扬麦4号/偃展1号RIL群体株高QTL挖掘及其对赤霉病抗性的效应分析与验证
赵 蝶 胡文静 程晓明 王书平 张春梅 李东升 高德荣
扬麦4号/偃展1号RIL群体株高QTL挖掘及其对赤霉病抗性的效应分析与验证
赵 蝶1,2胡文静2,3,*程晓明2王书平1张春梅2李东升2高德荣2,3,*
1长江大学农学院, 湖北荆州 434025;2江苏里下河地区农业科学研究所 / 农业农村部长江中下游小麦生物学与遗传育种重点实验室, 江苏扬州 225007;3扬州大学 / 江苏省粮食作物现代产业技术协同创新中心 / 江苏省植物功能基因组学重点实验室, 江苏扬州 225009
小麦的株高(plant height, PH)性状与赤霉病(fusarium head blight, FHB)抗性的关系密切。本研究利用扬麦4号/偃展1号(YM4/YZ1)杂交组合衍生的重组自交系(recombinant inbred lines, RIL)群体, 利用55K单核苷酸多态性(single-nucleotide polymorphism, SNP)芯片数据, 结合3年共6个环境下RIL群体及其亲本的株高数据, 挖掘株高性状的遗传位点。同时利用土壤表面赤霉病麦粒抛撒法和单花滴注法鉴定株高位点对赤霉病抗侵染(Type I)和抗扩展(Type II)两种类型的效应。在染色体2D、4B、4D、5A和7D上检测到7个与株高相关的数量性状位点(quantitative trait loci, QTL), 经过比对,可能是新的株高位点。、和的矮秆效应来源于扬麦4号, 其余4个QTL的矮秆效应来源于偃展1号。和均在6个环境下被检测到, 表型变异贡献率(PVE)范围分别为19.48%~44.11%和10.48%~13.71%。研究发现和位点上的高秆等位变异(YM4等位变异)分别降低侵染型平均病小穗率(average percentage of infected spikelets, PIS) 34.97%和19.09%,和位点上的矮秆等位变异(YM4等位变异)分别降低扩展型平均病小穗率(average percentage of diseased spikelets, PDS) 24.73%和14.56%。的矮秆等位变异来源于阿夫。利用小麦中国春2.1版本的参考基因组信息分析区间, 发现一共有146个有注释功能的高置信基因, 主要涉及合成细胞色素P450、脱水反应元件结合蛋白、乙烯响应转录因子、转录因子MYC2和细胞壁受体相关激酶等。进一步将位点紧密连锁SNP标记转化成育种可用分子标记KASP-5A, 并在126份小麦品种(系)中初步验证其对株高和赤霉病抗性的效应。研究结果可为的育种应用和精细定位奠定基础。
小麦; 株高; 赤霉病; QTL;; 候选基因
小麦(L.)是世界上最重要的粮食作物之一, 提供了我们日常饮食中20%以上的卡路里和蛋白质[1]。由于人口的不断增加和全球耕地面积的持续减少, 高产仍然是小麦育种计划的主要目标之一[2]。株高(plant height, PH)是一个与株型和产量潜力相关的关键性状[3]。因此, 利用矮秆基因提高产量已成为产量育种的主要策略之一。例如, 在小麦绿色革命期间, 将半矮秆品种引入小麦适当地降低株高值, 并对全球潜在粮食产量的提高做出了显著贡献[4]。众所周知, 小麦株高通常受多基因控制,其数量性状位点(quantitative trait loci, QTL)广泛分布在小麦21条染色体上[5]。其中两个矮秆基因和在小麦中分布最广并在小麦株高矮化过程中起到了重要作用[6-7],和均编码DELLA蛋白, 参与赤霉素(gibberellic acid, GA)信号的传导过程, 此类基因突变之后对GA信号不敏感而导致株高变矮, 这两个基因的特性及其功能标记对小麦矮化和高产育种起到了促进作用[8-11]。另一个在小麦育种中被广泛应用的矮秆基因是, 它被定位在染色体2D短臂上, 对GA3反应为敏感型[12]。编码核糖核酸酶h样结构域蛋白, 分别被Chai等[13]和Xiong等[14]证实为的候选基因。隐性基因来源于赤小麦, 位于7B染色体短臂上,为GA3敏感型, 对小麦有较强的矮化作用[15]。是一个对GA敏感的显性矮化基因, 位于染色体5A长臂上, 通过激活来降低植物高度, 显著缩短干细胞长度, 减少GA生物合成[16]。小麦半矮化是由于表达增加和GA含量降低造成的[17]。位于染色体7AS上, 其短茎性状可能与细胞数量减少有关[18]。位于6A染色体长臂上的一个GA敏感型矮秆基因,可能是其候选基因, 其过表达可显著降低GA水平并引起植株矮化, 但对产量性状无不利影响[19]。Borrill等[20]发现一个潜在替代传统矮化基因的新型矮化基因, 与传统矮化基因相比,的矮秆等位变异降低了植株高度, 并增加了茎秆强度, 其编码一个自激活活性NB-LRR基因, 而不是赤霉素信号或代谢的组成部分, 自激活活性的等位基因()引起致病相关基因的上调, 导致活性氧的产生, 活性氧能促进交联和细胞壁硬化, 导致生长减少。与小麦其他产量相关性状相似, 株高是复杂的数量性状, 受遗传和环境因素的交互作用影响较大。QTL定位为在遗传水平上分析这些复杂的数量性状提供了一种有效的方法, 同时也为小麦株高的分子标记辅助育种奠定基础[21-23]。