后GWAS时代结直肠癌致病SNP功能机制的研究进展
李以格,张丹丹
综 述
后GWAS时代结直肠癌致病SNP功能机制的研究进展
李以格1,2,3,张丹丹1,2,3
1. 浙江大学医学院病理学与病理生理学系,杭州 310058 2. 浙江大学医学院附属第二医院肿瘤内科,杭州 310058 3. 浙江省疾病蛋白质组学重点实验室,杭州 310058
结直肠癌(colorectal cancer, CRC)是受遗传与环境因素共同影响的复杂疾病,其中遗传因素发挥重要作用。至今,全基因组关联研究(genome-wide association studies, GWAS)已经发现了大量与结直肠癌风险相关的遗传变异。随之而来的后GWAS时代,越来越多的研究侧重于利用多组学数据和功能实验对潜在的致病位点进行解析。分析表明绝大多数风险单核苷酸多态性(single nucleotide polymorphism, SNP)位于非编码区,可能通过影响转录因子结合、表观遗传修饰、染色质可及性、基因组高级结构等,调控靶基因表达。本文对后GWAS时代结直肠癌致病位点的机制研究进行综述,阐述了后GWAS对于理解结直肠癌分子机制的重要意义,并探讨了结直肠癌GWAS的应用和前景,为实现GWAS成果转化提供参考。
结直肠癌;后全基因组关联研究;单核苷酸多态性;致病变异
结直肠癌(colorectal cancer, CRC)是常见的恶性肿瘤之一,严重威胁人类健康。据统计,2018年全球CRC新发病例超过180万,死亡病例约86万;位居发病瘤谱第3位,死亡瘤谱第2位[1]。在我国,2015年CRC新发病例估计有38.76万例,死亡病例18.71万例;位列发病瘤谱第4位,死亡瘤谱第5位[2,3]。吸烟、缺乏锻炼、不健康的饮食习惯等环境因素均会增加CRC患病风险[3];此外,遗传因素也影响着CRC的发生,大型双生子研究表明CRC的遗传力约占35%[4]。可见,CRC受遗传与环境因素共同作用。随着人类基因组计划等大型项目的开展以及测序技术的进步,利用全基因组关联研究(genome-wide association studies, GWAS)发现了大量结直肠癌易感位点,为了解和防治结直肠癌提供信息。
GWAS被认为是探索常见遗传变异主要是单核苷酸多态性(single nucleotide polymorphism, SNP)与复杂疾病相关性的“万能钥匙”[5],广泛应用于癌症、糖尿病和精神分裂症等疾病[6,7]。自2007年GWAS研究发现8q24.2, 18q21.1区域上的多态位点与结直肠癌风险显著相关[8~10]后,越来越多的位点被鉴定,然而由于连锁不平衡的存在以及基因与环境之间复杂的相互作用,GWAS识别的标签SNP不一定是真正的致病变异,因此迫切需要对GWAS结果进行深入解读。早前,Freedman等[11]和Edwards等[12]提出后GWAS(post-GWAS)研究策略,旨在筛选功能位点并阐明其潜在的分子机制(图1)。至今已有相当数量的研究对结直肠癌风险SNP进行功能解析。为了更好地理解功能SNP在结直肠癌发生发展过程中的作用,本文总结了致病位点的功能机制,并期望促进GWAS成果的临床转化,为疾病的预防、诊断寻找可靠的生物标志物和高效的治疗方法。
1 结直肠癌风险位点
截至到2020年11月,GWAS Catalog (http://www. genome.gov/gwastudies/)数据库收录了70多篇结直肠癌GWAS研究,共鉴定到821个相关位点,其中584个为非重复位点,241个SNP与结直肠癌风险显著相关(<5×10–8),绝大部分位于内含子、基因间等非编码区(表1)。从人群上看,影响东亚人群患病风险的SNP约有38个,与欧洲人群相关的SNP超过100个[14]。组学数据的累积、样本数量的增加和分析方法的改善,促进了新的风险位点不断被发现,这些位点包括位于已知风险位点上的新变异(rs6584283, 10q24.2[15]),甚至是一些稀有、低频变异(rs145364999, 频率为0.3%[16])。然而,这些变异的风险预测效应较弱(OR<1.5),但大部分SNP所在或邻近基因参与TGF-β/BMP (如、、)、Wnt (如、)等信号通路以及维持端粒生物功能(如、),暗示这些变异的功能效应赋予疾病易感性[17,18]。因此,有必要对风险变异进行进一步的功能分析。

图1 后GWAS研究策略
GWAS-SNP功能研究的一般策略是:(1)对结直肠癌相关位点进行基因型填补,获得连锁不平衡区域内的所有位点;(2)整合转录组、表观遗传等多组学数据对相关位点进行注释,筛选潜在功能位点并对候选位点做进一步的功能注释。如ENCODE、Roadmap等数据库提供了甲基化、组蛋白修饰、染色质开放程度等信息;表达数量性状基因座(expression quantitative trait loci, eQTL)数据有助于识别SNP可能影响的靶基因;Cistrome、JASPAR等数据库可用于预测SNP是否影响转录因子结合等;(3)利用体内外实验阐明风险位点的致病机制。常见的实验方法有:荧光素酶报告基因实验、ChIP-seq、染色体构象捕获技术、基因敲除等。参考文献[5]绘制。
2 结直肠癌后GWAS研究
2.1 功能注释
2.1.1 编码区SNP
