Improvement of the riboflavin production by engineering the precursor biosynthesis pathways in Escherichia coli
Zhibo Xu ,Zhenquan Lin ,Zhiwen Wang,Tao Chen*
Key Laboratory of Systems Bioengineering(Ministry of Education),Tianjin University,Tianjin 300072,China
SynBio Research Platform,Collaborative Innovation Center of Chemical Science and Engineering(Tianjin),School of Chemical Engineering and Technology,Tianjin University,Tianjin 300072,China
Edinburg-Tianjin Joint Research Centre for Systems Biology and Synthetic Biology,Tianjin University,Tianjin 300072,China
Keywords:Escherichia coli Riboflavin 3,4-Dihydroxy-2-butanone 4-phosphate synthase Purine pathway GTP Metabolic engineering
ABSTRACT 3,4-Dihydroxy-2-butanone 4-phosphate(DHBP)and GTP are the precursors for riboflavin biosynthesis.In this research,improving the precursor supply for riboflavin production was attempted by overexpressing ribB and engineering purine pathway in a riboflavin-producing Escherichia coli strain.Initially,ribB gene was overexpressed to increase the flux from ribulose 5-phosphate(Ru-5-P)to DHBP.Then ndk and gmk genes were overexpressed to enhance GTP supply.Subsequently,a R419L mutation was introduced into purA to reduce the flux from IMP to AMP.Finally,co-overexpression of mutant purF and prs genes further increased riboflavin production.The final strain RF18S produced 387.6 mg riboflavin·L-1 with a yield of 44.8 mg riboflavin per gram glucose in shake- flask fermentations.The final titer and yield were 72.2%and 55.6%higher than those of RF01S,respectively.It was concluded that simultaneously engineering the DHBP synthase and GTP biosynthetic pathway by rational metabolic engineering can efficiently boost riboflavin production in E.coli.
1.Introduction
Riboflavin(RF)is an essential substrate for the biosynthesis of flavin mononucleotide(FMN)and flavin adenine dinucleotide(FAD),both of which act as oxidation-reduction cofactors involved in a wide range of biological reaction[1].Riboflavin,as a technical bulk product,has been applied in many fields,such as pharmaceuticals,cosmetics,and human and animal nutrition[2].Riboflavin production in industry can be achieved by chemicalmanufacturing processorby biologicalprocess.Driven by costs,waste and energy considerations,biologicalprocess has replaced the chemical manufacturing process.
Riboflavin biosynthesis starts with GTP and Ru-5-P through a sevenstep synthetic pathway,which is similar in all organisms(Fig.1)[3].Previous reports have shown that overexpression of riboflavin biosynthetic genes was important for the riboflavin production.Examples included increasing the copy numbers of the rib operon and/or introducing an additional copy of the ribA gene(encoding DHBP synthase and GTP cyclohydrolase II)in Bacillus subtilis[4-6],co-overexpressing RIB1 gene(encoding GTP cyclohydrolase II)and RIB7 gene(encoding riboflavin synthase)in Candida famata[7].
According to previous reported results[8],pooravailability of purine nucleotides may be a limiting factor for further increase of riboflavin production in a riboflavin-producing strain.Several researches have focused on improving the flux of purine biosynthetic pathway to promote riboflavin production.In B.subtilis,modification of some key genes in purine biosynthetic pathway[8,9],pentose phosphate pathway[10]and gluconeogenic pathway[11]enhanced the supply of the precursor metabolites,which in turn facilitated riboflavin production.In Ashbya gossypii,overexpressing mutant deregulated isoforms of PRPP synthase significantly enhanced the riboflavin production[12].In addition,cooverexpressing purF and prs genes led to increase inosine production in E.coli and guanosine production in Bacillus amyloliquefaciens[13,14].Moreover,purA mutation was employed to increase the inosine production in E.coli[15]and B.subtilis[16,17].
