosome 7BL were predicted by blasting the sequences of the markers flanking the Yr041133 locus against the Chinese Spring reference genome sequence. All the genes were positioned more than 50 Mb away from Yr041133,indicating that they were unlikely to be identical to Yr041133 (Fig. 4C).
3.6. Micro-collinarity of the Yr041133 genomic region and qRT-PCR analysis of the candidate gene
The 1.7 cM genetic interval of Yr041133 between markers Xicst133 and Xicst338 corresponded to a ~0.8 Mb (608.9–609.7 Mb)genomic region on the Chinese Spring chromosome arm 7BL and was predicted to have 3 high confidence genes (Table S4). This region was syntenic with the T. durum cultivar Svevo 7BL(0.83 Mb, 593.7–594.5 Mb) and the T. turgidum ssp. dicoccoides accession Zavitan 7BL (1.4 Mb, 617.9–619.3 Mb) (Fig. S1). Microcollinearity analysis in the recently released 10+ wheat genomes revealed no significant presence and absence variations of the disease resistance related kinase.

Table 2 Genetic analysis of stripe rust resistance in line 041133 to Pst isolate CYR34.
One of annotated genes in the target genomic region,TraesCS7B01G352400.1, encodes a leucine-rich repeat (LRR) receptor like protein kinase,which may be associated with disease resistance.qRT-PCR analysis demonstrated that expression of this gene in the stripe rust resistant line 041133 was significantly higher than in the stripe rust susceptible landrace Qingxinmai at 12 and 24 hai with isolate CYR34(P <0.01 and P <0.05)(Fig.5).This indicates that TraesCS7B01G352400.1 is a potential candidate gene for Yr041133.
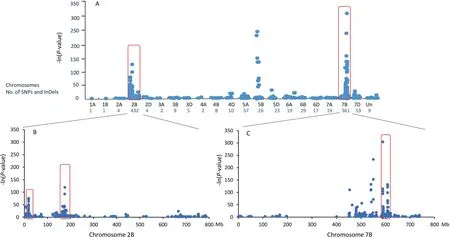
Fig. 2. BSR-Seq analysis identified single nucleotide polymorphisms (SNPs) putatively associated with Yr041133. (A) Distribution of SNPs on selected wheat chromosomes.Red rectangles indicate chromosomes regions on chromosomes 2B,5B,and 7B where SNPs may be associated with the resistance gene.Un,SNPs that were not anchored on any chromosomes. (B)SNP variants on chromosome 2B. SNP variants significantly enriched in the physical intervals of 17.0–18.5 Mb and 162–176 Mb are indicated by red rectangles. (C) SNP variants on chromosome 7B. SNP variants significantly enriched in the physical intervals of 600–620 Mb are indicated by the red rectangle.
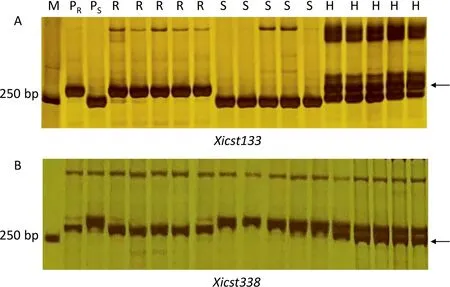
Fig. 3. Banding patterns of the closest Yr041133-flanking SSR markers Xicst133 (A) and Xicst338 (B) amplified in the parents and the selected RILs for cross Qingxinmai × 041133. M, DL2000 DNA ladder (Tiangen Biotech Co., Beijing); PR, resistant parent Line 041133; PS, susceptible parent Qingxinmai; R, resistant RILs; S,susceptible RILs. Arrows indicate the polymorphic bands that are specific for Yr041133.