位于编码区的SNP可分为同义和非同义突变,同义突变虽然不影响蛋白质的氨基酸序列,但可能通过影响转录后修饰、翻译速率等过程,改变蛋白的表达;而非同义SNP (non-synonymous SNP, nsSNP)会引起氨基酸的替换,造成蛋白结构、理化性质(稳定性、溶解性等)和功能发生改变。那些对蛋白结构和功能影响较大的非同义突变往往会在自然选择中被淘汰,推测剩下的非同义突变功能效应可能较小,这给nsSNP的研究带来一定的挑战。目前,已有大量的生物软件(如MUpro、INPS-MD、ModPred等)可用于预测nsSNP对蛋白结构和功能的影响,相较而言,nsSNP的功能机制相对简单,因此相关的研究也较多[19~22]。结合全外显子分析发现多个与结直肠癌发展相关的编码区SNP,如位于重要结构域上的错义突变rs3184504 (p.Trp263Arg)可能改变该蛋白对细胞分裂的调节功能;还有些编码变异可能影响可变剪切(rs16888728,)[23]。
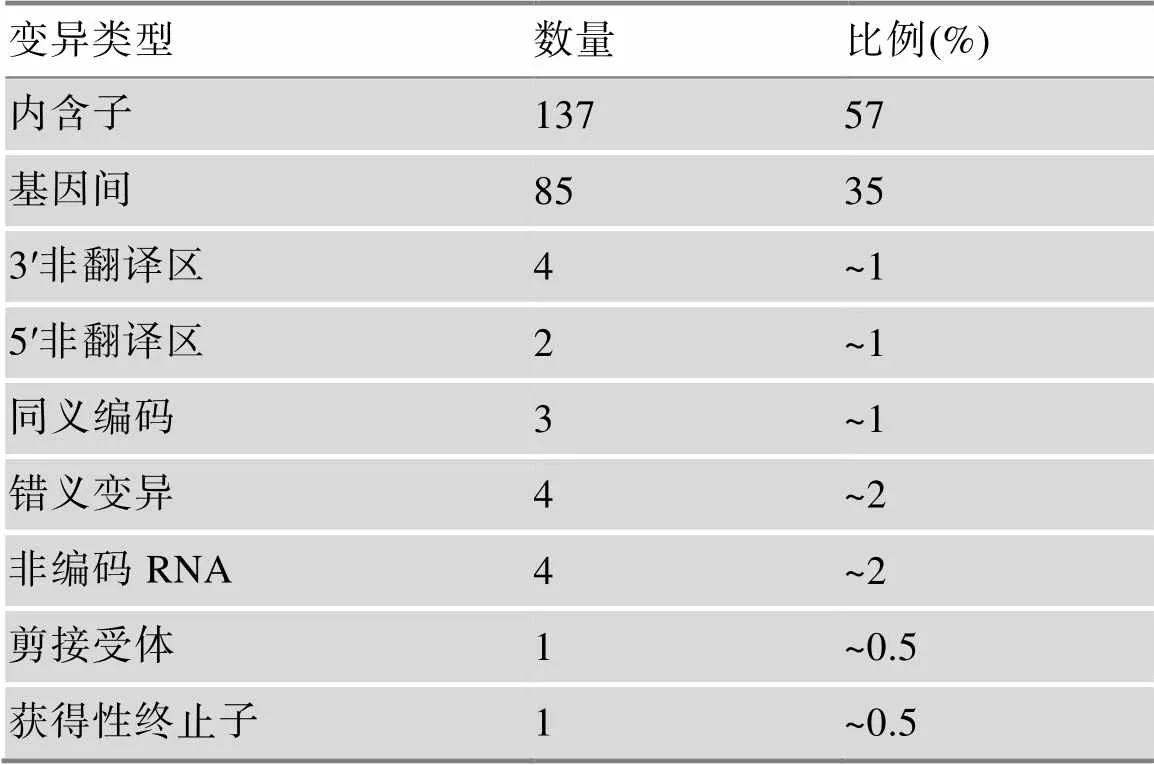
表1 GWAS鉴定的SNP的类型
2.1.2 非编码区SNP
目前GWAS发现的结直肠癌风险相关位点主要位于非编码区,这些位点可能参与基因转录、转录后加工、翻译和翻译后修饰等过程调控基因表达。在研究非编码区SNP时,首先需要明确这类SNP的靶基因,常用的方法是利用表达数量性状基因座(expression quantitative trait loci, eQTL)检测SNP与基因表达的关系,基于此策略,已发现大量非编码SNP可能影响的靶基因,包括、等以及一些与结直肠癌关系尚不明确的基因(如、)。非编码SNP可以通过近距离顺式或远距离反式作用调控靶基因的转录,研究发现这类风险SNP所在区域的组蛋白修饰特别丰富,尤其是与启动子、增强子活性相关的修饰(H3K4me3、H3K4me1、H3K27ac);并预测大部分SNP会破坏特定转录因子的结合基序,如rs6983267可能会改变与MYC、CTCF、TCF7L2等转录因子的结合[18]。有些非编码SNP可能影响增强子活性,通过远距离增强子与启动子相互作用改变靶基因的表达[14]。此外,基因组的3D结构在基因表达调控等过程中发挥重要作用[24,25],整合Hi-C等数据发现,一些非编码SNP所在区域与靶基因启动子区存在显著的染色质环相互作用[14,18,26],因此在对非编码SNP进行功能解析时,常常需要考虑染色质相互作用等。
非编码SNP功能多样,可参与到基因表达调控的各个进程中,可能位于不同的调控区,如miRNA种子序列结合位点区、可变剪切位点区等,还可以出现在非编码RNA上,包括长链非编码RNA和miRNA等。功能注释发现位于非翻译区的rs2279398可能改变与miRNA的结合效率[27];目前也识别到大量非编码RNA上的SNP,如rs2632159 (-)、rs6505162 ()等[28~31];位于的 rs1052918可能会引起Wnt信号通路的持续激活,导致细胞增殖失控和肿瘤发生[32]。可见,非编码SNP功能机制十分复杂,有待系统、深入的研究。
2.2 功能SNP机制的实验证据
2.2.1 编码区SNP的潜在功能机制
编码区SNP影响患病风险的机制离不开其所在基因编码蛋白的功能。由于这类SNP发生的频率相对较低,研究者往往聚焦在特定信号通路/基因或某种感兴趣的修饰方式,如6-甲基腺嘌呤(6-Methyladenosine, m6A)修饰,进行全外显子关联分析以发现效应较大的编码SNP。对参与TGFβ信号通路的12个基因进行外显子测序和关联性分析,筛选到上的低频错义变异rs3764482与中国汉族人群的结直肠癌风险显著相关,SMAD7能够抑制R-SMAD的磷酸化并在该通路中发挥负调控作用,鉴于此功能而设计的体外实验表明,该SNP通过影响R-SMAD磷酸化,改变TGFβ信号活性[33]。类似地,rs3750050 (, p.Thr573Ala)、rs149418249 (, p.Pro507Leu)通过破坏蛋白功能、蛋白与蛋白的相互作用,分别引起Ras/MEK/ERK通路和端粒功能异常,导致结直肠患病风险升高[34,35]。
编码区SNP还可能影响基因或蛋白的修饰。如m6A修饰主要发生在RNA上,参与mRNA稳定性的维持、mRNA前体剪切、翻译调控等过程,是近些年的研究热点。通过分析m6A相关SNP与结直肠癌风险的关系,发现在m6A编辑器的参与下,发生在外显子区的rs8100241[A]等位基因能够增加的m6A修饰水平和转录效率,促进该潜在抑癌蛋白的表达[36]。
值得注意的是,编码区SNP可能与其他SNP存在相互作用[23,37],发挥更强的功能效应。如:位于转录因子外显子区的rs138649767[A]等位基因,能激活含有rs6983267[G]的增强子,促进的表达[38];发生在外显子和内含子上的SNP可能存在调控与被调控的关系,也可能共同影响SMAD7的功能和TGFβ信号通路[33]。因此,在研究编码区SNP时,可以考虑SNP之间的相互作用,以更好的解析其功能机制。
2.2.2 非编码区SNP调控基因表达
结直肠癌风险SNP主要位于非编码区[39],根据所处位置发挥不同的机制,其所在区域可以是近端(启动子、增强子或超级增强子)或远端(基因间或基因内)应答元件。非编码SNP往往通过改变转录因子结合位点(transcription factor-binding site, TFBS)、表观遗传修饰和/或染色质结构,影响基因转录水平(表2,图2)。SNP造成的序列变化可能会产生新的TFBS或破坏已存在的TFBS,影响与转录因子(如SP1, NF1, GATA3; MYC, NFATC2, YY1等[40~44])的结合,调控靶基因的转录,参与细胞增殖、凋亡、迁移侵袭等过程。
(1)影响启动子活性。启动子区上的SNP一般通过影响与转录因子的结合,发挥调控作用。如:rs13278062和rs2243828分别位于和启动子区,体内体外实验表明,这两个SNP的[T]等位基因在结直肠癌发展中有着不同的作用,前者抑制克隆形成,后者促进细胞增殖,但它们的分子机制相似,都是通过增加与转录因子(Sp1/NF1、AP-2α)的结合亲和力,使DR4和MPO表达增加[40,47]。