Although wild-type E.coli does not accumulate riboflavin under natural conditions,it may be an efficient host for the production of riboflavin due to its fast-growing,abundant genetic information and tools for gene manipulation.The biosynthetic pathway and enzymes for riboflavin synthesis in E.coli have been studied in detail.In addition,previous studies have shown that the production of riboflavin required efficient energy generation and low maintenance metabolism in B.subtilis[18].Varying from 0.11 to 0.38 mmol(glucose)·g-1·h-1[19],the substrate maintenance coefficient of E.coli is obviously lower than that of wild-type B.subtilis(0.44 mmol(glucose)·g-1·h-1,[20]),which might contribute to a higher riboflavin yield when a high cell density fermentation process is conducted.

Fig.1.Diagram of the E.coli metabolic pathway of riboflavin biosynthesis,including EMP(Embden-Meyerhof-Parnas)pathway,TCA cycle,purine pathway and riboflavin biosynthetic pathway.Dashed lines indicate multiple enzymatic steps.The+indicated that the pathways were overexpressed.The-indicated that the pathways were reduced expression.Relevant reactions were represented by the genes encoding for corresponding enzymes:pgi,phosphoglucose isomerase;rpiA,ribose-5-phosphate isomerase A;rpiB,ribose-5-phosphate isomerase B;prs,PRPP synthetase;purF,PRPP amidotransferase;purA,adenylosuccinate synthetase;guaB,IMP dehydrogenase;guaA,GMP synthetase;gmk,guanylate kinase;guaC,GMP reductase;ndk,nucleoside diphosphate kinase;ribA,GTP cyclohydrolase II;ribB,3,4-dihydroxy-2-butanone 4-phosphate synthase;ribD,fused diaminohydroxyphosphoribosylaminopyrimidine deaminase/5-amino-6-(5-phosphoribosylamino)uracil reductase;ribE,6,7-dimethyl-8-ribityllumazine synthase;ribC,riboflavin synthase;ribF,bifunctional riboflavin kinase/FMN adenylyltransferase;appA,acid phosphatase;aphA,acid phosphatase/phosphotransferase.Abbreviations:G6P,glucose-6-phosphate;F6P,fructose-6-phosphate;PYR,pyruvic acid;Ru5P,ribulose-5-phosphate;R5P,ribose-5-phosphate;PRPP,phosphoribosylpyrophosphate;IMP,inosine monophosphate;Succinyl-AMP,adenylosuccinate;XMP,xanthosine monophosphate;GMP,guanosine monophosphate;GDP,guanosine diphosphate;GTP,guanosine triphosphate;DARPP,2,5-diamino-6-ribosylamino-4(3H)-pyrimidinone-5'-phosphate;ARPP,5-amino-6-(5'-phosphoribosylamino)uracil;ArPP,5-amino-6-(5'-phosphoribitylamino)uracil;ArP,4-(1-D-ribitylamino)-5-amino-2,6-dihydroxypyrimidine;DHPB,3,4-dihydroxy-2-butanone 4-phosphate;DRL,6,7-dimethyl-8-ribityl-lumazine;FMN, flavin mononucleotide;FAD, flavin adenine dinucleotide.
In this study,ribB and genes involved in de novo purine biosynthetic pathway,including ndk,gmk,purA,purF and prs,were simultaneously modified in a riboflavin-producing E.coli strain.Eventually,the final strain RF18S resulted in 387.6 mg riboflavin·L-1with a yield of 44.8 mg riboflavin·(g glucose)-1.
2.Materials and Methods
2.1.Strains,plasmids,media,and cultivation conditions
All of the bacteria strains and plasmids used in this study were listed in Table 1.E.coli K-12 MG1655 was used throughout this work.Plasmid p20C-EC10 used in this study was described in detail by Lin et al.[21].During strain construction,cultures were grown at 30°C in Luria-Bertani(LB)medium(per liter:10 g tryptone,5 g yeast extract,10 g NaCl)with the corresponding antibiotic when required.

