N6-腺苷甲基化修饰及其对LINE-1的调控机制
张傲,岑山,李晓宇
综 述
N-腺苷甲基化修饰及其对LINE-1的调控机制
张傲,岑山,李晓宇
中国医学科学院&北京协和医学院,医药生物技术研究所免疫生物学室,北京 100050
长散布元件-1 (long interspersed elements-1,LINE-1)是现今在人类基因组中唯一具有自主转座能力的转座子,其转座会引起细胞基因组结构和功能的改变,是导致多种严重疾病的重要因素。在转座过程中,LINE-1 mRNA是转座中间体的核心,宿主细胞对其进行相关修饰直接影响转座。N-腺苷甲基化修饰(m6A)是真核细胞RNA上最丰富且动态可逆的表观遗传修饰。目前发现m6A修饰也存在于LINE-1 mRNA上,参与LINE-1整个生命周期的调控,影响其转座和基因组中LINE-1相邻基因的表达,进而影响基因组稳定性、细胞自我更新与分化潜能,在人类发育和疾病中具有重要作用。本文介绍了LINE-1 m6A修饰的位置、功能以及相关机制,并总结了LINE-1的m6A修饰对其转座调控的研究进展,以期为相关疾病发生发展的机制研究和治疗提供新的思路。
m6A修饰;逆转录转座子;LINE-1;基因组;基因组稳定性
长散布元件(long interspersed elements,LINE-1)是一种非长末端重复序列(non-long terminal repeats,non-LTR)逆转录转座子。据统计大约45%的人类基因组衍生自转座子(transposable elements,TEs),其中LINE-1约占人基因组的17%,是目前人类基因组中唯一证实具有自主转座活性的转座子[1,2]。LINE-1以RNA为媒介进行转座,是一种RNA转座子[3],全长约6 kb,其编码的两个蛋白ORF1蛋白(ORF1p)和ORF2蛋白(ORF2p),在细胞质中与LINE-1 mRNA形成核糖核蛋白复合物(ribonucleoprotein complexes,RNPs),后利用ORF2p核酸内切酶及逆转录酶活性,以LINE-1 mRNA为模板逆转录产生cDNA,形成RNA:DNA杂交体,该过程被称为“靶点引导逆转录过程”(target-site primed reverse transcription,TPRT)[4,5],是LINE-1复制的关键步骤。基因组中大多数LINE-1 5′ UTR区缺失或倒置,丧失转座活性,仅80~100个LINE-1拷贝结构完整,是具有逆转座活性的LINE-1 (retrotransposition-competent LINE-1s,RC-L1s)。从物种进化上来看,活跃的逆转录转座子在生物进化、物种形成和胚胎发育、记忆形成等方面发挥生理学作用[6,7],但对个体而言,转座的发生会对宿主细胞基因组的结构和功能产生严重影响,LINE-1在基因组DNA中的插入、缺失和重组,会改变宿主基因的表达,导致衰老、癌症、基因疾病、代谢性疾病、神经退行性疾病和自身免疫性疾病等多种疾病的发生[8~11]。此外,LINE-1还可以协助不具有自主转座能力的非LTR转座子短散布元件(short interspersed elements,SINEs) Alu和加工后的假基因进行转座,进而诱发疾病[12]。因此宿主对正常体细胞中LINE-1的表达与转座活性是严格控制的,而且这种调控是多层次、多方面的,包括表观遗传修饰[13,14]、非编码小RNA[15,16]以及多种宿主限制因子[17~19]等调控。
除研究较多的DNA、组蛋白甲基化外,N-甲基化腺嘌呤(N-methylated adenine,m6A)陆续在细菌DNA、细菌和酵母的RNA和哺乳动物mRNA中被发现,m6A甲基化对RNA代谢和功能调控具有多样性[20~23]。随着对LINE-1转座调控机制的深入研究,研究者们发现在LINE-1上存在的m6A修饰对其转座调控也发挥着重要的作用。本文主要介绍m6A修饰的生物学功能,以及该修饰对LINE-1各阶段的调控机制和LINE-1周围染色质状态、基因表达的影响,以期对m6A修饰的生物学功能研究扩展及宿主对LINE-1调控网络的探究提供新的思路。
1 m6A修饰的生物学功能
m6A一般发生在RNA中腺苷酸的N位置上,是通过特定的甲基转移酶进行的甲基化修饰(图1),在mRNA和其他类型核内RNA,如转运RNA (transfer RNA,tRNA)、核糖体RNA(ribosomal RNA,rRNA)、小核RNA(small nuclear RNA,snRNA)均有分布。m6A甲基化具有RRACH共同识别序列(其中R表示A或G,H表示A、C或U),受多种调控因子调控,通过“编码器”m6A甲基转移酶装配,并可被“读码器”m6A结合蛋白识别或被“消码器”去甲基化酶移除[24,25]。m6A主要在终止密码子和3′非翻译区(3′UTR)附近富集,在内含子和5′非翻译区(5′UTR)也有低丰度的m6A。