(2)影响增强子活性。内含子区的SNP常位于增强子元件,也会改变与转录因子的结合,主要发挥远距离调控作用。发生在内含子区的rs7198799能够靶向转录因子NFATC2,远距离增强(距离致病位点超过200 kb)的表达,通过NFATC2- ZFP90-BMP4通路促进癌症发生[43];类似地,rs174575可以在转录因子E2F1的参与下,作为和位点特异的远距离增强子[50],有趣的是后者又能够促进FADS2的表达,形成环路,影响结直肠癌发生[50]。
单个SNP的效应可能较小,但是多个致病SNP对TFBS产生的累积效应可能对靶基因表达的影响很大。例如,rs61926301和rs7959129是分别发生在启动子区和内含子区上的SNP,这两个SNP的[T]风险等位基因能够分别增加与转录因子SP1和GATA3的结合能力,通过启动子与增强子相互作用的方式促进潜在癌基因的转录,影响细胞增殖、抑制细胞凋亡;基因表达的调控与染色质的高级结构密切相关,分析发现这两个SNP所在的区域富集活跃的组蛋白修饰峰和开放的染色质可及性[41]。
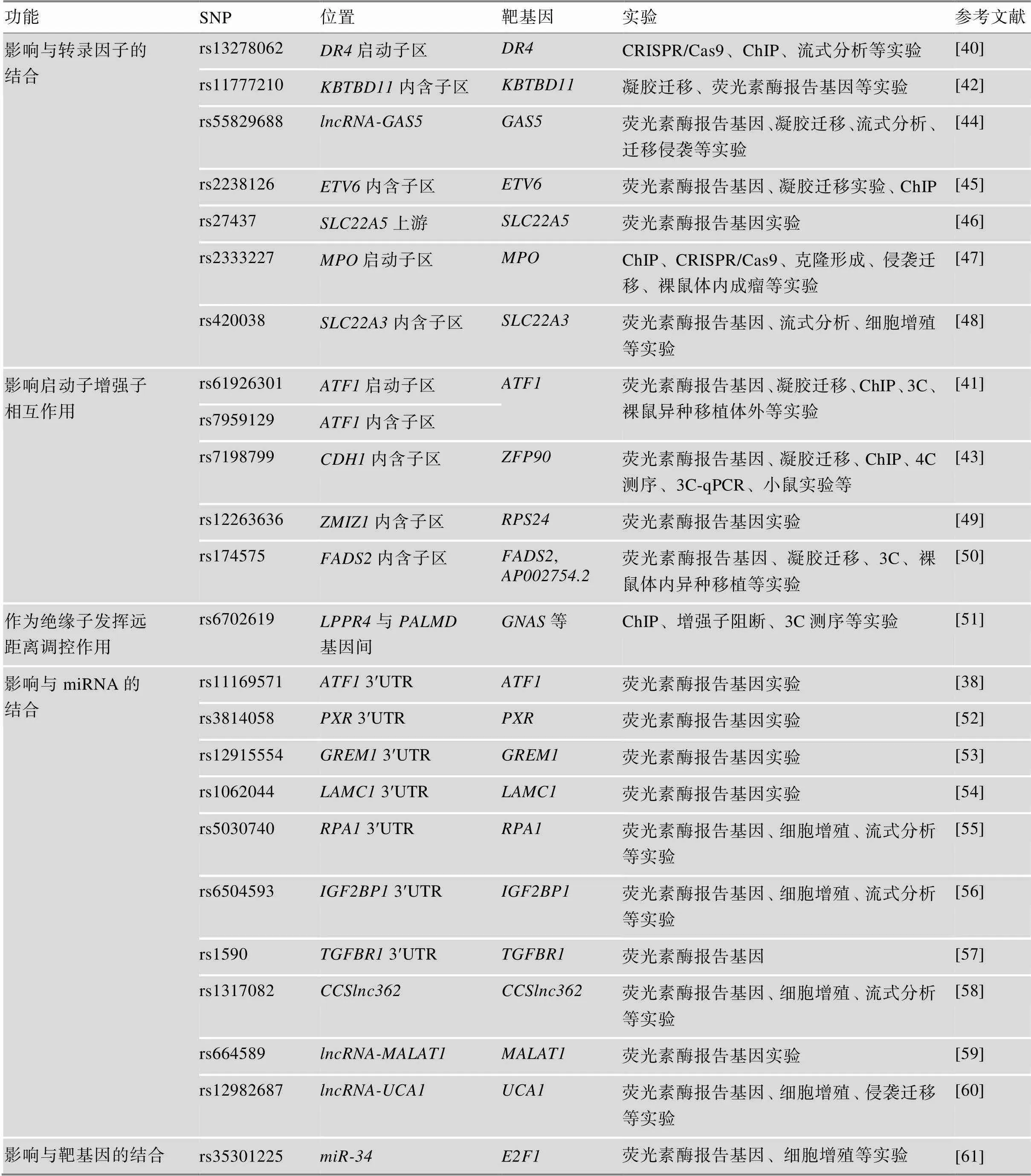
表2 后GWAS实验性研究阐明的非编码SNP作用机制
ChIP (chromatin immunoprecipitation):染色质免疫沉淀;3C (chromosome conformation capture):染色体构象捕获;4C (circular chromosome conformation capture):环形染色体构象捕获;UTR (untranslated region):非翻译区。
(3)其他。miRNA能够靶向基因3ʹ非翻译区(untranslated region, UTR),沉默基因表达。如,发生在3ʹUTR区的rs6504593突变位点减弱了与该区域的结合,使表达上调,引起结直肠癌发生[56]。发挥类似机制的还有位于、、等基因3ʹUTR区的SNP[38,53,54],此外,长链非编码RNA上的一些SNP也能通过改变与miRNA的结合发挥作用,如rs1317082、rs664589、rs12982687[58~60]等。若SNP发生在miRNA上,同样会影响其与靶基因的结合亲和力[61]。
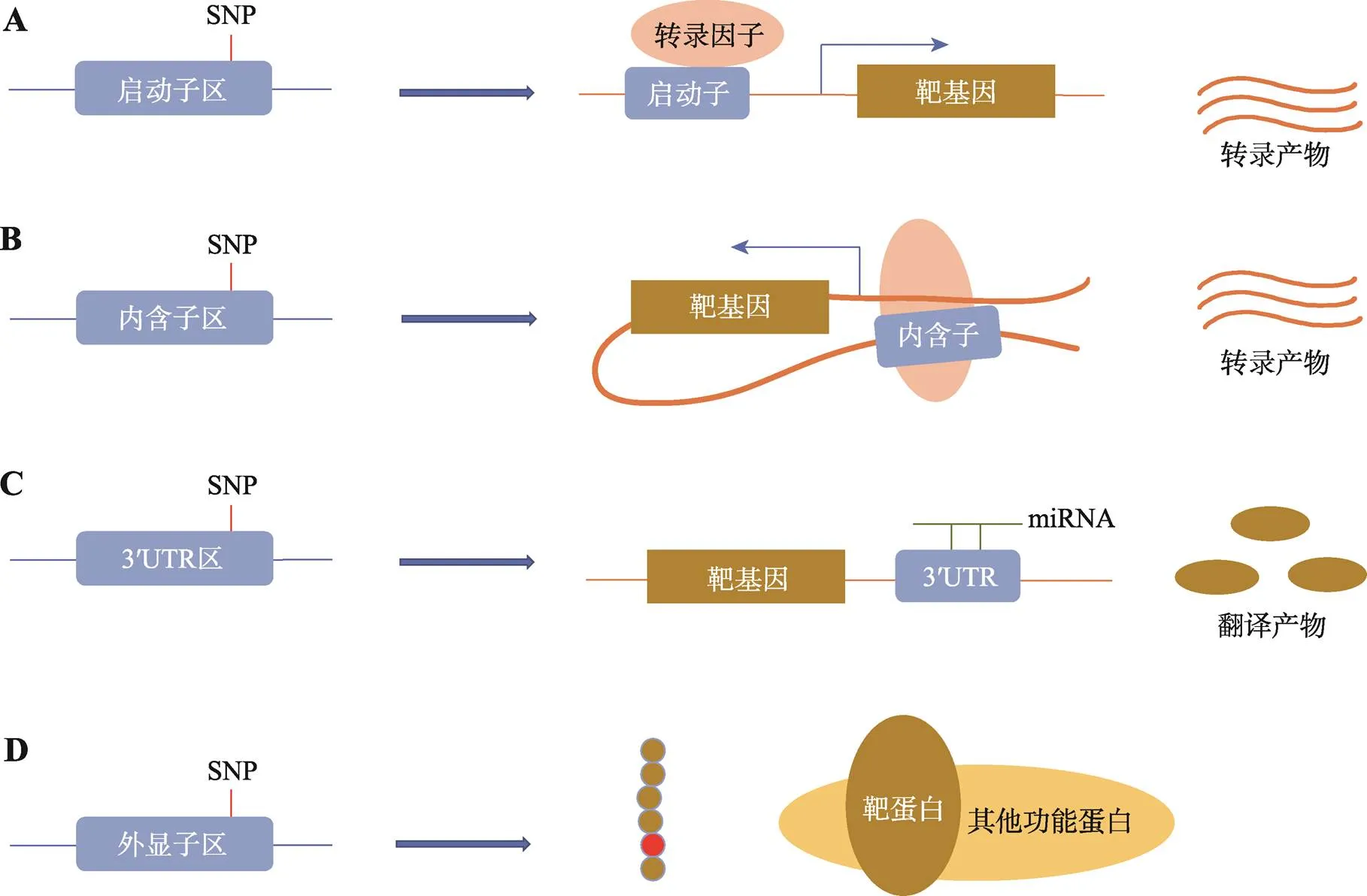
图2 致病SNP潜在功能机制总结
A:启动子区SNP的潜在功能机制。通过影响与转录因子的结合,调控靶基因的表达,影响结直肠癌发生;B:内含子区SNP的潜在功能机制。常在转录因子的参与下,通过远距离启动子增强子相互作用,影响靶基因表达;C:3ʹUTR区SNP的潜在功能机制。往往通过改变与miRNA的结合,影响靶基因转录后水平;D:外显子区SNP的潜在功能机制。可能通过改变氨基酸序列,影响蛋白与蛋白之间的相互作用。
3 结直肠癌GWAS的应用
GWAS和后GWAS研究不仅可以帮助人们更好地在遗传水平上理解结直肠癌的发病机制,也有助于筛查预防、风险分层和临床治疗等。
3.1 风险预测
通过组合已发现的结直肠癌风险位点计算遗传风险评分(genetic risk score, GRS)是GWAS-SNP重要的公共卫生价值之一[62],该方法对每个SNP的微弱效应进行叠加,大大提高了对疾病风险的预测能力,有潜力成为药物治疗、行为矫正的基础。基于37个已知CRC风险变异的GRS表明,与人群中位数相比,得分排在前1%的个体患CRC的风险增加了2.9倍[63];在中国南方汉族人群中,GRS结合传统风险因素构建的风险模型预测能力优于传统风险因素模型[64]。随着风险位点的数量不断增加,基于此建立的GRS风险模型的预测效能也将不断提高,有望实现肿瘤精准预防。
3.2 预后分析
rs5030740、rs9939049、rs11196172等结直肠癌风险SNP与患者的生存期显著相关,有可能发展成为可靠的预后标志物[55,65~67]。其中,rs5030740能够调控的表达,而低表达增加了结直肠癌细胞对奥沙利铂的敏感性,抑制了奥沙利铂治疗后的细胞增殖[55];另外,在接受贝伐单抗一线化疗的结直肠癌患者中开展的试验表明,携带rs699947-AA()和rs1799969-GA ()基因型的患者总生存期比其他患者更长[68]。上述研究表明风险SNP可用于分析预后、指导用药,实现个体化治疗。
4 结语与展望
尽管GWAS研究已经发现了大量结直肠癌风险相关位点,但大部分SNP的功能效应较小,更多高效力的位点有待发掘。相信随着测序技术的进步、人群研究规模的扩大、分析水平的提高,新的结直肠癌易感位点(包括一些低频、稀有变异)将会不断被发现[15,69~71]。目前,对结直肠癌潜在功能变异的筛选稍显不足,对其进行机制探索的实验更是屈指可数。各种组学、基因组结构等数据的涌现,以及孟德尔随机化的应用,为筛选潜在致病变异提供了可靠信息[72,73];实验技术的发展为阐明致病变异的生物学功能提供更可靠的证据,如:CRISPR/Cas9使单碱基编辑成为可能,染色体构象捕获及其衍生技术可探究SNP的远距离调控机制等等,相信未来将会有更多致病变异的分子机制被阐明。此外,已有研究表明几个不同区域的SNP同时突变时,结直肠癌的患病风险大大增加[38,49];SNP还与多种因素(如阿司匹林的服用、吸烟等)存在交互作用,影响结直肠癌风险[74,75]。可见癌症作为复杂疾病,遗传与遗传、遗传与环境之间的相互作用不可忽视[76]。因此在研究风险SNP的功能时,需要更多的关注SNP与SNP以及SNP与环境之间的作用。相信随着后GWAS研究的开展和深入,将会帮助我们更好地认识变异与结直肠癌发生发展之间的关系,推动个体化预防和精准治疗的发展。