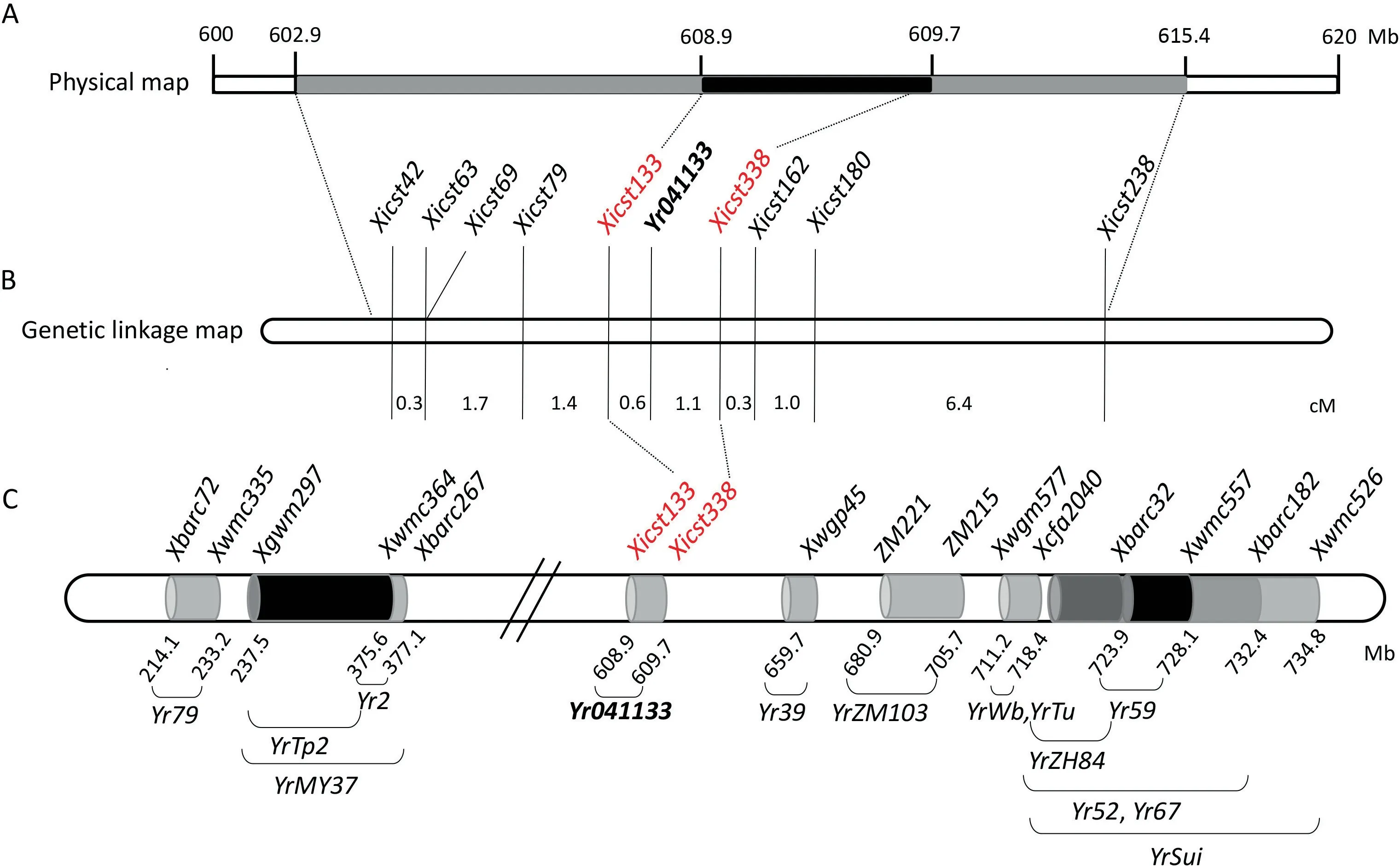
Fig. 4. Maps of SSR markers linked to Yr041133 on chromosome 7B. (A) Physical map of Yr041133. (B) Projection of (A) on the genetic map of the Yr041133 region. (C) Distribution of markers associated with other stripe rust resistance genes on chromosome 7B.
