Metabolic dysregulation in term infants from SARS-CoV-2-infected mothers
Mari Merce Cascant-Vilaplana · Inmaculada Lara-Cantón · Victoria Ramos-Garcia · Alejandro Pinilla-González ·Álvaro Solaz-García · Guillermo Quintás · Purificación Marín-Reina · Marta Aguar ·Laura Torrejón-Rodríguez · Máximo Vento · Julia Kuligowski · María Cernada
Severe acute respiratory syndrome coronavirus 2 (SARSCoV-2) is a novel beta coronavirus that causes coronavirus disease 2019 (COVID-19),a severe infectious respiratory disease.In January 2020,the World Health Organization(WHO) declared the outbreak a public health emergency of international concern,and in March 2020,it was declared a global pandemic [1].COVID-19 is the second pandemic of the twenty-first century,with over six hundred million infections and,to date,over six million deaths [2].
Pregnant women are one of the most vulnerable population requiring additional precautions against SARS-CoV-2 infection,as it has been associated with complications,such as preterm birth [3],fetal distress,and fetal growth restriction,due to maternal hypoxia [4].A range of viruses,such as varicella zoster virus,Epstein–Barr virus,herpes simplex virus,and cytomegalovirus (CMV),can be spread by intrauterine transmission.These viruses usually produce a mild form of illness in the mother but can have serious effects on the developing fetus [5,6],as fetuses and neonates suffer from inflammatory responses and metabolic dysregulation when their mothers are infected during late pregnancy.Although most of the neonates born to infected mothers are asymptomatic and negative for SARS-CoV-2,there are insufficient data available to evaluate the metabolic effects of SARS-CoV-2 infection in late pregnancy on the newborn[7,8].
The metabolome comprises all endogenous low-molecular-weight components (<2 kD) in a biological sample such as urine or plasma and reflects the status of an organism under determined environmental conditions,fluctuating according to physiological demands [9].Consequently,altered metabolic fingerprints could of fer novel avenues to better understand systems biology,detect or identify potential risks for various diseases,and ultimately help achieve the goals of personalized medicine [10].Several studies have identified alterations in endogenous metabolites in serum samples from adults with COVID-19 compared to healthy controls [11-14].Hence,metabolomics could be a useful tool to evaluate maternal–fetal/neonatal interactions following SARS-CoV-2 exposure.Early identification of metabolic disorders in the offspring could support risk assessment of potential long-term adverse outcomes,thus facilitating selection of appropriate monitoring approaches and timely application of preventive measures.To date,only one study has reported the use of untargeted metabolomics to investigate the metabolome of newborns from mothers with SARS-CoV-2 infection in comparison to newborns from mothers without SARS-CoV-2,focusing on plasma samples from a small cohort collected during the first 12 hours after birth [14].While differences in the relative levels of several lipid species and amino acids allowed discrimination between groups,it remained unclear whether these findings reflected maternal metabolic alterations due to viral infection or whether they actually showed infant sequelae.
We hypothesized that newborns from mothers infected with SARS-CoV-2 display early metabolic dysregulations,which could be mirrored in their urinary metabolome.In the present study,we compared the urinary metabolome of neonates from mothers with SARS-CoV-2 infection at the moment of hospital admission to those of a control group (i.e.,newborns from mothers without SARS-CoV-2 infection),with the aim of gaining insight into the potential impact of maternal infection on the newborn using a non-invasive approach.
The study was approved by the Ethics Committee for Biomedical Research of the Health Research Institute La Fe (Valencia,Spain) with approval number #2019-037-1 and 2020-269-1.All methods were performed in accordance with relevant guidelines and regulations,and informed consent was prospectively obtained from legal representatives of enrolled infants.