[1] Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries., 2018, 68(6): 394–424.
[2] Wu CX, Gu K, Gong YM, Zheng RS, Wang SM, Chen R, Zhang SW, Shi Y, Wei WQ, Fu C, He J. Analysis of incidence and mortality of colorectal cancer in China, 2015,, 2020, 30(4): 241–245.吴春晓, 顾凯, 龚杨明, 郑荣寿, 王少明, 陈茹, 张思维, 施燕, 魏文强, 付晨, 赫捷. 2015年中国结直肠癌发病和死亡情况分析. 中国癌症杂志, 2020, 30(4): 241–245.
[3] Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer., 2019, 394(10207): 1467–1480.
[4] Lichtenstein P, Holm NV, Verkasalo PK, Iliadou A, Kaprio J, Koskenvuo M, Pukkala E, Skytthe A, Hemminki K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland., 2000, 343(2): 78–85.
[5] Farashi S, Kryza T, Clements J, Batra J. Post-GWAS in prostate cancer: from genetic association to biological contribution., 2019, 19(1): 46–59.
[6] Liang WQ, Hou Y, Zhao CY. Schizophrenia-associated single nucleotide polymorphisms affecting microRNA function., 2019, 41(8): 677–685.梁文权, 侯豫, 赵存友. 精神分裂症相关单核苷酸多态性调控microRNA功能研究进展. 遗传, 2019, 41(8): 677–685.
[7] Cao L, Li ZQ, Shi YY, Liu Y. Telomere length and type 2 diabetes: Mendelian randomization study and polygenic risk score analysis., 2020, 42(9): 882–888.曹岚, 李志强, 师咏勇, 刘赟. 端粒长度与2型糖尿病:孟德尔随机化研究与多基因风险评分分析. 遗传, 2020, 42(9): 882–888.
[8] Tomlinson I, Webb E, Carvajal-Carmona L, Broderick P, Kemp Z, Spain S, Penegar S, Chandler I, Gorman M, Wood W, Barclay E, Lubbe S, Martin L, Sellick G, Jaeger E, Hubner R, Wild R, Rowan A, Fielding S, Howarth K, Consortium C, Silver A, Atkin W, Muir K, Logan R, Kerr D, Johnstone E, Sieber O, Gray R, Thomas H, Peto J, Cazier JB, Houlston R. A genome-wide association scan of tag SNPs identifies a susceptibility variant for colorectal cancer at 8q24.21., 2007, 39(8): 984–988.
[9] Zanke BW, Greenwood CMT, Rangrej J, Kustra R, Tenesa A, Farrington SM, Prendergast J, Olschwang S, Chiang T, Crowdy E, Ferretti V, Laflamme P, Sundararajan S, Roumy S, Olivier JF, Robidoux F, Sladek R, Montpetit A, Campbell P, Bezieau S, O'Shea AM, Zogopoulos G, Cotterchio M, Newcomb P, McLaughlin J, Younghusband B, Green R, Green J, Porteous MEM, Campbell H, Blanche H, Sahbatou M, Tubacher E, Bonaiti-Pellie C, Buecher B, Riboli E, Kury S, Chanock SJ, Potter J, Thomas G, Gallinger S, Hudson TJ, Dunlop MG. Genome-wide association scan identifies a colorectal cancer susceptibility locus on chromosome 8q24., 2007, 39(8): 989–994.