Table 1 Strains and plasmids used in this study
2.2.Genome engineering

Fig.2.Strategy of fragment construction and scarless gene manipulation based onλRed mediated recombination and I-Sce Inuclease.(A)Construction of promoter swapping ribB,ndk,gmk fragments and point mutational purA fragment.(B)Construction of desensitized prs and purF fragments.
All primers used in this study were listed in Supplementary Table 1.The strategy of fragment construction and genome manipulation is outlined in Fig.2.All transformation constructs were prepared according to the method previously reported[21].Since a large number of primers were required for these constructs,a schematic diagram for overlap PCR primer position previously reported by Goswami[22]was used to indicate the location of each primer using a simple numbering system.pTKS/CS was employed to amplify the tetA cassette.As shown in Fig.2,PCR fragment constructs strategies using tetA as a selection marker with and without an additional fragment 2(mutant purF or prs genes).The numbers in Fig.2 are included in the primernames listed in Supplementary Table 1 under the overlap heading.The mutant purF and prs genes was constructed by splicing with SOE-PCR with the primer pairs purF 2-F1/purF 2-R1,purF 2-F2/purF 2-R2,purF 2-F3/purF 2-R3 and prs 2-F1/prs 2-R1,prs 2-F2/prs 2-R2,respectively[23].All primers used during construction were listed in Supplementary Table 1.
The final DNA fragment was transformed into E.coli cells carrying pTKRED by electroporation,according to the method described previously[21,24].In the first recombination,fragment consisting of tetA cassette,two duplication regions(DRs) flanking tetA and target sequence,was used to replace or insert into target site.The tetracycline resistance mutants were selected and confirmed by using primer tests 1-F/3-R.In the second recombination,arabinose was added to induce I-Sce Iendonuclease expression for cleaving the tetA and facilitating the recombination between DRs.The loss of tetA in the desired recombinantstrains was further verified by colony PCR using primer tests 1-F/4-R or 1-F/5-R.This strategy was used to replace promoter(ribB,ndk and gmk genes),introduce pointmutation(purA gene)and insertan additional copy of mutant gene(mutant purF and prs genes)into the genome at target loci.
2.3.Shaking- flask fermentation for riboflavin production
Single E.coli colonies from transformed strains were inoculated into 5 ml LB medium and incubated overnight at 37°C as the first seed cultures.Then the cells were inoculated into 50 ml LB medium at a starting OD600of 0.01 and cultured to mid-exponential growth phase at 250 r·min-1,37 °C as the secondary seed cultures.An appropriate amount of the secondary seed cultures was used to yield an initial OD600of approximately 0.01 in LBG medium(LB medium with 1%glucose)and 3-morpholinopropanesulfonic acid(MOPS)20.9 g·L-1(pH 7.4)at37°C as indicated.When appropriate,IPTG was used at a concentration of 2 mmol·L-1.When necessary,a final concentration of 100 μg · ml-1ampicillin was added.The cultivation of each strain was performed in triplicate.
2.4.Analytical methods
Cell growth was determined by measuring the optical density at 600 nm(OD600)and one unit of absorbance at 600 nm corresponded to 0.38 g cell dry weight(DCW)per liter.For riboflavin measurements,culture samples were diluted with 0.05 mol·L-1NaOH to the linear range of the spectrophoto meter and the A444was immediately measured.Glucose concentration was measured enzymatically using Model-SBA40 Biosensor analyzer(Institute of Microbiology,Shandong,China).The culture samples were centrifuged and the resulting supernatant was used for measurement of the concentration of acetate by using a HPLC system(Agilent,HP1100)equipped with an Aminex HPX-87H anion exchange column(Bio-Rad Laboratories,Richmond,CA,USA)and refractive index detector.The column was run at a constant flow rate of 0.4 ml·min-1,with 0.005 mol·L-1H2SO4as the mobile phase.The results represented the means±S.D.of three independent experiments.