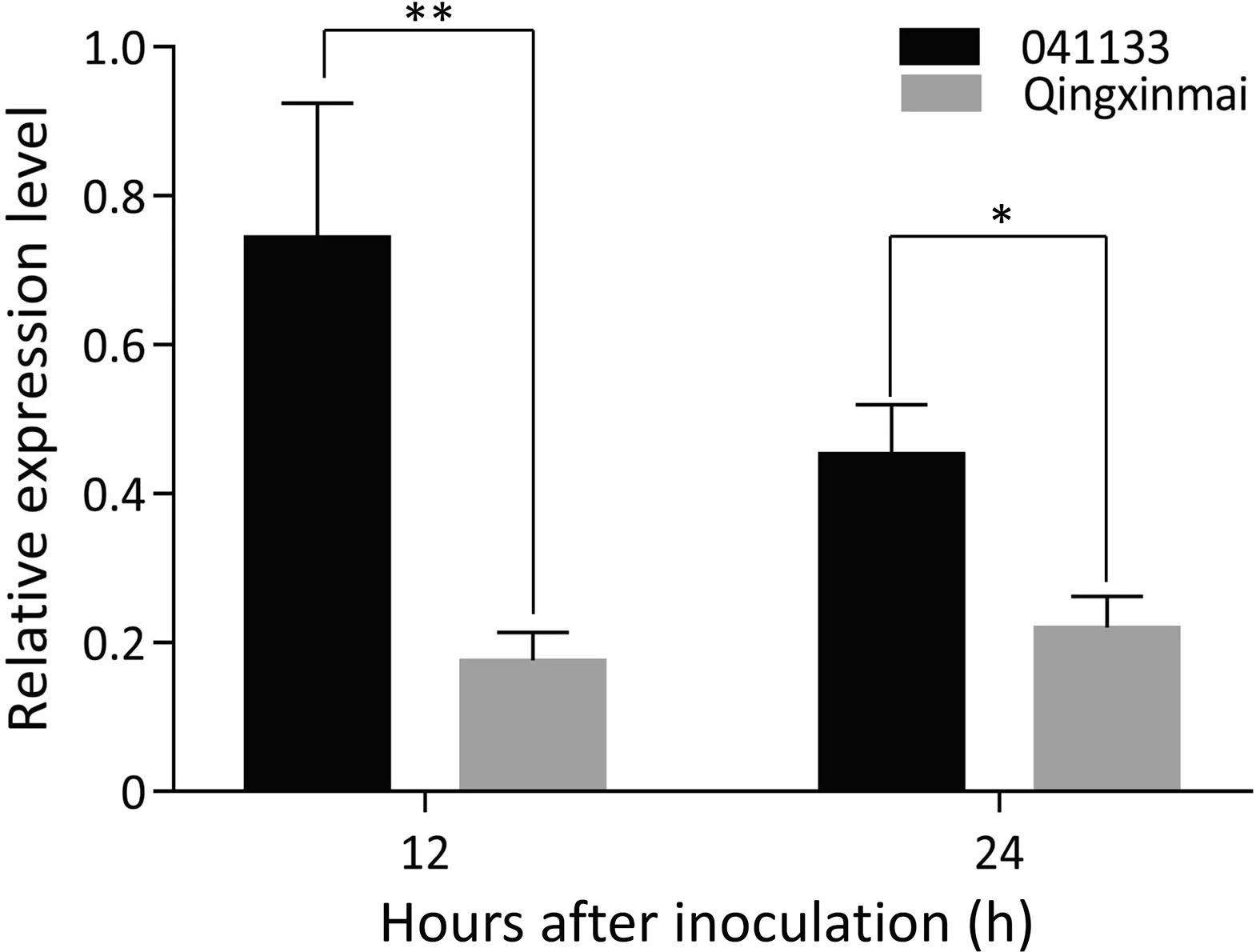
Fig. 5. qRT-PCR analysis of the candidate gene TraesCS7B01G352400.1 in Line 041133 and Qingxinmai 12 and 24 h after inoculation with CYR34. Bars indicate standard variation (n = 6). *, **, significant difference at P <0.05 and P <0.01,respectively, based on the Fisher’s least significant difference method.