m6A甲基化对RNA代谢过程的多个环节均具有重要的影响,包括RNA剪接[26,27]、核输出[28]、降解[29,30]和翻译[31,32]。在RNA剪接过程中,m6A被相应蛋白识别并结合,通过招募YTHDC1蛋白、抑制剪接因子或改变RNA局部构象来调节mRNA前体(pre-mRNA)选择性剪接[26,27]。在核输出过程中,m6A被YTHDC1蛋白识别,促进RNA与核输出组分的相互作用,调控mRNA的亚细胞定位[28]。m6A还可以被YTHDF2蛋白识别进而降解m6A修饰的靶转录本[29,30]。此外,m6A对mRNA转录后调控也具有一定的作用,mRNA的翻译方式与m6A在转录本的位置有关。正常生理条件下,m6A修饰主要位于RNA 的3′UTR区,被YTHDF1蛋白或YTHDF3蛋白识别,招募真核细胞翻译起始因子eIF3,促进帽依赖性翻译[31,32]。而在应激条件下,5′UTR区的m6A作为m6A诱导的核糖体进入位点(m6A-induced ribosome engagement site,MIRES),促进mRNA进行帽非依赖性翻译,这种m6A介导的帽非依赖性翻译同样需要m6A“读码器”eIF3的识别[33]。另有研究表明,mRNA上的m6A可影响转录本与tRNAs的相互作用进而抑制翻译[34]。
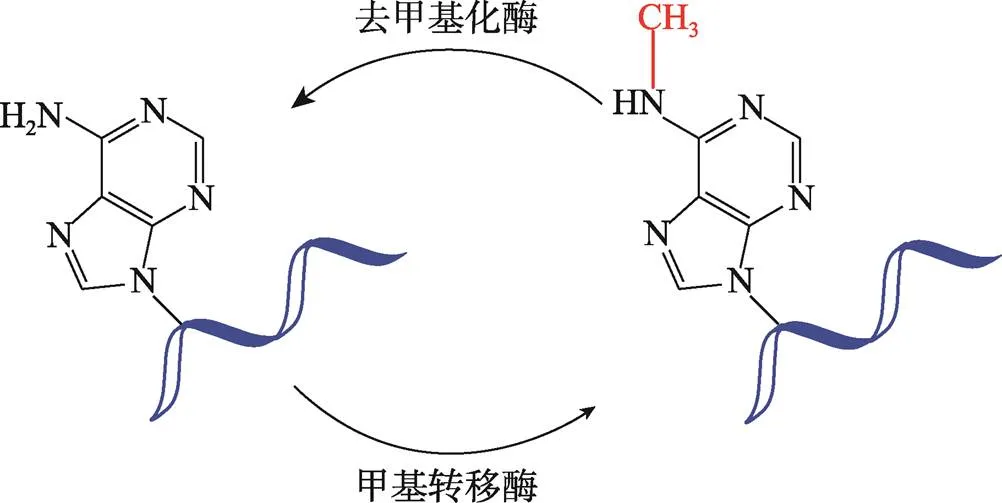
图1 腺苷酸甲基化修饰结构示意图
RNA腺苷酸N位置的甲基化修饰通过m6A甲基转移酶装配,被m6A结合蛋白识别或被去甲基化酶移除。
m6A还参与哺乳动物多种病理生理学过程,包括胚胎发育[35]、神经发生[36]、昼夜节律[37]、应激反应[38]、肿瘤发生[24,39]和病毒感染[40]等。随着m6A甲基化组学分析的发展,m6A在肿瘤发生发展的相关机制研究取得了主要突破。RNA m6A甲基化水平改变影响细胞的增殖、分化与自我更新[41]。m6A是肿瘤代谢的重要调节因子,肿瘤代谢应激反应可导致异常的m6A甲基化,调节代谢重组相关的信号通路、转录因子和代谢酶[42]。目前已有多项研究发现,m6A修饰异常与多种癌症的发生发展相关,不同底物的m6A修饰会促进或抑制肿瘤的发展,具有促癌和抑癌的双重作用,是一把双刃剑[24]。机体对LINE-1的调控影响基因组稳定性,含有m6A修饰的LINE-1 mRNA具有宿主逃逸机制,被正向选择并表达,从而促进疾病的发生发展。研究发现,是口腔鳞状细胞癌(oral squamous cell carcinoma,OSCC)的原癌基因,其编码的HNRNPA2B1蛋白可能作为m6A“读码器”促进LINE-1 mRNA翻译,进而通过LINE-1/TGF-β1/Smad2/Slug信号通路靶向上皮细胞-间充质转化(epithelial-mesenchymal transition,EMT),促进肿瘤细胞增殖、迁移和侵袭[43]。这提示m6A修饰的LINE-1可能与多种癌症致病机制均相关,为癌症预防及治疗提供了新的思路。根据m6A修饰的不同位置,将其分为4个部分:具有转座活性的LINE-1 5′ UTR、位于宿主基因内含子区域和形成R-环的LINE-1的m6A修饰,以及LINE-1 DNA的6mA修饰(图2A)。近年来,研究人员发现m6A对LINE-1的整个复制周期均有调控作用,“编码器”对各阶段LINE-1进行m6A修饰,该修饰被“读码器”识别,或被“消码器”移除,影响LINE-1的转座活性及转录与翻译水平(图2B)。
2 m6A修饰在LINE-1复制周期不同阶段的调控机制
2.1 具有转座活性的LINE-1 5′ UTR的m6A修饰
最新的一项研究表明,LINE-1转录本是人类细胞中主要的m6A修饰RNA。与DNA和一些组蛋白甲基化的抑制作用相反,如组蛋白H3K9me3,RC-L1s的RNA m6A修饰可促进其表达与转座。m6A偏向于修饰年轻的LINE-1,这些LINE-1结构完整,并具有丰富的RRACH序列。除了帽依赖性翻译外,m6A还启动LINE-1 RNA帽非依赖性翻译。该研究发现,在LINE-1 5′ UTR第333位发生m6A获得性突变后,形成m6A共识别序列,使得第332位腺苷上发生m6A修饰,eIF3识别该修饰位点后,提高ORF1的翻译速率,刺激ORF2p合成,产生具有逆转录活性的LINE-1 核糖核蛋白RNP,促进LINE-1逆转录转座[44,45]。