2.5.Reverse transcription(RT)-PCR analysis
Cells were harvested from LBG medium at 37°C during exponential growth phase,when they reached an OD600of~1.Total RNA was extracted from E.coli cells by using RNAprep pure Cell/Bacteria Kit(Tiangen,Beijing,China)following the manufacturer's protocol.Single-stranded cDNA was synthesized using Fast Quant RTKit(Tiangen,Beijing,China)with the total RNA as the templates.Quantity real-time PCR(qPCR)was performed on a Light Cycler 480 II(Roche,Basel,Switzerland)with Real Master Mix(SYBR Green I)(Tiangen,Beijing,China).All primers used for qPCR amplification were designed and shown in Supplementary Table 1.The fold change of each transcript in each sample relative to the control sample was measured in triplicates,normalized to internal control gene rrsA and calculated according to the 2(-Delta Delta C(T))method[25].The relative transcription levels were expressed as means±S.D.
2.6.Intracellular metabolite concentration measurement
To measure intracellular concentrations of metabolites(GTP,GDP,etc.),the preparation of samples were carried out by perchloric acid extraction[26].The resulting samples were immediately analyzed using the procedure of Müller et al.[27].
3.Results and Discussion
3.1.Effect of overexpressing ribB on riboflavin production
In our previous study,plasmid p20C-EC10 was constructed by inserting an artificial rib operon(including ribA,ribB,ribD,ribE and ribC genes)in vectorp20C.Introducing the plasmid into wild-type E.coli MG1655,namely RF01S,resulted in riboflavin accumulation,and a titer of 225.1 mg·L-1riboflavin was produced by the resulting strain E.coli RF01S[21](Table 2).To further improve DHBP availability,we replaced the native promoter of ribB gene(encoding DHBP synthase)with a constitutive promoter tac in E.coli RF01,creating RF07.RT-PCR assays showed that the transcription level of ribB gene was 7-fold higher than that of parent strain RF01,suggesting that ribB was successfully overexpressed.Strain RF07 was transformed with plasmid p20C-EC10,resulting in strain RF07S.Under aerobic conditions,RF07S produced 246.1 mg riboflavin·L-1with a yield of 31.6 mg riboflavin·g-1glucose(Table 2).In B.subtilis,ribA gene encodes a bifunctional enzyme which has both DHBP synthase and GTP cyclohydrolase II activity.A previous report has shown that expression of a truncated B.subtilis ribA gene which only encoded the DHBP synthase in a riboflavinproducing B.subtilis led to lower riboflavin productivity[5].However,this study indicated that overexpressing the DHBP synthase in E.coli caused a slight increase in riboflavin titer and yield.This phenomenon might be due to different hosts and genetic backgrounds.
The biosynthesis of riboflavin requires DHBP and GTP as precursors which are synthesized through two branched pathways,respectively(Fig.1).The formation of DHBP is catalyzed by DHBP synthase.GTP is formed in the cellthrough the de novo purine pathway.Due to the in distinctive effect on riboflavin accumulation by overexpressing ribB gene,it could be deduced that further improving GTP supply might further facilitate riboflavin production.To verify this hypothesis,some key genes in purine biosynthetic pathway were modified to enhance the flux from Ru-5-P to GTP.
3.2.Effect of overexpressing GTP biosynthetic genes on riboflavin production
Firstly,GTP biosynthetic genes(including ndk,gmk)were selected to overexpress.The native promoter of ndk gene was replaced with a synthetic promoter PJ23100 in strains RF07S and RF01S,creating strains RF09S and RF08S.Analogously,gene gmk(encoding guanylate kinase)was overexpressed by promoter replacement in RF09S and RF01S,resulting in strains RF11S and RF10S.Quantitative real-time PCR analysis demonstrated that these modifications indeed enhanced the transcription levels of the ndk and gmk genes.The replacement of the native promoter resulted in the up-regulation of the transcription levels of the ndk and gmk genes 7.2-and 6.5-fold,respectively.