[10] Broderick P, Carvajal-Carmona L, Pittman AM, Webb E, Howarth K, Rowan A, Lubbe S, Spain S, Sullivan K, Fielding S, Jaeger E, Vijayakrishnan J, Kemp Z, Gorman M, Chandler I, Papaemmanuil E, Penegar S, Wood W, Sellick G, Qureshi M, Teixeira A, Domingo E, Barclay E, Martin L, Sieber O, CORGI Consortium, Kerr D, Gray R, Peto J, Cazier JB, Tomlinson I, Houlston RS. A genome-wide association study shows that common alleles ofinfluence colorectal cancer risk., 2007, 39(11): 1315–1317.
[11] Freedman ML, Monteiro ANA, Gayther SA, Coetzee GA, Risch A, Plass C, Casey G, De Biasi M, Carlson C, Duggan D, James M, Liu PY, Tichelaar JW, Vikis HG, You M, Mills IG. Principles for the post-GWAS functional characterization of cancer risk loci., 2011, 43(6): 513–518.
[12] Edwards SL, Beesley J, French JD, Dunning AM. Beyond GWASs: illuminating the dark road from association to function., 2013, 93(5): 779–797.
[13] Liu JY, Zhang LN, Zheng H. Research strategy of the case-control post-genome-wide association study., 2015, (7): 810–812,813.刘佳宇, 张丽娜, 郑红. 后全基因组病例对照研究时代的功能研究策略. 天津医药, 2015, (7): 810–812,813.
[14] Lu YC, Kweon SS, Cai QY, Tanikawa C, Shu XO, Jia WH, Xiang YB, Huyghe JR, Harrison TA, Kim J, Shin A, Kim DH, Matsuo K, Jee SH, Guo XY, Wen WQ, Shi JJ, Li BS, Wang N, Shin MH, Li HL, Ren ZF, Oh JH, Oze I, Ahn YO, Jung KJ, Gao J, Gao YT, Pan ZZ, Kamatani Y, Chan AT, Gsur A, Hampe J, Le Marchand L, Li L, Lindblom A, Moreno V, Newcomb PA, Offit K, Pharoah PDP, van Duijnhoven FJB, Van Guelpen B, Vodicka P, Weinstein SJ, Wolk A, Wu AH, Hsu L, Zeng YX, Long JR, Peters U, Matsuda K, Zheng W. Identification of novel loci and new risk variant in known loci for colorectal cancer risk in east Asians., 2020, 29(2): 477–486.
[15] Lu YC, Kweon SS, Tanikawa C, Jia WH, Xiang YB, Cai QY, Zeng CJ, Schmit SL, Shin A, Matsuo K, Jee SH, Kim DH, Kim J, Wen WQ, Shi JJ, Guo XY, Li BS, Wang N, Zhang B, Li XX, Shin MH, Li HL, Ren ZF, Oh JH, Oze I, Ahn YO, Jung KJ, Conti DV, Schumacher FR, Rennert G, Jenkins MA, Campbell PT, Hoffmeister M, Casey G, Gruber SB, Gao J, Gao YT, Pan ZZ, Kamatani Y, Zeng YX, Shu XO, Long JR, Matsuda K, Zheng W. Large-scale genome-wide association study of East Asians identifies loci associated with risk for colorectal cancer., 2019, 156(5): 1455–1466.
[16] Huyghe JR, Bien SA, Harrison TA, Kang HM, Chen S, Schmit SL, Conti DV, Qu CH, Jeon J, Edlund CK, Greenside P, Wainberg M, Schumacher FR, Smith JD, Levine DM, Nelson SC, Sinnott-Armstrong NA, Albanes D, Alonso MH, Anderson K, Arnau-Collell C, Arndt V, Bamia C, Banbury BL, Baron JA, Berndt SI, Bézieau S, Bishop DT, Boehm J, Boeing H, Brenner H, Brezina S, Buch S, Buchanan DD, Burnett-Hartman A, Butterbach K, Caan BJ, Campbell PT, Carlson CS, Castellví-Bel S, Chan AT, Chang-Claude J, Chanock SJ, Chirlaque MD, Cho SH, Connolly CM, Cross AJ, Cuk K, Curtis KR, de la Chapelle A, Doheny KF, Duggan D, Easton DF, Elias SG, Elliott F, English DR, Feskens EJM, Figueiredo JC, Fischer R, FitzGerald LM, Forman D, Gala M, Gallinger S, Gauderman WJ, Giles GG, Gillanders E, Gong J, Goodman PJ, Grady WM, Grove JS, Gsur A, Gunter MJ, Haile RW, Hampe J, Hampel H, Harlid S, Hayes RB, Hofer P, Hoffmeister M, Hopper JL, Hsu WL, Huang WY, Hudson TJ, Hunter DJ, Ibañez-Sanz G, Idos GE, Ingersoll R, Jackson RD, Jacobs EJ, Jenkins MA, Joshi AD, Joshu CE, Keku TO, Key TJ, Kim HR, Kobayashi E, Kolonel LN, Kooperberg C, Kühn T, Küry S, Kweon SS, Larsson SC, Laurie CA, Le Marchand L, Leal SM, Lee SC, Lejbkowicz F, Lemire M, Li CI, Li L, Lieb W, Lin Y, Lindblom A, Lindor NM, Ling H, Louie TL, Männistö S, Markowitz SD, Martín V, Masala G, McNeil CE, Melas M, Milne RL, Moreno L, Murphy N, Myte R, Naccarati A, Newcomb PA, Offit K, Ogino S, Onland-Moret NC, Pardini B, Parfrey PS, Pearlman R, Perduca V, Pharoah PDP, Pinchev M, Platz EA, Prentice RL, Pugh E, Raskin L, Rennert G, Rennert HS, Riboli E, Rodríguez-Barranco M, Romm J, Sakoda LC, Schafmayer C, Schoen RE, Seminara D, Shah M, Shelford T, Shin MH, Shulman K, Sieri S, Slattery ML, Southey MC, Stadler ZK, Stegmaier C, Su YR, Tangen CM, Thibodeau SN, Thomas DC, Thomas SS, Toland AE, Trichopoulou A, Ulrich CM, Van Den Berg DJ, van Duijnhoven FJB, Van Guelpen B, van Kranen H, Vijai J, Visvanathan K, Vodicka P, Vodickova L, Vymetalkova V, Weigl K, Weinstein SJ, White E, Win AK, Wolf CR, Wolk A, Woods MO, Wu AH, Zaidi SH, Zanke BW, Zhang Q, Zheng W, Scacheri PC, Potter JD, Bassik MC, Kundaje A, Casey G, Moreno V, Abecasis GR, Nickerson DA, Gruber SB, Hsu L, Peters U. Discovery of common and rare genetic risk variants for colorectal cancer., 2019, 51(1): 76–87.
[17] Qu XF, Wang MY, Cai SJ, Wei QY. Research progress and prospect of genome-wide association analysis of colorectal cancer., 2019, 11(1): 5–12.屈晓飞, 王梦筠, 蔡三军, 魏庆义. 结直肠癌全基因组关联分析研究进展及展望. 中国癌症防治杂志, 2019, 11(1): 5–12.