LINE-1的5′ UTR m6A修饰是其产生逆转录转座功能所必需的,只有具有完整5′ UTR,m6A修饰相关酶才可调控LINE-1的表达[46]。此外,m6A修饰可以改变RNA-蛋白相互作用或RNA二级结构,这可能影响LINE-1 ORF2p的酶活性[45]。目前已发现,m6A甲基化酶METTL3使LINE-1 m6A水平升高,促进其逆转座[45]。相反,m6A去甲基化酶ALKBH5使LINE-1 m6A水平降低,抑制其逆转座[45]。m6A甲基化酶METTL14和ZC3H13或其识别蛋白YTHDC1缺失将降低宿主中m6A标记的年轻LINE-1的水平[46]。虽然m6A修饰提高LINE-1 RNA的翻译效率,但不改变LINE-1 RNA在细胞内定位[45]。此外,m6A仅对年轻LINE-1的表达和逆转座活性有促进作用,在较老或低甲基化的LINE-1中有抑制作用,当m6A识别蛋白缺陷时,古老的LINE-1转座活性反而增加[46]。
2.2 基因内含子中无转座活性的 LINE-1 的m6A修饰
基因组中多数LINE-1 5′ UTR区域缺失或突变,失去逆转座活性。研究发现,基因内含子中经m6A修饰后的无逆转座活性的LINE-1 (m6A-marked intronic LINE-1s,MILs)是一种新的调控元件,优先驻留在长基因中,作为转录“障碍”阻碍宿主基因的表达,但具体机制尚不清楚[46]。这些长基因在DNA损伤修复(DNA damage repair,DDR)等生理过程中发挥关键作用。研究发现,m6A识别蛋白SAFB/SAFB2复合体以m6A增强的方式结合RC-L1s和MILs RNA来抑制其表达[46]。此外,SAFB/SAFB2还可纠正MILs对重要宿主基因的转录阻断作用,以保护宿主基因的转录,但SAFB并不与m6A发生特异性结合,可能通过m6A改变局部RNA结构以实现RNA-RBP (RNA结合蛋白)相互作用(即“m6A开关”),形成的LINE-1 RNA高级结构允许更强的L1-SAFB结合[46]。MILs通过影响长基因转录,使m6A调节的L1-宿主相互作用在基因调控、基因组完整性、人类发育和疾病中发挥广泛作用[46,47]。
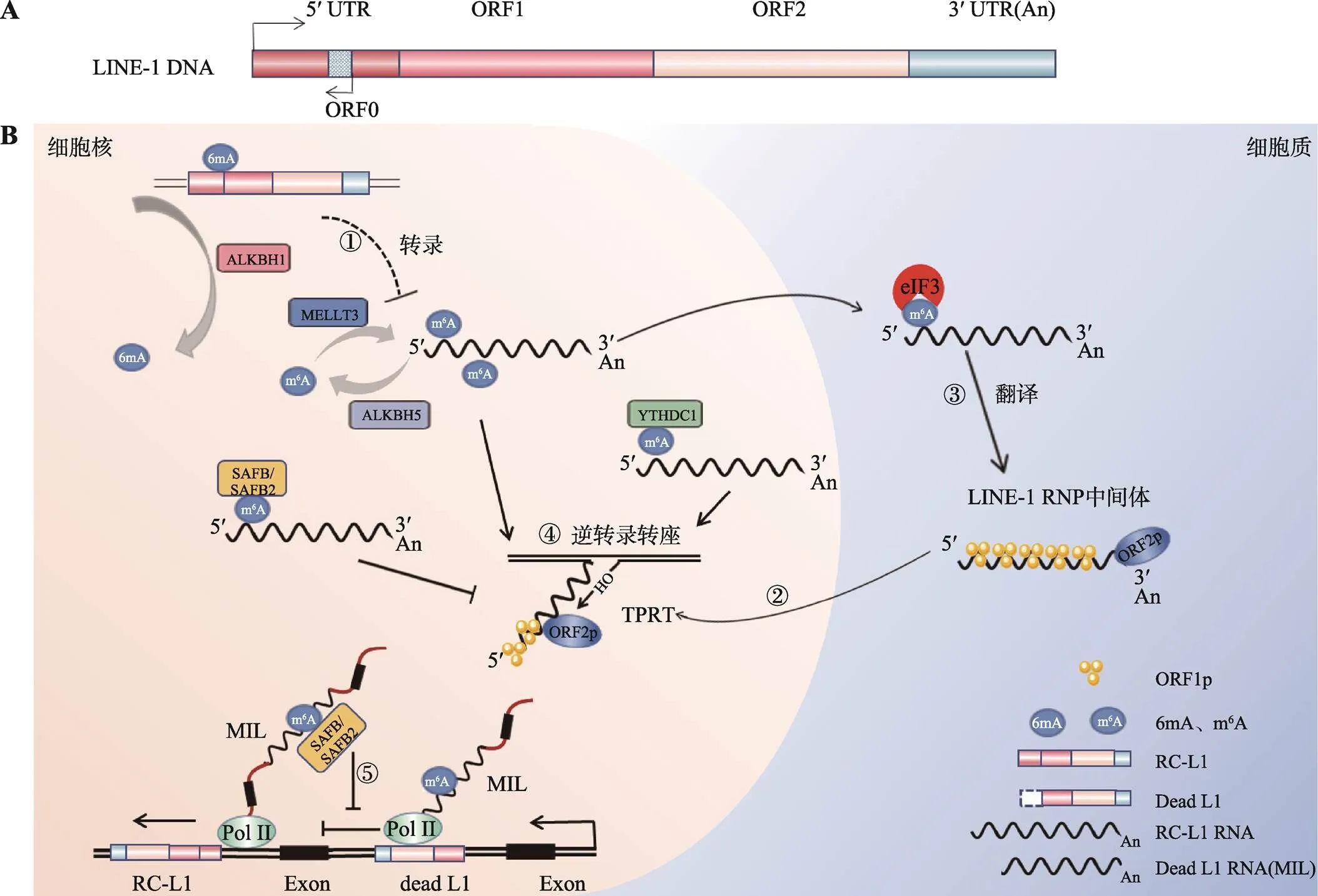
图2 m6A修饰对LINE-1的影响
A:LINE-1的结构。LINE-1由开放阅读框ORF0、ORF1、ORF2和非编码区5′UTR、3′UTR构成,5′UTR 有两个启动子,是双向的:正义启动子活性可转录 ORF1、ORF2,反义启动子(ASP)能够启动与LINE-1方向相反的转录物转录。B:m6A修饰酶影响LINE-1复制周期模式图。①LINE-1 DNA可能富集6mA甲基化修饰,抑制mRNA转录;②LINE-1 mRNA与ORF1p、ORF2p结合生成LINE-1 RNP复合物,入核后进行“TPRT”生成cDNA,插入宿主基因组;③在细胞质中,翻译起始因子eIF3与m6A特异性相互作用,提高翻译水平;④METTL3、YTHDC1促进LINE-1逆转座,ALKBH5、SAFB/SAFB2抑制LINE-1逆转座;⑤SAFB/SAFB2可纠正MILs对重要宿主基因的转录阻断。
2.3 LINE-1 RNA:DNA杂交分子(R-环)的m6A修饰
R-环普遍存在于高转录基因中,并在重复序列中积累,其中包括逆转录转座子LINE-1[48]。LINE-1逆转录转座过程中,RNP剪切基因组DNA双链,形成由LINE-1 RNA:DNA杂交分子和未配对单链DNA组成的R-环,R环在细胞分裂S期达到顶峰,参与了从转录调控到DNA修复等诸多重要生物学过程[49]。Abakir等[50]发现,在人多能性干细胞(human pluripotent stem cells,hPSCs)RNA:DNA杂交体中有大量m6A修饰,m6A修饰存在于RNA:DNA杂交体的RNA链上,含有m6A的R环在细胞周期G2/M期积累,在G0/G1期耗尽。在正常生理条件下,R-环在基因启动子区和终止区富集,参与mRNA转录起始和终止,调控基因表达。当R-环没有被正常分解时,其积累会导致DNA损伤和/或复制叉停滞,破坏基因组的稳定性[51~53]。m6A修饰可调控R环的积累,不同的m6A结合蛋白识别R环,维持基因组的稳定性。目前已发现甲基转移酶METTL3、识别蛋白HNRNPA2B1、促进mRNA翻译的YTHDF1以及促进mRNA降解的YTHDF2均与富集R环的位点相互作用[50]。已有研究表明,YTHDF2可阻止含有m6A的LINE-1 RNA:DNA杂交体积累,有助于修复哺乳动物中R-环依赖性DNA损伤,维护基因组稳定性[50]。