In the shake flask experiments,the riboflavin titer of RF09S was increased to 285.8 mg·L-1,with a 26.7%increase compared with that of RF01S(Table 2).Further,strain RF11S produced a 34.2%higher riboflavin titer(302.3 mg·L-1)and a 19.8%higher riboflavin yield[34.5 mg riboflavin·(g glucose)-1]compared to RF01S(Table 2).
3.3.Effect of purA 419 mutation expression on riboflavin production
In B.subtilis,a purA60 mutation was used to block the flow of IMP to AMP and increase the riboflavin production[4],but these strains required to supply adenosine for fermentation.Thus,we introduced a purA(R419L)mutation[28]instead of knockout to reduce the activity of adenylosuccinate synthase.The purA gene of strain RF11S was modified,and the resulting strain RF13S exhibited 315.7 mg riboflavin·L-1with a yield of 35.9 mg riboflavin·(g glucose)-1(Table 2).The titer and yield increased 40.2%and 24.7%compared to that of RF01S,respectively(Table 2).The purA mutant strain RF13S was defective in adenylosuccinate synthase activity due to a reduced kinetic behavior[27],which led to a decreased flux from IMP to AMP and an enhanced flux into GTP biosynthetic pathway,and subsequently resulted in a higher riboflavin production in strain RF13S.
3.4.Effect of pur F and prsmutation overexpression on riboflavin production
To improve the supply of IMP and relieve feedback regulation of some end-products in purine biosynthetic pathway,overexpression of two mutant enzymes involved in purine biosynthesis was conducted and their effects on riboflavin production were investigated.
PRPP is an important metabolite for purine biosynthesis as it is required in both the de novo and salvage pathways ofGTP[12].Accordingly,we focused on PRPP amidotransferase(encoded by purF gene)and PRPP synthase(encoded by prs gene)and introduced an additional purF expression cassette at mrr locus of RF13S and RF01S chromosome,including a tac-promoter,a proper RBS(Ribosomal Binding Site)followed by a codon mutant purF(K326G,P410W)gene.The resulting strains were designated as RF15S and RF14S.The tac-promoter was used to abolish the inherent regulation of the purF gene.Two codon replacements K326G and P410W were also introduced into the purF to remove the amino group that might be important for GTP binding and attenuate the inhibitory effect of ATP[29].Similarly,we constructed a modified prs expression cassette,which was under the control of tac-promoter and with a D128A codon mutation to desensitize allosteric endproduct inhibition[13].The mutant prs gene was inserted into araD locus of strains RF15S and RF01S,creating strains RF18S and RF16S.Quantitative RT-PCR experiments were performed to detect these two genes transcript abundance.Integrating an additional copy of these two genes further upregulated the transcription levels of the purF and prs genes about 4.7-and 5.8-fold,respectively,implying successful overexpression.
Under our experimental conditions,RF15S produced 340.0 mg riboflavin·L-1,which was 51.0%higher than that of RF01S(Table 2).The riboflavin titer of strain RF18S reached 387.6 mg·L-1,about 14.0%higher than that of RF15S and 72.2%higher than RF01S,respectively(Table 2,Fig.3).The highestyield of riboflavin was 44.8 mg riboflavin·g-1of consumed glucose,increasing 55.8%compared to the control strain RF01S(Table 2).