[18] Law PJ, Timofeeva M, Fernandez-Rozadilla C, Broderick P, Studd J, Fernandez-Tajes J, Farrington S, Svinti V, Palles C, Orlando G, Sud A, Holroyd A, Penegar S, Theodoratou E, Vaughan-Shaw P, Campbell H, Zgaga L, Hayward C, Campbell A, Harris S, Deary IJ, Starr J, Gatcombe L, Pinna M, Briggs S, Martin L, Jaeger E, Sharma-Oates A, East J, Leedham S, Arnold R, Johnstone E, Wang HT, Kerr D, Kerr R, Maughan T, Kaplan R, Al-Tassan N, Palin K, Hänninen UA, Cajuso T, Tanskanen T, Kondelin J, Kaasinen E, Sarin AP, Eriksson JG, Rissanen H, Knekt P, Pukkala E, Jousilahti P, Salomaa V, Ripatti S, Palotie A, Renkonen-Sinisalo L, Lepistö A, Böhm J, Mecklin JP, Buchanan DD, Win AK, Hopper J, Jenkins ME, Lindor NM, Newcomb PA, Gallinger S, Duggan D, Casey G, Hoffmann P, Nöthen MM, Jöckel KH, Easton DF, Pharoah PDP, Peto J, Canzian F, Swerdlow A, Eeles RA, Kote-Jarai Z, Muir K, Pashayan N, consortium P, Harkin A, Allan K, McQueen J, Paul J, Iveson T, Saunders M, Butterbach K, Chang-Claude J, Hoffmeister M, Brenner H, Kirac I, Matosevic P, Hofer P, Brezina S, Gsur A, Cheadle JP, Aaltonen LA, Tomlinson I, Houlston RS, Dunlop MG. Association analyses identify 31 new risk loci for colorectal cancer susceptibility., 2019, 10(1): 2154.
[19] George Priya Doss C, Rajasekaran R, Arjun P, Sethumadhavan R. Prioritization of candidate SNPs in colon cancer using bioinformatics tools: an alternative approach for a cancer biologist., 2010, 2(4): 320–346.
[20] Steck SE, Butler LM, Keku T, Antwi S, Galanko J, Sandler RS, Hu JJ. Nucleotide excision repair gene polymorphisms, meat intake and colon cancer risk., 2014, 762: 24–31.
[21] Theodoratou E, Farrington SM, Timofeeva M, Din FV, Svinti V, Tenesa A, Liu T, Lindblom A, Gallinger S, Campbell H, Dunlop MG. Genome-wide scan of the effect of common nsSNPs on colorectal cancer survival outcome., 2018, 119(8): 988–993.
[22] Zhang MZ, Huang C, Wang ZY, Lv HB, Li XM. In silico analysis of non-synonymous single nucleotide polymorphisms (nsSNPs) in the humangene associated with congenital cataract., 2020, 21(1): 12.
[23] Timofeeva MN, Kinnersley B, Farrington SM, Whiffin N, Palles C, Svinti V, Lloyd A, Gorman M, Ooi LY, Hosking F, Barclay E, Zgaga L, Dobbins S, Martin L, Theodoratou E, Broderick P, Tenesa A, Smillie C, Grimes G, Hayward C, Campbell A, Porteous D, Deary IJ, Harris SE, Northwood EL, Barrett JH, Smith G, Wolf R, Forman D, Morreau H, Ruano D, Tops C, Wijnen J, Schrumpf M, Boot A, Vasen HFA, Hes FJ, van Wezel T, Franke A, Lieb W, Schafmayer C, Hampe J, Buch S, Propping P, Hemminki K, Försti A, Westers H, Hofstra R, Pinheiro M, Pinto C, Teixeira M, Ruiz-Ponte C, Fernández-Rozadilla C, Carracedo A, Castells A, Castellví-Bel S, Campbell H, Bishop DT, Tomlinson IPM, Dunlop MG, Houlston RS. Recurrent coding sequence variation explains only a small fraction of the genetic architecture of colorectal cancer., 2015, 5: 16286.
[24] Huang QT, Li Q, Zhang YB. Linking chromatin conformation to gene function., 2020, 42(1): 1–17.黄其通, 李清, 张玉波. 染色质构象与基因功能. 遗传, 2020, 42(1): 1–17.
[25] Gorkin DU, Qiu YJ, Hu M, Fletez-Brant K, Liu T, Schmitt AD, Noor A, Chiou J, Gaulton KJ, Sebat J, Li Y, Hansen KD, Ren B. Common DNA sequence variation influences 3-dimensional conformation of the human genome., 2019, 20(1): 255.
[26] Lv HQ, Hao LL, Liu EH, Wu ZF, Han JQ, Liu Y. Current status and future perspectives in bioinformatical analysis of Hi-C data., 2020, 42(1): 87–99.吕红强, 郝乐乐, 刘二虎, 吴志芳, 韩九强, 刘源. 基于生物信息学的Hi-C研究现状与发展趋势. 遗传, 2020, 42(1): 87–99.
[27] Kang BW, Jeon HS, Chae YS, Lee SJ, Park JS, Choi GS, Kim JG. Impact of genetic variation in microRNA-binding site on susceptibility to colorectal cancer., 2016, 36(7): 3353–3361.
[28] Yang ML, Huang Z, Wu LN, Wu R, Ding HX, Wang BG.rs2632159 polymorphism could be a biomarker for colorectal cancer susceptibility., 2019, 39(7): BSR20190708.
[29] Jia WR, Zeng LY, Luo SQ, Bai F, Zhong R, Wu L, Huang GL, Pu XX. Association ofrs6505162 C>A polymorphism with susceptibility and metastasis of colorectal carcinoma., 2018, 97(6): e9846.
[30] Chen YT, Du ML, Chen W, Zhu LJ, Wu CY, Zhang ZD, Wang ML, Chu HY, Gu D, Chen JF. Polymorphism rs2682818 inis associated with colorectal cancer susceptibility in a Han Chinese population., 2018, 7(4): 1194–1200.
[31] He HJ, Lei L, Chen EF, Xu XN, Wang LL, Pan JQ, Yang FF, Wang M, Dong J, Yang J. The screening of the functional microRNA binding site SNPs in sporadic colorectal cancer genes., 2017, 18(6): 407–413.
[32] Qiu QC, Liu J, Shao JF, Lou XY, Chen C, Lin BY. Target-resequencing to identify microRNA-associated SNP and predict the effect of SNP on microRNA function in colorectal cancer patients., 2015, 37(10): 759–763.
[33] Li JY, Zou L, Zhou Y, Li L, Zhu Y, Yang Y, Gong YJ, Lou J, Ke JT, Zhang Y, Tian JB, Zou DY, Peng XT, Chang J, Gong J, Zhong R, Zhou XB, Miao XP. A low-frequency variant inmodulates TGF-β signaling and confers risk for colorectal cancer in Chinese population., 2017, 56(7): 1798–807.
[34] Shen N, Li L, Xu W, Tian JB, Yang Y, Zhu Y, Gong YJ, Ke JT, Gong J, Chang J, Zhong R, Miao XP. A missense variant inassociated with the risk of colorectal cancer by modifying Ras/MEK/ERK signaling., 2019, 59: 109–114.
[35] Li JY, Chang J, Tian JB, Ke JT, Zhu Y, Yang Y, Gong YJ, Zou DY, Peng XT, Yang N, Mei SF, Wang XY, Cheng LM, Hu WG, Gong J, Zhong R, Miao XP. A rare variant P507L ininterrupts TPP1-TIN2 interaction, influences telomere length, and confers colorectal cancer risk in Chinese population., 2018, 27(9): 1029–1035.
[36] Tian JB, Ying PT, Ke JT, Zhu Y, Yang Y, Gong YJ, Zou DY, Peng XT, Yang N, Wang XY, Mei SF, Zhang YX, Wang CY, Zhong R, Chang J, Miao XP.N6-Methyladenosine-related variant is associated with colorectal cancer risk by maintaining the genomic stability., 2020, 146(12): 3281–3293.
[37] Sud A, Kinnersley B, Houlston RS. Genome-wide association studies of cancer: current insights and future perspectives., 2017, 17(11): 692–704.