2.4 LINE-1 DNA的6mA修饰
DNAN-甲基化腺嘌呤(6mA)修饰在原核生物中广泛分布,而在哺乳动物细胞中丰度极低[54,55]。早期研究人员利用SMRT-ChIP在小鼠胚胎干细胞(mouse embryonic stem cells,mESCs)中发现6mA修饰,证明6mA修饰与LINE-1转座子的进化年龄呈负相关,在年轻、完整的LINE-1元件中强烈富集。与LINE-1 RNA的m6A甲基化沉积位置相似,6mA大多数富集在年轻全长LINE-1的5′ UTR和ORF1上。在6mA去甲基化酶ALKBH1缺陷的细胞中,DNA 6mA水平增加导致转录沉默。6mA修饰与LINE-1转座子及其邻近增强子和基因的表观遗传沉默相关,在胚胎干细胞分化过程中抵抗基因激活信号[56]。与其他常染色体相比,较年轻的全长LINE-1在X染色体上强烈富集,经6mA修饰后沉默位于X染色体上的基因[56,57]。不同于6mA在其他生物基因中的激活作用,它在哺乳动物进化中表现出表观遗传沉默的新作用。然而,该研究结果存在很大争议,其他研究者对真核生物中DNA 6mA的存在表示怀疑,认为已有方法受污染源的影响容易产生假阳性结果。故作者使用6mASCOPE方法对6mA定量去卷积,结果排除非特异性偏倚后,不支持HEK293中年轻LINE-1具有6mA富集特点[54]。但这项研究仍存在局限性,需要进一步优化检测方法。
3 LINE-1的m6A修饰对染色质状态和基因表达的调控
LINE-1上修饰的m6A不仅调控其自身的复制过程,对其相邻基因的表观遗传调控、塑造基因组结构和维持基因组稳定性方面也具有广泛的作用。染色体相关调控RNA (chromat-in-associated regulatory RNAs,carRNAs)上的m6A修饰可以全局调控染色质状态和转录,依赖于METTL3甲基化的carRNAs包括启动子相关RNA、增强子RNA和重复序列RNA(如LINE-1)。carRNAs m6A修饰可以维持基因间区域染色质浓缩,而YTHDC1识别m6A后,通过核外泌体靶向(nuclear exosome targeting,NEXT)复合物促进carRNAs降解。m6A甲基化缺失导致染色质开放和转录本富集,这与活性组蛋白H3K4me3和H3K27ac修饰增加相关,后续招募表观遗传因子如组蛋白乙酰转移酶(EP-300)来维持开放的染色质构象和下游转录。此外,carRNAs中“重复序列RNA”在m6A高甲基化和转录下调之间表现出强相关性,其中LINE-1受影响最大,影响细胞自我更新和分化潜能[58~60]。
另有研究发现,识别蛋白YTHDC1通过多种机制参与逆转录转座子的调控和染色质修饰。在mESCs中,YTHDC1与m6A修饰的LINE-1转录本结合,募集组蛋白甲基转移酶SETDB1、TRIM28和核仁素(nucleolin,NCL),共同形成沉默复合物,促进H3K9me3的富集,沉默逆转录转座子[59,60]。此外,YTHDC1识别细胞核中LINE-1 RNA上的m6A,招募转录调控因子KAP1,并调控LINE1-NCL复合物的形成和KAP1在染色质上的募集,形成LINE1-NCL-KAP1复合物,抑制2细胞期(two-cell stage,2C)胚胎特异性转录的主要激活因子Dux,关闭2C基因表达程序。同时,LINE1-NCL-KAP1复合物可与核糖体DNA(rDNA)结合,促进rRNA合成和mESCs自我更新[59,61]。KAP1在LINE-1上的富集同样也促进H3K9me3沉积,导致在mESCs和内细胞团(ICM)细胞中组蛋白修饰位点的转录沉默,降低染色质开放状态,有助于识别mESCs并促进胚胎发育,调节mESCs从2C样状态退出[62]。另一项研究发现,在mESCs中发现肥胖蛋白FTO是LINE-1 m6A去甲基化酶,促进LINE-1相邻基因位点的染色质开放。FTO与LINE-1 RNA和LINE-1 RNA-DNA相互作用的消失导致染色质浓缩、抑制性组蛋白标记富集,顺式调控相邻基因,降低相邻基因表达。有趣的是,与YTHDC1作用相反,FTO敲除后,LINE-1 RNA反式调节不相邻的2C基因,使2C基因去抑制,导致类似2C状态发生和mESCs状态丢失,使得多功能性基因的表达减少,细胞分化和自我更新受损,因此FTO-LINE-1轴对于胚胎发育是必不可少的[63,64]。
4 结语与展望