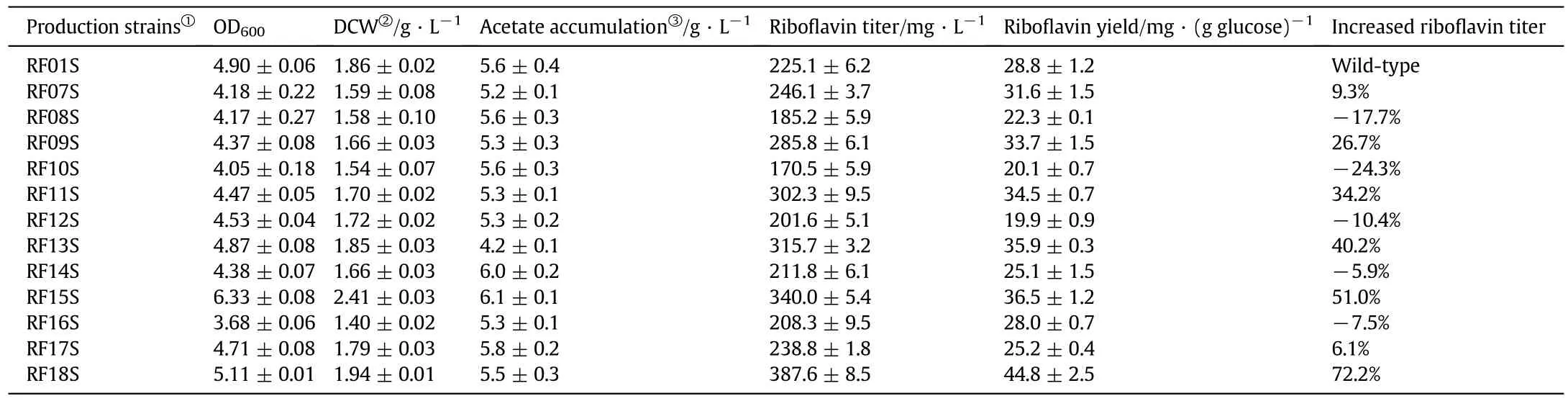
Table 2 Metabolic characterization of the various strains constructed in the study

Fig.3.Time-course profiles of key fermentation parameters:OD600,glucose and riboflavin titer in LBG medium during shake batch cultivations.Open,gray symbols and solid,black symbols represent RF01S and RF18S,respectively.Triangle,OD600;circle,riboflavin titer;square,glucose.The values shown represent the means of three independent experiments and the error bars represent standard deviations of three values.
Being consistent with previous research,our result displayed a significant increase in riboflavin production by overexpression of mutant purF and prs genes.For instance,overexpression of mutant PRPP synthase in A.gossypii greatly enhanced the riboflavin production[12].Co-overexpression of prs and purF gene in B.amyloliquefaciens led to a 14.4%improvement of guanosine production[14].In E.coli,coamplification of desensitized-purF and desensitized-prs increased the usage and supply of PRPP,thereby greatly elevating the production of inosine[13].
However,modulating or co-modulating the five purine pathway genes without overexpressing the ribB(strains RF17S and RF18S)had little or negative effect on riboflavin production(Table 2).In contrast,as much as 72.2%increase of the riboflavin production could be obtained when ribB gene and purine pathway genes were simultaneously modified.These results suggested that coordination of gene modifications between DHBP and GTP synthesis pathways by rational metabolic engineering can efficiently boost riboflavin production in E.coli.
3.5.Intracellular purine nucleotides and acetate concentrations
Some of the related intracellular purine nucleotides,such as GTP,GMP,IMP,AMP,and acetate concentrations were also measured.As shown in Table 3,compared to RF07S,GMP had a lower level as expected,but GTP also had a lower level in RF11S.A possible explanation of GTP decrease is that more GTP entered into riboflavin biosynthetic pathway for higher riboflavin production.Due to modulation of purA gene,AMP concentration was about 8-fold lower and IMP concentration was 16%higher in RF13S than those of RF11S,which implied that the flux from IMP to AMP was greatly reduced and more flux was redirected into GTP biosynthetic pathway.Moreover,overexpression of mutant purF and prs genes further elevated IMP concentration,but lowered the GTP concentration unexpectedly.We speculated that the GTP concentration was also affected by other reactions involving the conversion between GTP and GDP/GMP,or other pathways consuming GTP such as folate biosynthesis pathway.