4. Discussion
Boom and bust cycles of stripe rust epidemics following introduction and widespread use of single genes for resistance havebeen a repetitive experience in China.The main problem is that the resistance gene(s) being used are often not identified until they become ineffective. In reality the same line is used by multiple breeding programs because of its widespread effectiveness or alternatively a series of presumed different sources of resistance are used as parents only to later find that the same gene was involved as was the case with the gene variously named Yr24/Yr26/YrCH42. CYR34 is now the prevailing pathotype in China following its identification in 2009[11].Part of its success as a pathotype is its obvious selective ability on cultivars with Yr24/Yr26/YrCH42, but it also has virulence for other genes used in Chinese cultivars and appears to be highly aggressive allowing it to compete with other pathogypes on mutually susceptible cultivars.The frequency of this pathotype is increasing rapidly,from 16.1%in 2014 to 34.9%in 2016 in Gansu province,China[28,29].It is so virulent that only a few known genes, such as Yr5, Yr15, and Yr32,remain to be effective [30]. The break-down of single rust resistance genes Yr24/Yr26/YrCH42 to CYR34 in many newly released commercial wheat cultivars has caused an urgent need to search for new stripe rust resistance genes against this dangerous Pst pathotype.Although a large quantities of wheat cultivars or breeding lines were tested resulting in the identification of a number of wheat genotypes resistant to CYR34 [31–36], genetic and molecular localization of the resistance genes is needed to facilitate their use in breeding programs.
Line 041133 was resistant to several Pst isolates, including the recently emerged virulent CYR34. We identified a new dominant gene Yr041133 conferring ASR to CYR34.BSR-Seq analysis enriched the disease resistance-associated SNPs in an about 20 Mb (600–620 Mb) genomic interval on the chromosome arm 7BL. Genetic mapping by newly developed SSR markers reduced the target genomic interval of Yr041133 to an about 800 kb (608.9–609.7 Mb)genomic region.However,several Pst isolates,including CYR29, were found to be virulence to 041133(Table 1). Therefore,it is necessary to use the stripe rust resistance gene Yr041133 in pyramiding with other effective Yr genes in the breeding programe.
In the integrated genetic linage map of the identified Yr genes on chomosome 7B(Fig.4C),Yr6 and Yr63 were located on the short arm of chromosome 7B [37–39], while Yr79 [40], YrTp2 [41], and yrMY37[42]were located proximal to Yr041133 with physical distances of more than 230 Mb. To the distal side of Yr041133, there are Yr39 [43], Yr52 [44], Yr59 [45], Yr67 [46,47], YrZH84 [48],YrSuj[49], YrZM103 [50], YrTu and YrWb [51]. The closest gene in terms of physical distance was Yr39,about 50 Mb from Yr041133.Yr39 in USA spring wheat cultivar Alpowa [43], Yr52 in Indian line PI 183527[44],Yr59 in Iraqi line PI 178759[45],and Yr79 in Pakistani landrace PI 182103 [40] are high-temperature adult plant (HTAP)resistance genes and hence unlikely to be identical to Yr041133.Based on its physical position Yr041133 appears to be located at unique locus that is distinct from the projected postions of those 13 Yr genes mapped to chromosome 7B (Fig. 4C).
Mu et al. [52] recently examined the virulence of Pst isolate CYR34, as well as isolates CYR32 and CYR33, on Kalyansona(Yr2), Heinese Kolben (Yr6), Alpowa (Yr39), PI 183527 (Yr52), PI 178759 (Yr59), C591 (Yr67), PI 182103 (Yr79), and Zhou 8425B(YrZH84). The three Pst isolates were virulent on these genes in a seedling test, except for the avirulence of CYR32 on Yr67 and of CYR32 and CYR33 on YrZH84. Virulence of CYR34 on genotypes with Yr2, Yr6, and Yr67 was observed in other studies [11,34,42].These results provide further evidence for differences between Yr041133 and other genes on the chromosome arm 7BL.