m6A修饰对LINE-1的调控机制目前正在深入研究中,一些问题仍需要进一步探究阐述,如LINE-1 RNA上的m6A被YTHDC1识别后促进抑制性组蛋白富集,抑制基因表达。但另有研究发现,m6A“读码器”YTHDC1协同转录使组蛋白H3K9me2去甲基化,促进基因表达[65]。多种表观遗传信号共同调节基因的表达,故仍需进一步探究LINE-1不同表观转录组修饰间的相互影响,以及与染色质修饰的相互作用关系。此外,LINE-1 DNA 6mA是否具有显著性富集特点也有待进一步探讨。若LINE-1 DNA 上6mA修饰富集且抑制其活性,而LINE-1 RNA m6A修饰促进其转座,那么两者是否在发育或疾病中相互干扰,以及如何介导LINE-1活性或宿主基因表达,是未来研究的重要内容。此外,LINE-1 m6A修饰调控组蛋白修饰,阻止染色质开放状态及相邻基因的表达。但由不同m6A相关酶介导调控的2C基因表达作用相反,出现这种差异是由于m6A调控相关蛋白具有特异性还是其他调控系统参与其中仍不可知。另外,值得注意的是,m6A对不同的逆转录转座子家族具有截然相反的影响:YTHDC1识别某些TEs上m6A修饰后破坏其稳定性,如IAPs[60];m6A通过招募YTHDF家族缩短IAP mRNA半衰期[66]。这表明在TEs可能发生了额外的依赖于m6A的调控,如依赖于其他m6A甲基转移酶(METTL5、METTL16和ZCCHC4)或识别结合蛋白的活性,这些蛋白可以通过翻译后修饰或与其他分子相互作用进行调控[60]。随着高通量测序等新技术的发展,研究人员对m6A的研究有望发现新的生物调节系统,LINE-1的m6A修饰也有望成为未来疾病治疗与诊断的新靶点。
在肿瘤疾病研究方面,LINE-1可作为诊断癌症的生物标志物和潜在的治疗靶点[67]。其中,LINE-1 DNA或组蛋白的大量低甲基化,被认为是大多数恶性转化的标志,是一种很有前途的癌症发展的候选生物标志物[8]。而LINE-1虽通常被认为具有促癌功能,但在急性髓系粒细胞白血病(AML)中发挥抑癌作用[68]。这是宿主不同调控机制的作用结果。LINE-1 m6A甲基化修饰研究的突破性进展或许将有助于解开LINE-1相关疾病研究的许多未解之谜。
[1] Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann Y, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Raymond C, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blöcker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowki J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ, Szustakowki J, International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome, 2001, 409(6822): 860–921.
[2] Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and in health, 2008, 18(3): 343–358.
[3] Goodier JL, Kazazian HH. Retrotransposons revisited: the restraint and rehabilitation of parasites, 2008, 135(1): 23–35.
[4] Babushok DV, Kazazian HH. Progress in understanding the biology of the human mutagen LINE-1, 2007, 28(6): 527–539.
[5] Zhang X, Zhang R, Yu JP. New understanding of the relevant role of LINE-1 retrotransposition in human disease and immune modulation, 2020, 8: 657.
[6] Jachowicz JW, Bing XY, Pontabry J, Bošković A, Rando OJ, Torres-Padilla ME. LINE-1 activation after fertilization regulates global chromatin accessibility in the early mouse embryo, 2017, 49(10): 1502– 1510.