Recruitment started in October 2020 and ended in January 2021 and was performed at the Maternity Ward of University and Polytechnic Hospital La Fe (Valencia,Spain).The study group consisted of full-term,asymptomatic infants born from mothers infected by SARS-CoV-2.The control group included healthy,full-term neonates born from non-infected mothers during the study period.All mothers were tested at admission in the delivery room to determine whether they had symptoms or not.A polymerase chain reaction (PCR) test for SARS-CoV-2 was performed from a nasopharyngeal sample using the Focus Symplexa COVID-19 kit from DiaSorin Molecular(Cypress,California,USA).Neonates born from positive mothers were also tested for SARS-CoV-2.
Urine samples were collected using sterile cotton pads placed in the diaper during the first 48–72 hours of life.Cotton pads were placed in the diaper and collected within four hours.They were placed in a dry polypropylene container and then stored at − 80 °C until further processing.
Cotton pads containing urine were thawed on ice and extracted using a sterile syringe.Urine samples were homogenized on a Vortex®mixer for 10 seconds and centrifuged at 16,000 g and 4 °C for 15 minutes.A volume of 50 μL of supernatant was added to 50 μL of an internal standard mixture containing reserpine,L-phenyl-D5-alanine,leucine enkephalin,caffeine-D9,betaine-D11,2-deoxyguanosine-13C,15N,8-hydroxy-2’-deoxyguanosine-13C,15N,taxifolin,p-tyrosine-D2,and creatinine-D3at a concentration of 2 μmol/L each in 0.1% v/v HCOOH.
Samples were analyzed using two ultra-performance liquid chromatography coupled to quadrupole time-of-flight mass spectrometry (UPLC-QTOF-MS) methods.Detailed information on data acquisition,statistics,and data analysis is available at https:// zenodo.org under accessory number https:// doi.org/ 10.5281/ zenodo.75851 14 and in the Supplementary Material.