[38] Chang J, Tian JB, Yang Y, Zhong R, Li JY, Zhai K, Ke JT, Lou J, Chen W, Zhu BB, Shen N, Zhang Y, Gong YJ, Zhu Y, Zou DY, Peng XT, Huang K, Miao XP. A rare missense variant inassociates with colorectal cancer risk by interacting with a GWAS-identified regulatory variant in theenhancer., 2018, 78(17): 5164– 5172.
[39] Moreno V, Alonso MH, Closa A, Vallés X, Diez- Villanueva A, Valle L, Castellví-Bel S, Sanz-Pamplona R, Lopez-Doriga A, Cordero D, Solé X. Colon-specific eQTL analysis to inform on functional SNPs., 2018, 119(8): 971–977.
[40] Wu SS, Meng QT, Zhang CC, Sun H, Lu RZ, Gao N, Yang HB, Li XB, Aschner M, Chen R.mediates the progression, invasion, metastasis and survival of colorectal cancer through the Sp1/NF1 switch axis on genomic locus., 2018, 143(2): 289–297.
[41] Tian JB, Chang J, Gong J, Lou J, Fu MP, Li JY, Ke JT, Zhu Y, Gong YJ, Yang Y, Zou DY, Peng XT, Yang N, Mei SF, Wang XY, Zhong R, Cai KL, Miao XP. Systematic functional interrogation of genes in GWAS loci identifiedas a key driver in colorectal cancer modulated by a promoter-enhancer interaction., 2019, 105(1): 29–47.
[42] Gong J, Tian J, Lou J, Wang X, Ke J, Li J, Yang Y, Gong Y, Zhu Y, Zou D, Peng X, Yang N, Mei S, Zhong R, Chang J, Miao X. A polymorphic MYC response element ininfluences colorectal cancer risk, especially in interaction with an MYC-regulated SNP rs6983267., 2018, 29(3): 632–639.
[43] Yu CY, Han JX, Zhang JF, Jiang PL, Shen CQ, Guo FF, Tang JY, Yan TT, Tian XL, Zhu XQ, Ma D, Hu Y, Xie YH, Du W, Zhong M, Chen JX, Liu Q, Sun DF, Chen YX, Zou WP, Hong J, Chen HY, Fang JY. A 16q22.1 variant confers susceptibility to colorectal cancer as a distal regulator of., 2020, 39(6): 1347–1360.
[44] Wang YJ, Wu SS, Yang X, Li XB, Chen R. Association between polymorphism in the promoter region of lncRNAand the risk of colorectal cancer., 2019, 39(4): BSR20190091.
[45] Wang ML, Gu DY, Du ML, Xu Z, Zhang SZ, Zhu LJ, Lu JC, Zhang R, Xing JL, Miao XP, Chu HY, Hu ZB, Yang L, Tang CJ, Pan L, Du HN, Zhao J, Du J, Tong N, Sun J, Shen H, Xu J, Zhang Z, Chen J. Common genetic variation inis associated with colorectal cancer susceptibility., 2016, 7: 11478.
[46] Zou DY, Lou J, Ke JT, Mei SF, Li JY, Gong YJ, Yang Y, Zhu Y, Tian JB, Chang J, Zhong R, Gong J, Miao XP. Integrative expression quantitative trait locus-based analysis of colorectal cancer identified a functional polymorphism regulatingexpression., 2018, 93: 1–9.
[47] Meng QT, Wu SS, Wang YJ, Xu J, Sun H, Lu RZ, Gao N, Yang HB, Li XB, Tang BP, Aschner M, Chen R.promoter polymorphism rs2333227 enhances malignant phenotypes of colorectal cancer by altering the binding affinity of AP-2α., 2018, 78(10): 2760–2769.
[48] Ren AJ, Sun SW, Li SW, Chen T, Shu YQ, Du ML, Zhu LJ. Genetic variants incontribute to the susceptibility to colorectal cancer., 2019, 145(1): 154–163.
[49] Zou DY, Zhang HL, Ke JT, Li JY, Zhu Y, Gong YJ, Yang Y, Tian JB, Zhang Y, Peng XT, Cai KL, Zhong R, Chang J, Miao XP. Three functional variants were identified to affectexpression and significantly associated with risk of colorectal cancer., 2020, 94(1): 295– 303.
[50] Tian JB, Lou J, Cai YM, Rao ML, Lu ZQ, Zhu Y, Zou DY, Peng XT, Wang HX, Zhang M, Niu SY, Li Y, Zhong R, Chang J, Miao XP. Risk SNP-mediated enhancer-promoter interaction drives colorectal cancer through bothand., 2020, 80(9): 1804–1818.
[51] Statkiewicz M, Maryan N, Kulecka M, Kuklinska U, Ostrowski J, Mikula M. Functional analyses of a low-penetrance risk variant rs6702619/1p21.2 associating with colorectal cancer in Polish population., 2019, 66(3): 329–336.
[52] Ni HZ, Su BF, Pan LM, Li XY, Zhu XX, Chen XJ. Functional variants inare associated with colorectal cancer susceptibility in Chinese populations., 2015, 39(6): 972–977.
[53] Li JY, Liu H, Zou L, Ke JT, Zhang Y, Zhu Y, Yang Y, Gong YJ, Tian JB, Zou DY, Peng XT, Gong J, Zhong R, Huang K, Chang J, Miao XP. A functional variant inconfers risk for colorectal cancer by disrupting abinding site., 2017, 8(37): 61318–61326.
[54] Ke JT, Tian JB, Li JY, Gong YJ, Yang Y, Zhu Y, Zhang Y, Zhong R, Chang J, Gong J. Identification of a functional polymorphism affecting microRNA binding in the susceptibility locus 1q25.3 for colorectal cancer., 2017, 56(9): 2014–2021.
[55] Li SW, Xu KL, Gu DY, He L, Xie LS, Chen ZX, Fan ZM, Zhu LJ, Du ML, Chu HY, Zhang ZD, Wu Y, Ni M, Wang ML. Genetic variants inassociated with the response to oxaliplatin-based chemotherapy in colorectal cancer., 2019, 54(11): 939–949.
[56] Xie LS, Li SW, Jin J, He L, Xu KL, Zhu LJ, Du ML, Liu YQ, Chu HY, Zhang ZD, Wang ML, Shi DN, Gu DY, Ni M. Genetic variant inbinding sites is associated with colorectal cancer risk., 2019, 23(3): 2012–2019.
[57] Gu DY, Li SW, Du ML, Tang CJ, Chu HY, Tong N, Zhang ZD, Wang ML, Chen JF. A genetic variant located in the-binding site ofis associated with the colorectal cancer risk., 2019, 54(2): 141– 148.
[58] Shen CQ, Yan TT, Wang ZH, Su HC, Zhu XQ, Tian XL, Fang JY, Chen HY, Hong J. Variant of SNP rs1317082 at() creates a binding site forand diminishes the susceptibility to CRC., 2018, 9(12): 1177.
[59] Wu S, Sun H, Wang Y, Yang X, Meng Q, Yang H, Zhu H, Tang W, Li X, Aschner M, Chen R.rs664589 polymorphism inhibits binding to, contributing to colorectal cancer risk, growth, and metastasis., 2019, 79(20): 5432–41.
[60] Fu Y, Zhang YZ, Cui JY, Yang G, Peng SF, Mi WN, Yin XY, Yu Y, Jiang JW, Liu Q, Qin YY, Xu W. SNP rs12982687 affects binding capacity of lncRNAwith: involvement in smoking-triggered colorectal cancer progression., 2020, 18(1): 37.
[61] Jiang HQ, Ge FY, Hu BN, Wu LM, Yang HJ, Wang HY. rs35301225 polymorphism inpromotes development of human colon cancer by deregulation of 3'UTR inin Chinese population., 2017, 17: 39.
[62] Jin GF, Shen HB. Cancer risk prediction in post-genome- wide association study era., 2015, 36(10): 1045–1046.靳光付, 沈洪兵. 后全基因组关联研究时代的肿瘤风险预测. 中华流行病学杂志, 2015, 36(10): 1045–1046.