[7] Mao Y, Li XY. Advances in the study of LINE-1 retrotransposition in nervous system, 2019, 16(5): 27–29, 46.毛洋, 李晓宇. 神经系统中LINE-1转座的研究进展. 中国医药导报, 2019, 16(5): 27–29, 46.
[8] Ponomaryova AA, Rykova EY, Gervas PA, Cherdyntseva NV, Mamedov IZ, Azhikina TL. Aberrant methylation of LINE-1 transposable elements: a search for cancer biomarkers, 2020, 9(9): 2017.
[9] Burns KH. Our conflict with transposable elements and its implications for human disease, 2020, 15: 51–70.
[10] Gorbunova V, Seluanov A, Mita P, McKerrow W, Fenyö D, Boeke JD, Linker SB, Gage FH, Kreiling JA, Petrashen AP, Woodham TA, Taylor JR, Helfand SL, Sedivy JM. The role of retrotransposable elements in ageing and age- associated diseases, 2021, 596(7870): 43–53.
[11] Liu Q, Wang JH, Li XY, Cen S. The connection between LINE-1 retrotransposition and human tumorigenesis, 2016, 38(2): 93–102.刘茜, 王瑾晖, 李晓宇, 岑山. 逆转录转座子LINE-1与肿瘤的发生和发展. 遗传, 2016, 38(2): 93–102.
[12] Ostertag EM, Goodier JL, Zhang Y, Kazazian HH. SVA elements are nonautonomous retrotransposons that cause disease in humans, 2003, 73(6): 1444– 1451.
[13] Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond, 2012, 13(7): 484–492.
[14] Fukuda K, Shinkai Y. SETDB1-mediated silencing of retroelements, 2020, 12(6): 596.
[15] Hamdorf M, Idica A, Zisoulis DG, Gamelin L, Martin C, Sanders KJ, Pedersen IM. miR-128 represses L1 retrotransposition by binding directly to L1 RNA, 2015, 22(10): 824–831.
[16] De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O’Carroll D. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements, 2011, 480(7376): 259–263.
[17] Choi J, Hwang SY, Ahn K. Interplay between RNASEH2 and MOV10 controls LINE-1 retrotransposition, 2018, 46(4): 1912–1926.
[18] Goodier JL. Restricting retrotransposons: a review, 2016, 7: 16.
[19] Hu SQ, Li J, Xu FW, Mei S, Le Duff Y, Yin LJ, Pang XJ, Cen S, Jin Q, Liang C, Guo F. SAMHD1 inhibits LINE-1 retrotransposition by promoting stress granule formation, 2015, 11(7): e1005367.
[20] Dunn DB, Smith JD. Occurrence of a new base in the deoxyribonucleic acid of a strain of, 1955, 175(4451): 336–337.
[21] Littlefield JW, Dunn DB. Natural occurrence of thymine and three methylated adenine bases in several ribonucleic acids, 1958, 181(4604): 254–255.
[22] Adler M, Weissmann B, Gutman AB. Occurrence of methylated purine bases in yeast ribonucleic acid, 1958, 230(2): 717–723.
[23] Desrosiers R, Friderici K, Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells, 1974, 71(10): 3971–3975.
[24] Sun T, Wu RY, Ming L. The role of m6A RNA methylation in cancer, 2019, 112: 108613.
[25] Shi HL, Wei JB, He C. Where, when, and how: context- dependent functions of RNA methylation writers, readers, and erasers, 2019, 74(4): 640–650.
[26] Xiao W, Adhikari S, Dahal U, Chen YS, Hao YJ, Sun BF, Sun HY, Li A, Ping XL, Lai WY, Wang X, Ma HL, Huang CM, Yang Y, Huang N, Jiang GB, Wang HL, Zhou Q, Wang XJ, Zhao YL, Yang YG. Nuclear m6A reader YTHDC1 regulates mRNA splicing, 2016, 61(4): 507–519.
[27] Bartosovic M, Molares HC, Gregorova P, Hrossova D, Kudla G, Vanacova S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3′-end processing, 2017, 45(19): 11356–11370.
[28] Roundtree IA, Luo GZ, Zhang ZJ, Wang X, Zhou T, Cui YQ, Sha JH, Huang XX, Guerrero L, Xie P, He E, Shen B, He C. YTHDC1 mediates nuclear export ofN- methyladenosine methylated mRNAs, 2017, 6: e31311.
[29] Wang X, Lu ZK, Gomez A, Hon GC, Yue YN, Han DL, Fu Y, Parisien M, Dai Q, Jia GF, Ren B, Pan T, He C.N-methyladenosine-dependent regulation of messenger RNA stability, 2014, 505(7481): 117–120.
[30] Yang Y, Hsu PJ, Chen YS, Yang YG. Dynamic transcriptomic m6A decoration: writers, erasers, readers and functions in RNA metabolism, 2018, 28(6): 616–624.
[31] Wang X, Zhao BS, Roundtree IA, Lu ZK, Han DL, Ma HH, Weng XC, Chen K, Shi HL, He C.N-methyladenosine modulates messenger RNA translation efficiency, 2015, 161(6): 1388–1399.
[32] Shi HL, Wang X, Lu ZK, Zhao BS, Ma HH, Hsu PJ, Liu C, He C. YTHDF3 facilitates translation and decay ofN-methyladenosine-modified RNA, 2017, 27(3): 315–328.
[33] Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 5′ UTR m6A promotes cap-independent translation, 2015, 163(4): 999–1010.
[34] Boulias K, Greer EL. Biological roles of adenine methylation in RNA, 2023, 24(3): 143– 160.