A total of 49 newborns were enrolled in this study: 23 in the control group and 26 in the study group.Demographic details and characteristics of the study population are shown in Table 1.No statistically significant differences were found between infants born to mothers with SARS-CoV-2 infection and controls,except for some SARS-CoV-2-positive mothers(23%) who had mild symptoms of infection (fever,cough,dyspnea,fatigue,myalgia,anosmia,headache,vomiting,or diarrhea).Nasal and pharyngeal PCR for SARS-CoV-2 was negative in all enrolled neonates,and none of them presented COVID-19 symptoms.Placentas of SARS-CoV-2-positive mothers were analyzed after delivery,and all PCR tests were negative for SARS-CoV-2.Both groups were well balanced in terms of antibiotics administered to mothers and of the newborn's type of feeding.Neonates receiving human milk were breastfed,while those receiving formula used bottle feeding.
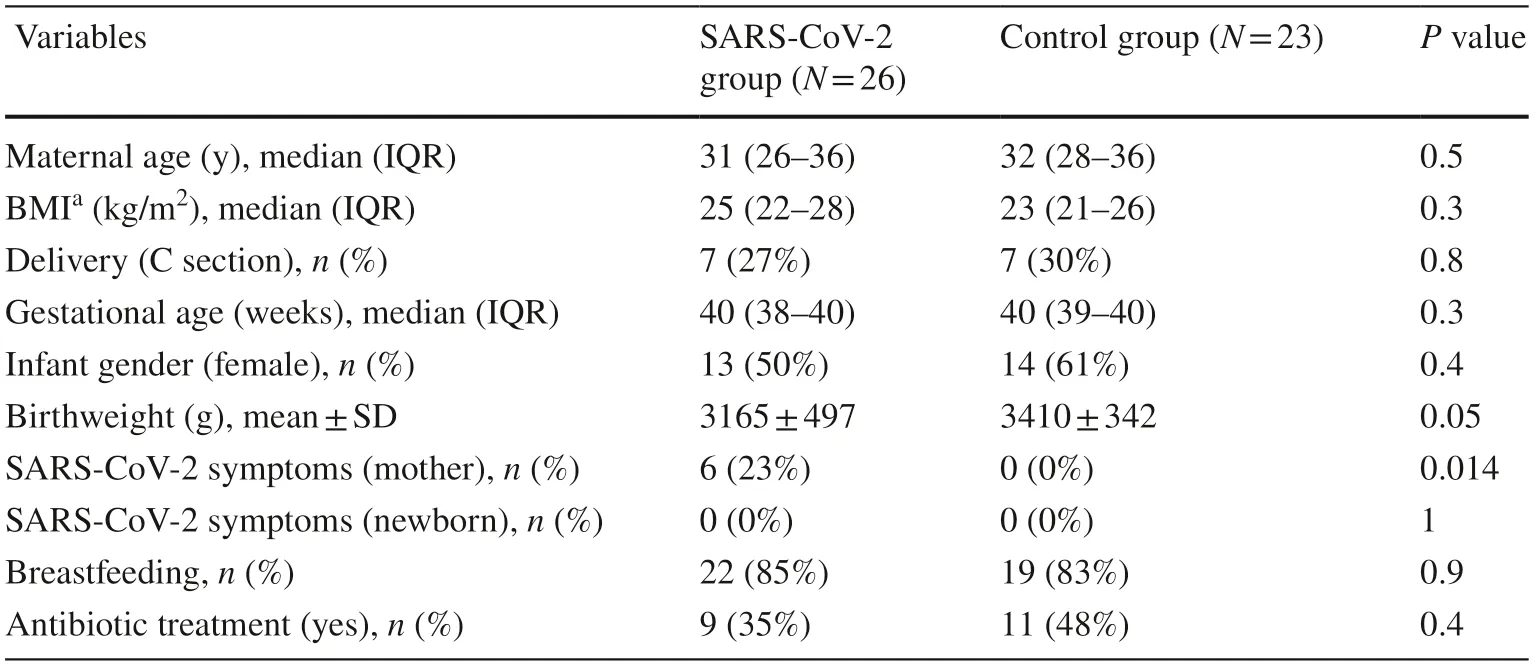
Table 1 Demographic data and characteristics of patients included in the study
Annotated features from untargeted UPLC-QTOF-MS experiments are shown in Supplementary Fig.1.The Principal Component Analysis (PCA) scores plot from the analysisof cleaned-up data is shown in Fig. 1 a.Both groups of urine samples (i.e.,urine from neonates of mothers with SARSCoV-2 infectionvs.healthy controls) overlapped,indicating that the most prominent sources of variance captured were unrelated to maternal SARS-CoV-2 infection.A comparison between the metabolic profiles of both sample groups was carried out by multivariable analysis (i.e.,partial least squares discriminant analysis,PLSDA).Figure 1 b showed a statistically significant discrimination between the control and SARS-CoV-2 groups,assessed by permutation testing(Wilcoxon rank-sum test,Pvalue=0.007).The discriminant performance of the PLSDA model is demonstrated by the receiver operating characteristic (ROC) curve with an area under curve (AUC) of 0.82 estimated by cross-validation(CV) (Fig. 1 c).Furthermore,no statistically significant differences were found between the urinary metabolome of infants with symptomaticvs.asymptomatic mothers (Wilcoxon rank-sum test,500 permutations,Pvalue=0.4).