[63] Frampton MJE, Law P, Litchfield K, Morris EJ, Kerr D, Turnbull C, Tomlinson IP, Houlston RS. Implications of polygenic risk for personalised colorectal cancer screening., 2016, 27(3): 429–434.
[64] Li JY, Chang J, Zhu Y, Yang Y, Gong YJ, Ke JT, Lou J, Zhong R, Gong J. Risk prediction of colorectal cancer with common genetic variants and conventional non— genetic factors in a Chinese Han population., 2015, 36(10): 1053–1057.李娇元, 常江, 朱颖, 杨洋, 龚雅洁, 柯俊涛, 娄娇, 钟荣, 龚静. 基于常见遗传变异和传统风险因素的中国南方汉族人群结直肠癌风险预测模型研究. 中华流行病学杂志, 2015, 36(10): 1053–1057.
[65] Summers MG, Maughan TS, Kaplan R, Law PJ, Houlston RS, Escott-Price V, Cheadle JP. Comprehensive analysis of colorectal cancer-risk loci and survival outcome: A prognostic role forvariants., 2020, 124: 56–63.
[66] Penney KL, Banbury BL, Bien S, Harrison TA, Hua XW, Phipps AI, Sun W, Song MY, Joshi AD, Alberts SR, Allegra CJ, Atkins J, Colangelo LH, George TJ, Goldberg RM, Lucas PC, Nair SG, Shi Q, Sinicrope FA, Wolmark N, Yothers G, Peters U, Newcomb PA, Chan AT. Genetic variant associated with survival of patients with stage II-III colon cancer., 2020, 18(12): 2717–2723.e3.
[67] Song N, Kim K, Shin A, Park JW, Chang HJ, Shi JJ, Cai QY, Kim DY, Zheng W, Oh JH. Colorectal cancer susceptibility loci and influence on survival., 2018, 57(12): 630–637.
[68] Papachristos A, Kemos P, Katsila T, Panoilia E, Patrinos GP, Kalofonos H, Sivolapenko GB.andgene polymorphisms as predictors of clinical outcome to first-line bevacizumab-based treatment in metastatic colorectal cancer., 2019, 20(22): 5791
[69] Wang HS, Haiman CA, Burnett T, Fortini BK, Kolonel LN, Henderson BE, Signorello LB, Blot WJ, Keku TO, Berndt SI, Newcomb PA, Pande M, Amos CI, West DW, Casey G, Sandler RS, Haile R, Stram DO, Le Marchand L. Fine- mapping of genome-wide association study-identified risk loci for colorectal cancer in African Americans., 2013, 22(24): 5048–5055.
[70] Schmit SL, Schumacher FR, Edlund CK, Conti DV, Ihenacho U, Wan P, Van Den Berg D, Casey G, Fortini BK, Lenz HJ, Tusié-Luna T, Aguilar-Salinas CA, Moreno- Macías H, Huerta-Chagoya A, Ordóñez-Sánchez ML, Rodríguez-Guillén R, Cruz-Bautista I, Rodríguez-Torres M, Muñóz-Hernández LL, Arellano-Campos O, Gómez D, Alvirde U, González-Villalpando C, González-Villalpando ME, Le Marchand L, Haiman CA, Figueiredo JC. Genome-wide association study of colorectal cancer in Hispanics., 2016, 37(6): 547–556.
[71] Wang HS, Schmit SL, Haiman CA, Keku TO, Kato I, Palmer JR, van den Berg D, Wilkens LR, Burnett T, Conti DV, Schumacher FR, Signorello LB, Blot WJ, Zanetti KA, Harris C, Pande M, Berndt SI, Newcomb PA, West DW, Haile R, Stram DO, Figueiredo JC, Hispanic Colorectal Cancer Study, Le Marchand L. Novel colon cancer susceptibility variants identified from a genome-wide association study in African Americans., 2017, 140(12): 2728–2733.
[72] Collins RL, Brand H, Karczewski KJ, Zhao XF, Alföldi J, Francioli LC, Khera AV, Lowther C, Gauthier LD, Wang H, Watts NA, Solomonson M, O'Donnell-Luria A, Baumann A, Munshi R, Walker M, Whelan CW, Huang YQ, Brookings T, Sharpe T, Stone MR, Valkanas E, Fu J, Tiao G, Laricchia KM, Ruano-Rubio V, Stevens C, Gupta N, Cusick C, Margolin L, Genome Aggregation Database Production Team, Genome Aggregation Database Consortium, Taylor KD, Lin HJ, Rich SS, Post WS, Chen YDI, Rotter JI, Nusbaum C, Philippakis A, Lander E, Gabriel S, Neale BM, Kathiresan S, Daly MJ, Banks E, MacArthur DG, Talkowski ME. A structural variation reference for medical and population genetics., 2020, 581(7809): 444–451.
[73] Zhou D, Jiang Y, Zhong X, Cox NJ, Liu CY, Gamazon ER. A unified framework for joint-tissue transcriptome-wide association and Mendelian randomization analysis., 2020, 52(11): 1239–1246.
[74] Song N, Lee J, Cho S, Kim J, Oh JH, Shin A. Evaluation of gene-environment interactions for colorectal cancer susceptibility loci using case-only and case-control designs., 2019, 19(1): 1231.
[75] Zhang K, Li SW, Gu DY, Xu KL, Zheng R, Xin JY, Meng YX, Ben S, Chu HY, Zhang ZD, Shu YQ, Du ML, Liu LX, Wang ML. Genetic variants ininteracting with smoking can enhance colorectal cancer risk., 2020, 94(1): 325–333.
[76] Jung SY, Papp JC, Sobel EM, Zhang ZF. Post Genome- wide gene-environment interaction study using random survival forest: insulin resistance, lifestyle factors, and colorectal cancer risk., 2019, 12(12): 877–890.
Progress on functional mechanisms of colorectal cancer causal SNPs in post-GWAS
Yige Li1,2,3, Dandan Zhang1,2,3
Colorectal cancer (CRC) is caused by genetic and environmental factors, and the genetic component plays a significant role in CRC development. Currently, genome-wide association studies (GWAS) have identified a large number of genetic loci associated with CRC risk. In the post-GWAS era, more and more efforts focus on deciphering the biological mechanisms behind these potential causal variants by using multi-omics data and functional experiments. Many analyses have revealed that most risk single nucleotide polymorphisms (SNPs) are located in non-coding regions and these variants may regulate the expression of target genes by altering the transcription factor-binding motif, epigenetic modification, chromatin accessibility or 3D genome conformation. Results obtained from post-GWAS era have highlighted the possibility of moving from association to function. In this review, we summarize the current status of CRC post-GWAS studies and discusses the clinical application as well as future directions of CRC GWAS, in order to better gain insight into the molecular basis of CRC and provide evidence for prevention.
colorectal cancer; post-GWAS; SNP; casual variant
2020-12-01;
2021-01-11
国家自然科学基金项目(编号:81773027,81101640),浙江省自然科学基金项目(编号:LY21H160027),中央高校基本科研业务费专项资金资助[Supported by the National Natural Science Foundation of China (Nos. 81773027, 81101640), Natural Science Foundation of Zhejiang Province of China (No. LY21H160027) , and Fundamental Research Funds for the Central Universities]
李以格,在读硕士研究生,专业方向:病理学与病理生理学。E-mail: yigelee@zju.edu.cn
张丹丹,博士,副教授,研究方向:复杂疾病遗传学及分子机制。E-mail: dandanz@zju.edu.cn
10.16288/j.yczz.20-320
2021/1/22 11:06:54
URI: https://kns.cnki.net/kcms/detail/11.1913.R.20210122.1052.005.html
(责任编委: 周钢桥)