[35] McGraw S, Vigneault C, Sirard MA. Temporal expression of factors involved in chromatin remodeling and in gene regulation during early bovineembryo development, 2007, 133(3): 597–608.
[36] Deng JH, Chen XH, Chen AD, Zheng XC. m6A RNA methylation in brain injury and neurodegenerative disease, 2022, 13: 995747.
[37] Xu ZJ, Lv BB, Qin Y, Zhang B. Emerging roles and mechanism of m6A methylation in cardiometabolic diseases, 2022, 11(7): 1101.
[38] Wilkinson E, Cui YH, He YY. Context-dependent roles of RNA modifications in stress responses and diseases, 2021, 22(4): 1949.
[39] Deng LJ, Deng WQ, Fan SR, Chen MF, Qi M, Lyu WY, Qi Q, Tiwari AK, Chen JX, Zhang DM, Chen ZS. m6A modification: recent advances, anticancer targeted drug discovery and beyond, 2022, 21(1): 52.
[40] Loh D, Reiter RJ. Melatonin: regulation of viral phase separation and epitranscriptomics in post-acute sequelae of COVID-19, 2022, 23(15): 8122.
[41] Pan YT, Ma P, Liu Y, Li W, Shu YQ. Multiple functions of m6A RNA methylation in cancer, 2018, 11(1): 48.
[42] An YY, Duan H. The role of m6A RNA methylation in cancer metabolism, 2022, 21(1): 14.
[43] Zhu FY, Yang TR, Yao MF, Shen T, Fang CY. HNRNPA2B1, as a m6A reader, promotes tumorigenesis and metastasis of oral squamous cell carcinoma, 2021, 11: 716921.
[44] Dmitriev SE, Andreev DE, Terenin IM, Olovnikov IA, Prassolov VS, Merrick WC, Shatsky IN. Efficient translation initiation directed by the 900-nucleotide-long and GC-rich 5′ untranslated region of the human retrotransposon LINE-1 mRNA is strictly cap dependent rather than internal ribosome entry site mediated, 2007, 27(13): 4685–4697.
[45] Hwang SY, Jung H, Mun S, Lee S, Park K, Baek SC, Moon HC, Kim H, Kim B, Choi Y, Go YH, Tang WXF, Choi J, Choi JK, Cha HJ, Park HY, Liang P, Kim VN, Han K, Ahn K. L1 retrotransposons exploit RNA m6A modification as an evolutionary driving force, 2021, 12(1): 880.
[46] Xiong F, Wang RY, Lee JH, Li SL, Chen SF, Liao ZA, Hasani LA, Nguyen PT, Zhu XY, Krakowiak J, Lee DF, Han L, Tsai KL, Liu Y, Li WB. RNA m6A modification orchestrates a LINE-1-host interaction that facilitates retrotransposition and contributes to long gene vulnerability, 2021, 31(8): 861–885.
[47] Billon V, Cristofari G. Nascent RNA m6A modification at the heart of the gene-retrotransposon conflict, 2021, 31(8): 829–831.
[48] Niehrs C, Luke B. Regulatory R-loops as facilitators of gene expression and genome stability, 2020, 21(3): 167–178.
[49] Mita P, Wudzinska A, Sun XJ, Andrade J, Nayak S, Kahler DJ, Badri S, LaCava J, Ueberheide B, Yun CY, Fenyö D, Boeke JD. LINE-1 protein localization and functional dynamics during the cell cycle, 2018, 7: e30058.
[50] Abakir A, Giles TC, Cristini A, Foster JM, Dai N, Starczak M, Rubio-Roldan A, Li MM, Eleftheriou M, Crutchley J, Flatt L, Young L, Gaffney DJ, Denning C, Dalhus B, Emes RD, Gackowski D, Corrêa IR, Garcia-Perez JL, Klungland A, Gromak N, Ruzov A. N6-methyladenosine regulates the stability of RNA:DNA hybrids in human cells, 2020, 52(1): 48–55.
[51] Skourti-Stathaki K, Proudfoot NJ. A double-edged sword: R loops as threats to genome integrity and powerful regulators of gene expression, 2014, 28(13): 1384–1396.
[52] García-Muse T, Aguilera A. R loops: from physiological to pathological roles, 2019, 179(3): 604–618.
[53] Duda KJ, Ching RW, Jerabek L, Shukeir N, Erikson G, Engist B, Onishi-Seebacher M, Perrera V, Richter F, Mittler G, Fritz K, Helm M, Knuckles P, Bühler M, Jenuwein T. m6A RNA methylation of major satellite repeat transcripts facilitates chromatin association and RNA:DNA hybrid formation in mouse heterochromatin, 2021, 49(10): 5568–5587.
[54] Kong YM, Cao L, Deikus G, Fan Y, Mead EA, Lai WY, Zhang YZ, Yong R, Sebra R, Wang HL, Zhang XS, Fang G. Critical assessment of DNA adenine methylation in eukaryotes using quantitative deconvolution, 2022, 375(6580): 515–522.
[55] Chen LQ, Zhang Z, Chen HX, Xi JF, Liu XH, Ma DZ, Zhong YH, Ng WH, Chen T, Mak DW, Chen Q, Chen YQ, Luo GZ. High-precision mapping reveals rareN- deoxyadenosine methylation in the mammalian genome, 2022, 8(1): 138.
[56] Wu TP, Wang T, Seetin MG, Lai YQ, Zhu SJ, Lin KX, Liu YF, Byrum SD, Mackintosh SG, Zhong M, Tackett A, Wang GL, Hon LS, Fang G, Swenberg JA, Xiao AZ. DNA methylation onN-adenine in mammalian embryonic stem cells, 2016, 532(7599): 329–333.