The release of genome sequences for common wheat and its diploid and tetraploid progenitors and related species has revolutionized methodology and greatly speeded up the discovery of disease resistance genes. High-throughput mining of SNP by sequencing either DNA or RNA and various versions of SNP arrays,in combination with BSA and accurate disease resistance phenotyping,allows rapid and large scale detection of disease resistance genes in wheat [53,54]. Because it is easily applied and cost effective, this strategy has been widely used in identifying stripe rust genes and QTL, for example, YrZM103 [50], YrZH22 [55], YrMM58 and YrHY1 [56], and QYrcen.nwafu-7BL [52]. BSR-Seq can usually enrich SNPs and InDels associated with the traits of interest in potential genomic regions on single chromosomes. We obtained multiple genomic regions with significant enrichment of SNPs and InDels on chromosomes 2B and 7B. Similarly, Zhang et al.[50] detected stripe rust resistance-associated SNPs on chromosomes 2B and 7B when mapping stripe rust resistance gene YrZM103. They also found that the enriched SNPs on chromosome 2B were not associated with YrZM103 and the gene was finally localized on 7BL. The reasons that SNPs enriched on the nontargeted genomic regions between the two contrasting RNA bulks could not be explained in this study.There is a possibility that difference in chromosome structures between the wheat genotypes studied and the reference genome of Chinese Spring may exist since chromosome segmental translocation, inversion and rearrangement were frequently reported in wheat genomes [26].Another explanation is false detection of SNPs between homeologous and paralogous transcripts in the hexaploid wheat genome with the current BSR-Seq data analysis algorithm. Other possibilities may exist for minor effect stripe rust resistance QTL and enhancers in those chromosomal regions that need further characterization.
Because chromosome 7B harbours several genes for resistance to stripe rust, it is difficult to precisely determine their relationships only based on the molecular mapping or allelism test. Feng et al. [40] investigated the relationships among six Yr genes,namely, Yr39, Yr52, Yr59, Yr67, Yr79, and YrZH84, using allelisms test with F2populations in their study on Yr79. They concluded that these genes were linked rather than allelic. However, since F2is a very inefficiency way to peform allelism test and phenotypes of the F3progenies need to be confirmed, the ultimate determination of their relationships requires fine genetic mapping and positional cloning. We identified an annotated gene that encode a LRR receptor like kinase protein associated with disease resistance in the genomic interval to which Yr041133 was mapped (Table S4).This gene was significantly induced up-regulation by Pst isolate CYR34 in Line 041133. This candidate for the Yr041133 resistance allele must be verified by mutation,gene editing,genetic transformation,and functional analyses in future.Previously,Yr5,Yr7, and YrSP [57], Yr10 [58], YrAS2388R [59], and YrU1 [60] proved to encode different forms of nucleotide-binding and leucine-rich repeat proteins(NLRs).Yr15[61]and Yr36[62]belong to the kinase proteins.The ultimate cloning of Yr041133 will be helpful in illucidating its function and also the relationship between the genes on chromosome 7B.
CRediT authorship contribution statement
Hongjie Li:Funding acuisition, Conceptualization, Writing -original draft.Zhingyong Liu:Conceptulation, Writing - review &editing.Yahui Li:Investigation, Validation, Visualization.Jinghuang Hu:Investigation, Validation, Visualization.Xiaohan Shi:Investigation, Validation, Visualization.Dan Qiu:Investigation,Validation, Visualization.Peipei Wu:Investigation, Validation,Visualization.Gebremedhin Habteab Goitom:Investigation,Validation, Visualization.Siqi Wang:Investigation, Validation,Visualization.Ruiming Lin:Investigation,Writing-review&editing.Qiuhong Wu:Software.Jingzhong Xie:Software.Hongjun Zhang:Formal analysis, Methodology, Investigation, and Resources.Li Yang:Formal analysis, Methodology, Investigation,and Resources.Hongwei Liu:Formal analysis,Methodology,Investigation,and Resources.Yang Zhou:Formal analysis,Methodology,Investigation, and Resources.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Financial support of this research by the National Key Research and Development Program of China (2017YFD0101000) and the Agricultural Science and Technology Innovation Program of CAAS(CAAS-ZDRW202002) are gratefully appreciated. The authors thank Professor S.Y. Hou of Qinhai Agricultural and Forestry Sciences for providing seeds of Line 041133.
Appendix A. Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2021.06.009.
- The Crop Journal的其它文章
- Origin, evolution, and molecular function of DELLA proteins in plants
- Far-red light: A regulator of plant morphology and photosynthetic capacity
- A rice XANTHINE DEHYDROGENASE gene regulates leaf senescence and response to abiotic stresses
- Dissection of heterotic loci for grain yield using interconnected chromosome segment substitution lines in rice
- A soybean NAC homolog contributes to resistance to Phytophthora sojae mediated by dirigent proteins
- The boron transporter SiBOR1 functions in cell wall integrity, cellular homeostasis, and panicle development in foxtail millet