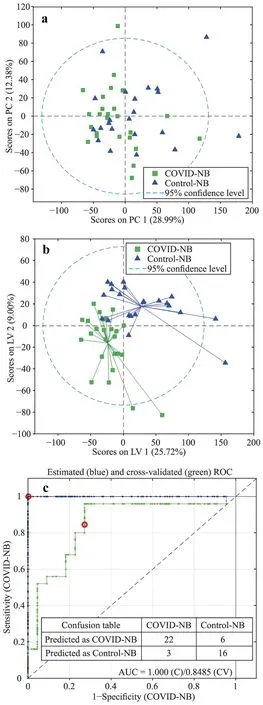
Fig.1 Multivariable analysis of metabolomics data. a PCA scores of pre-processed data after normalization to creatinine and auto-scaling; b and c PLSDA scores and ROC curve from a PLSDA model comparing COVID-19 vs.control groups with three latent variables and using leave one out CV.Data were normalized and auto-scaled.Blue and green lines in the ROC plot represent the estimated and CV ROC curves,respectively,and the red circle is the model threshold.NB newborn,AUC area under the curve,PC principal component,LV latent variable,COVID coronavirus disease 2019,PCA principal component analysis,PLSDA partial least squares discriminant analysis,ROC receiver operating characteristic,C calibration,CV crossvalidation
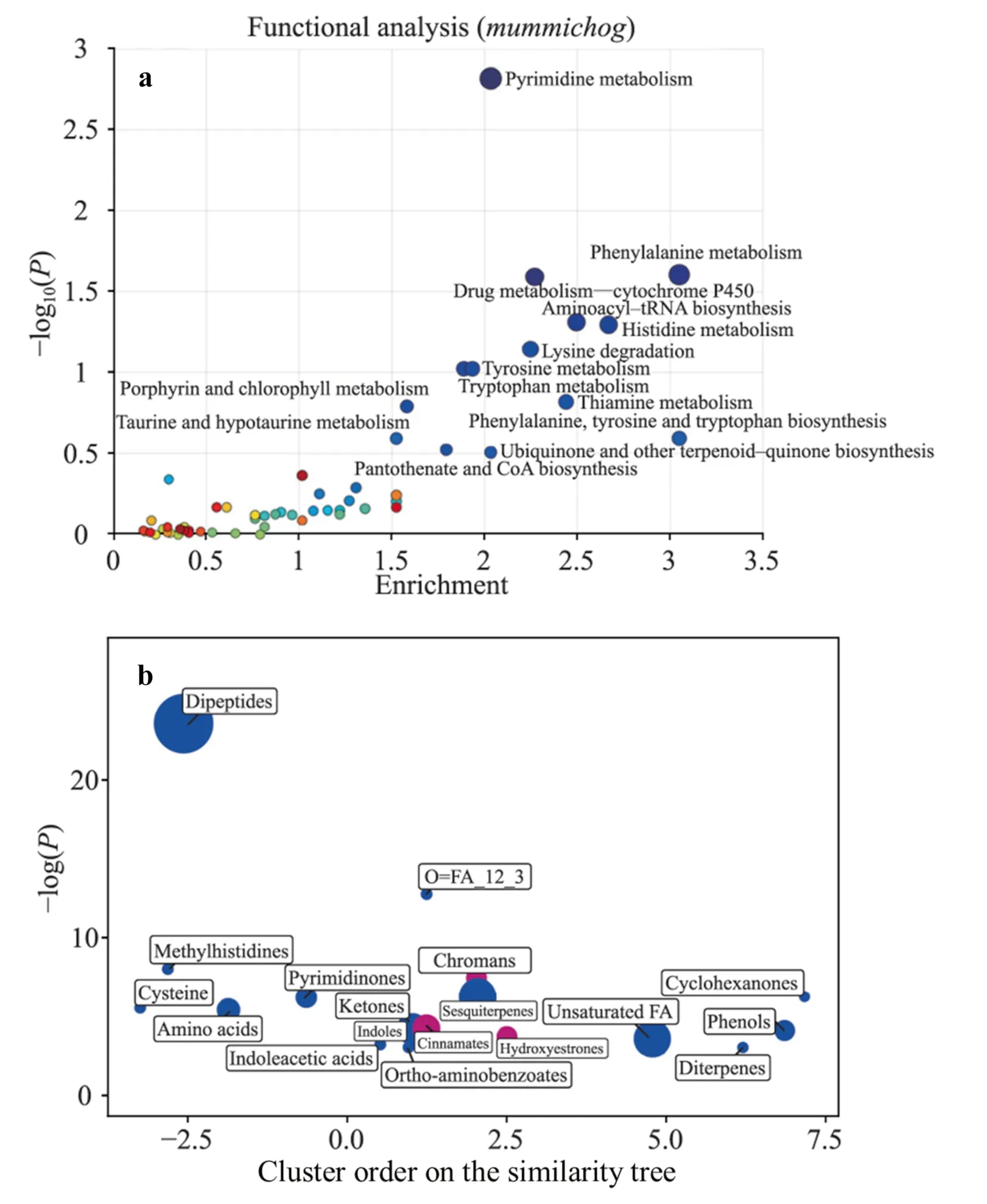
Fig.2 Enrichment analysis of urinary fingerprints of infants from mothers with SARSCoV-2 infection compared to control infants. a Functional enrichment analysis using the mummichog algorithm; b ChemRICH set enrichment statistics plot.Notes: a significantly altered pathways with more than two significant hits are labeled; b each node reflects a significantly altered cluster of metabolites,and enrichment P values were obtained using the Kolmogorov–Smirnov test;node sizes are proportional to the total number of metabolites in each cluster set.Blue nodes indicate a relative decrease in compound levels in infants of mothers with SARS-CoV-2 infection compared to control infants;purple nodes contain both increased and decreased metabolites. CoA coenzyme A,tRNA transfer ribonucleic acid,SARS-CoV-2 severe acute respiratory syndrome coronavirus 2,FA fatty acid
Functional pathway analysis revealed alterations in a total of 14 metabolic pathways in newborns with mothers with SARS-CoV-2 infection in comparison to neonates from the control group (Fig. 2 a and Supplementary Table 1).Chemical similarity enrichment analysis (ChemRICH),based on information on the chemical structures of significantly altered annotated compounds,was employed to define sets of related molecules in a unique and non-overlapping way.The results are visualized in a two-dimensional scatter plot in Fig. 2 b,and the enrichment results for individual cluster sets are shown in Supplementary Table 2.