[57] Bailey JA, Carrel L, Chakravarti A, Eichler EE. Molecular evidence for a relationship between LINE-1 elements and X chromosome inactivation: the Lyon repeat hypothesis, 2000, 97(12): 6634–6639.
[58] Liu J, Dou XY, Chen CY, Chen C, Liu C, Xu MM, Zhao SQ, Shen B, Gao YW, Han DL, He C.N-methyladenosine of chromosome-associated regulatory RNA regulates chromatin state and transcription, 2020, 367(6477): 580– 586.
[59] Liu JD, Gao MW, He JP, Wu KX, Lin SY, Jin LM, Chen YP, Liu H, Shi JJ, Wang XW, Chang L, Lin YY, Zhao YL, Zhang XF, Zhang M, Luo GZ, Wu GM, Pei DQ, Wang J, Bao XC, Chen JK. The RNA m6A reader YTHDC1 silences retrotransposons and guards ES cell identity, 2021, 591(7849): 322–326.
[60] Selmi T, Lanzuolo C. Driving chromatin organisation through N6-methyladenosine modification of RNA: what do we know and what lies ahead?, 2022, 13(2): 340.
[61] Percharde M, Lin CJ, Yin YF, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen XH, Ramalho-Santos M. A LINE1-Nucleolin partnership regulates early development and ESC identity, 2018, 174(2): 391–405.e19.
[62] Chen C, Liu WQ, Guo JY, Liu YY, Liu XL, Liu J, Dou XY, Le RR, Huang YX, Li C, Yang LY, Kou XC, Zhao YH, Wu Y, Chen JY, Wang H, Shen B, Gao YW, Gao SR. Nuclear m6A reader YTHDC1 regulates the scaffold function of LINE1 RNA in mouse ESCs and early embryos, 2021, 12(6): 455-474.
[63] Sommerkamp P. Substrates of the m6A demethylase FTO: FTO-LINE1 RNA axis regulates chromatin state in mESCs, 2022, 7(1): 212.
[64] Wei JB, Yu XB, Yang L, Liu XL, Gao BY, Huang BX, Dou XY, Liu J, Zou ZY, Cui XL, Zhang LS, Zhao XS, Liu QZ, He PC, Sepich-Poore C, Zhong N, Liu WQ, Li YH, Kou XC, Zhao YH, Wu Y, Cheng XJ, Chen C, An YM, Dong XY, Wang HY, Shu Q, Hao ZY, Duan T, He YY, Li XK, Gao SR, Gao YW, He C. FTO mediates LINE1 m6A demethylation and chromatin regulation in mESCs and mouse development, 2022, 376(6596): 968–973.
[65] Li Y, Xia LJ, Tan KF, Ye XD, Zuo ZX, Li MC, Xiao R, Wang ZH, Liu XN, Deng MQ, Cui JR, Yang MT, Luo QZ, Liu S, Cao X, Zhu HR, Liu TQ, Hu JX, Shi JF, Xiao S, Xia LX.N-methyladenosine co-transcriptionally directs the demethylation of histone H3K9me2, 2020, 52(9): 870–877.
[66] Chelmicki T, Roger E, Teissandier A, Dura M, Bonneville L, Rucli S, Dossin F, Fouassier C, Lameiras S, Bourc’his D. m6A RNA methylation regulates the fate of endogenous retroviruses, 2021, 591(7849): 312–316.
[67] Rodic N. LINE-1 activity and regulation in cancer, 2018, 23(9): 1680–1686.
[68] Gu ZM, Liu YX, Zhang Y, Cao H, Lyu JH, Wang X, Wylie A, Newkirk SJ, Jones AE, Lee M, Botten GA, Deng M, Dickerson KE, Zhang CC, An WF, Abrams JM, Xu J. Silencing of LINE-1 retrotransposons is a selective dependency of myeloid leukemia, 2021, 53(5): 672–682.
N-adenosine methylation and the regulatory mechanism on LINE-1
Ao Zhang, Shan Cen, Xiaoyu Li
Long interspersed elements-1(LINE-1) is the only autonomous transposon in human genome,and its retrotransposition results in change of cellular genome structure and function, leading occurrence of various severe diseases. As a central key intermediated component during life cycle of LINE-1 retrotransposition, the host modification of LINE-1 mRNA affects the LINE-1 transposition directly.N-adenosine methylation(m6A), the most abundant epigenetic modification on eukaryotic RNA, is dynamically reversible. m6A modification is also found on LINE-1 mRNA, and it participants regulation of the whole LINE-1 replication cycle, with affecting LINE-1 retrotransposition as well as its adjacent genes expression, followed by influencing genomic stability, cellular self-renewal, and differentiation potential, which plays important roles in human development and diseases. In this review, we summarize the research progress in LINE-1 m6A modification, including its modification positions, patterns and related mechanisms, hoping to provide a new sight on the mechanism research and treatment of related diseases.
m6A modification; retrotransposon; LINE-1; genome; genome stability
2023-11-10;
2023-12-28;
2024-01-19
国家自然科学基金面上项目(编号:31870164)资助[Supported by the National Natural Science Foundation of China (No.31870164)]
张傲,硕士研究生,专业方向:LINE-1与肿瘤维持机制的研究。E-mail: za1632649341@163.com
岑山,博士,研究员,研究方向:病毒学。E-mail: shancen@hotmail.com
李晓宇,博士,研究员,研究方向:病毒学。E-mail: xiaoyulik@hotmail.com
10.16288/j.yczz.23-248
(责任编委: 宋旭)