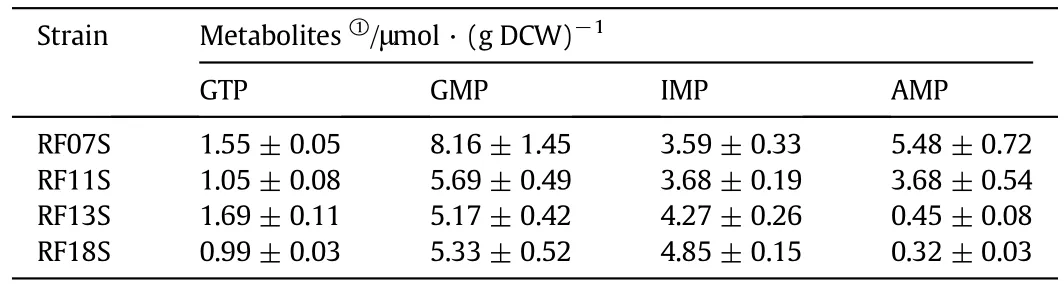
Table 3 Intracellular metabolite concentration in cell extracts of E.coli
Stepwise improvement on riboflavin production led to a strain producing 387.6 mg riboflavin·L-1in shake- flask cultures.The riboflavin production of constructed strain RF18S was 1.8-fold higher as compared to that of recombinant strains of B.subtilis in flask batch culture,387.6 mg riboflavin·L-1versus220 mg riboflavin·L-1[9]and higher than that of A.gossypii in flask batch culture was 228 mg riboflavin·L-1[12],though still lower than that of C.famata[7].As most recombinant E.coli,a major problem existed with aerobic cultivation on glucose at high growth rates—formation and accumulation of considerable amounts of acetate i.e.over flow metabolism,all riboflavin producing strains excreted amounts of acetate(4.2-6.1 g·L-1)as a major byproduct,which led to growth retardation(Table 2).Thus,reducing over flow metabolism by slowing the glucose uptake rate and reusing the acetate would be a potential strategy to further improve the cell growth,riboflavin production and yield.
4.Conclusions
In this work,we overexpressed ribB and engineered the genes of purine biosynthetic pathway in a riboflavin-producing E.coli strain.Riboflavin titer and yield were enhanced gradually during this gene manipulation.In shake- flask fermentations,the final strain RF18S accumulated 387.6 mg riboflavin·L-1with a yield on glucose substrate of 44.8 mg riboflavin·g-1,as much as a 72.2%increase of in riboflavin titer and a 55.6%increase in riboflavin yield compared to the wildtype RF01S.This demonstrated that simultaneously engineering the DHBP synthase and GTP biosynthetic pathway by rational metabolic engineering can significantly facilitate riboflavin production in E.coli.Further manipulations might be performed to modulate central carbon metabolism,reduce the over flow metabolism(acetate)and improve Ru-5-P supply for higher riboflavin production.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.cjche.2015.08.013.
Acknowledgments
The authors thank Dr.Thomas E.Kuhlman for kindly providing requested plasmids.This work was supported by National High-tech R&D Program of China[2012AA02A702,2012AA022103].
 Chinese Journal of Chemical Engineering2015年11期
Chinese Journal of Chemical Engineering2015年11期
- Chinese Journal of Chemical Engineering的其它文章
- N-methyl-2-(2-nitrobenzylidene)hydrazine carbothioamide—A new corrosion inhibitor for mild steel in 1 mol·L-1 hydrochloric acid
- A dual-scale turbulence model for gas-liquid bubbly flows☆
- Gas-liquid hydrodynamics in a vessel stirred by dual dislocated-blade Rushton impellers☆
- Convective mass transfer enhancement in a membrane channel by delta winglets and their comparison with rectangular winglets☆
- Cobalt-free gadolinium-doped perovskite Gd x Ba1-x FeO3-δas high-performance materials for oxygen separation☆
- Synthesis and adsorption property of zeolite FAU/LTA from lithium slag with utilization of mother liquid☆