This study is the first to report changes in postnatal urinary metabolites of healthy newborns born to SARS-CoV-2-positive mothers compared to a control group of SARSCoV-2-negative mothers.A previous study compared the plasma metabolites of newborns from infected and healthy mothers during the first 12 hours after birth [14].Although the relative levels of certain lipid species and amino acids differed between the two groups,it was unclear whether these findings reflected maternal metabolic changes due to viral infection or actual effects on the infants themselves.
The present study showed that maternal infection appears to affect infant metabolism for >24 hours after birth.However,presence or absence of SARS-CoV-2 symptoms in mothers did not affect the urinary metabolome of infants.
Despite testing negative for SARS-CoV-2,several metabolic pathways in the infants showed changes commonly seen in patients infected with the virus.These changes include altered pyrimidine metabolism,specifically changes in cytosine,which has been linked to cell metabolism in SARS-CoV-2 [15].Additionally,3′-deoxy-3′,4′-didehydrocytidine,a molecule associated with antiviral activity [16],has been found in human serum and suggested as a biomarker for acute viral infections,including SARS-CoV-2[17].The metabolic fingerprints also revealed significant changes in drug metabolism by cytochrome P450,which can be impacted by cytokine increases and inflammation related to infection [18].
Amino acid and related metabolic pathway changes have been observed in healthy newborns born to SARS-CoV-2-positive mothers.Similar alterations have been found in SARS-CoV-2-positive adult and pediatric populations [13,15,19-21] and non-hospitalized patients during the postacute phase [22].SARS-CoV-2 infection has been linked to a unique metabolic profile and complex systemic response,including significant disruptions in amino acid and tryptophan metabolic pathways,potentially causing neurotoxicity,neurological disruption,and liver dysfunction [20,21].Immuno-metabolic disorders in response to SARS-CoV-2 infection,such as tryptophan metabolism alterations,have been observed and correlate with clinical laboratory markers of inflammation and renal status [13,21].
We observed changes in metabolic pathways involving cofactors,vitamins,and chromans.In an in vitro model,transcriptional and metabolomic analyses were performed eight hours after SARS-CoV-2 infection [23],showing that SARS-CoV-2 remodels host folate and one-carbon metabolism at the post-transcriptional level,thereby supporting de novo purine synthesis.In SARS-CoV-2-infected cells,intracellular glucose and folate levels were depleted,and viral replication was exquisitely sensitive to inhibitors of folate and one-carbon metabolism.
Dipeptides were reduced in newborn urine samples from SARS-CoV-2-infected mothers,consistent with a prior study[12],in which the levels of four dipeptides were significantly lower in mild and severe COVID-19 patients and most of the dysregulated dipeptides did not recover until hospital discharge.
Our study found that infants of SARS-CoV-2-infected mothers had lower levels of medium-chain and unsaturated fatty acids in their urine.These fatty acids have been shown to have antiviral properties [24,25],and their deficiency may make humans more susceptible to viral infections,including SARS-CoV-2 [26,27].Analysis of lipid mediators in hospitalized COVID-19 patients also revealed differences in immunoregulatory and pro-inflammatory lipid mediators between moderate and severe disease states.In addition,our findings are consistent with a previous study [14] that found decreased levels of long-and very long-chain free fatty acids in plasma samples from neonates of COVID-19-infected mothers,along with increased levels of certain amino acids.
In conclusion,we found that the metabolic fingerprints of newborns differ depending on whether their mothers were infected with SARS-CoV-2 during late pregnancy,as shown through non-invasive urine samples.Our results suggest that even though there is no evidence of transmission of the virus across the placenta,there may be metabolic disruptions in newborns whose mothers were infected with SARS-CoV-2.Interestingly,no clinical or analytical abnormalities were observed in these babies during their stay at the maternity care.Further studies are needed to determine the relationship between the acute metabolic disruptions caused by SARS-CoV-2 in newborns and their long-term outcomes.
Supplementary InformationThe online version contains supplementary material available at https:// doi.org/ 10.1007/ s12519-023-00735-5.
AcknowledgementsThe authors are grateful to parents who agreed to participate in this study and the nursery staffwho helped to collect the samples.
Author contributionsMMCV and ILC contributed equally to the study.ILC,APG,ASG,PMR,LTR,and MAG: data curation,writing–review and editing.MMCV,VRG,and GQ: formal analysis,writing–review and editing.MV,JK,and MC: conceptualization,supervision,writing–review and editing.All authors approved the final manuscript as submitted and agree to be accountable for all aspects of the work.
FundingThis work was supported by the Health Research Institute Carlos III (Spanish Ministry of Economy,Industry and Innovation)and co-funded by the European Union [PI18/01292,CD19/00037,CM20/00187,and CPII21/00003] and the Health Research Institute La Fe,Spain [grant number 2019-050-1_CRC].
Data availabilityRaw data from metabolomics experiments are available at https:// zenodo.org under accessory number https:// doi.org/ 10.5281/ zenodo.75851 14 .XCMS software ( http:// metlin.scrip ps.edu/xcms/ ).
Declarations
Conflict of interestNo financial or non-financial benefits have been received or will receive from any party related directly or indirectly to the subject of this article.
Ethical approvalThe study was approved by the Ethics Committee for Biomedical Research of the Health Research Institute La Fe (Valencia,Spain) with approval number #2019-037-1 and 2020-269-1.
 World Journal of Pediatrics2023年9期
World Journal of Pediatrics2023年9期
- World Journal of Pediatrics的其它文章
- Fecal microbiota transplantation in childhood: past,present,and future
- Pediatric endocrinopathies related to COVID-19: an update
- Mapping the quality of prenatal and postnatal care and demographic differences on child mortality in 26 low to middle-income countries
- Clinical epidemiology and disease burden of bronchiolitis in hospitalized children in China: a national cross-sectional study
- Serum levels of the novel adipokine isthmin-1 are associated with obesity in pubertal boys
- Effects of fentanyl and sucrose on pain in retinopathy examinations with pain scale,near-infrared spectroscopy,and ultrasonography:a randomized trial