赤霉病(fusarium head blight, FHB), 主要由禾谷镰刀菌(Schwabe)引起, 严重危害小麦或其他谷物的生产和粮食安全。中国的长江中下游麦区和东北春麦区常遭受到赤霉病的危害[24-26], 近年来, 由于以小麦-玉米为主的耕作制度和少耕留茬, 赤霉病已成为黄淮流域小麦的主要病害之一[27-28]。小麦对赤霉病的抗性具有多种表现形式, 其中抗侵染(Type I)和抗扩展(Type II)是两种最主要的类型, 抗侵染反映的是小麦抵抗赤霉菌初侵染的能力, 而抗扩展反映的是在赤霉菌侵染后宿主抵御其通过菌丝生长沿着穗轴延伸的能力[29]。小麦抗赤霉病育种瓶颈之一是同时提高小麦赤霉病抗性和产量。已知的一些赤霉病抗源例如苏麦3号、望水白的产量潜力太低, 株高达到130~150 cm, 易倒伏, 应用起来较困难[30]。相关研究发现小麦的一些农艺性状, 如芒的长短、小穗密度、株高、抽穗期、穗长等对赤霉病的侵染和扩展均有不同程度的影响, 其中, 株高等对赤霉病的侵染影响较大, 有研究表明在自然条件下植株高可使小麦减轻受病原菌感染程度[31-35]。控制株高的QTL/基因中矮秆基因和对赤霉病的侵染能力具有显著的效应[30,36-39], 而株高位点对赤霉病的扩展严重度具有显著效应的相关报道罕见[30]。
扬麦4号(Yangmai 4, YM4)是长江中下游流域20世纪80年代育成的第一个赤霉病抗性达到中抗的小麦品种, 在赤霉病大流行年份依然可以抵抗赤霉病的危害, 其亲本是南大2419、胜利麦(1-3-2系)和阿夫选系[26]。偃展1号(Yanzhan 1, YZ1)是一个矮秆、早熟、高产、抗旱、抗寒但高感赤霉病的小麦品种。扬麦4号株高为90~100 cm, 偃展1号的株高为70~80 cm, 经过前期分子标记检测表明扬麦4号不携有和两个主效矮秆基因, 偃展1号携有基因。目前对扬麦4号和偃展1号的株高遗传基础尚未有解析, 本研究的目的是: (1) 利用扬麦4号/偃展1号(YM4/YZ1)重组自交系(recombinant inbred lines, RIL)群体解析扬麦4号和偃展1号的株高遗传基础; (2) 利用RIL群体研究所定位到的株高位点对赤霉病抗侵染和抗扩展的效应; (3) 开发主效稳定的株高QTL相应的KASP (kompetitive allele specific PCR)分子标记, 并在自然群体中验证其对株高和赤霉病抗性的效应, 为小麦分子标记辅助育种奠定基础。
1 材料与方法
1.1 试验材料及田间试验
将扬麦4号与黄淮麦区的优良冬小麦品种偃展1号进行杂交, 利用单粒传方法获得F10代的RIL群体, 共151个家系。扬麦4号/偃展1号种群在小麦生长季分别种植于2018—2019年间荆州试验点(海拔35~45 m, 北纬33.46度, 东经118.22度, 年降水量910 mm, 简称19JZ)、扬州试验点西区和扬州试验点东区(海拔10~20 m, 北纬32.24度, 东经119.26度, 年降水量1020 mm, 简称19YZW和19YZE), 2019—2020年间扬州试验点西区和扬州试验点东区(简称20YZW和20YZE), 2020—2021年间扬州试验点东区(简称21YZE)。田间试验分为3组进行, 分别是株高评价、Type I型赤霉病抗性评价和Type II型赤霉病抗性评价。田间试验采用完全随机设计, 2个重复。每个RIL和2个亲本平均播撒30粒种子, 每行133 cm, 双行间隔25 cm。田间试验和疾病控制的管理遵循当地标准做法, 2种抗赤霉病评价苗圃中未使用任何杀菌剂。采用与RIL相同的方案, 于2019—2020年间和2020—2021年间的扬州试验点西区种植126个小麦品种(系)。待到成熟期, 在扬麦4号/偃展1号群体和2个亲本中随机选取生长状况相近的每个家系的10株代表性植株, 从主茎穗植株基部到主茎穗顶部(不包括芒)测量株高。田间赤霉病评价在2019—2020年间和2020—2021年间的扬州试验站进行, 采用土壤表面赤霉病麦粒抛撒法和单花滴注法分别对扬麦4号/偃展1号群体和2个亲本的抗侵染型(Type I)和抗扩展型(Type II)的赤霉病抗性进行鉴定。Type I型接种在孕穗前5 d进行第1次接种, 孕穗期再进行第2次接种, 等到开花后10 d开始调查, 记录至少一个小穗出现明显赤霉病症状的病小穗数和每穗总小穗数, 计算出每个RIL家系每次重复(共20穗)的平均病小穗率(average percentage of infected spikelets, PIS)。Type II型接种是在花期用Hu等[40]描述的4种禾谷镰孢()菌株的混合物接种, 在接种21 d后, 记录每个RIL家系(共20穗)的病小穗的平均百分比(average percentage of diseased spikelets, PDS), 作为赤霉病严重程度的衡量标准。接种后根据胡文静等[30]方法进行田间保湿和调查赤霉病。从每天早上07:00—18:00, 每30 min喷雾5 min, 以提供有利于赤霉病感染的高度湿润条件。接种4周后, 观察接种穗的病小穗数和每穗总小穗数。计算每个RIL的平均病小穗率作为赤霉病严重程度的度量。2019— 2021两年间的126个小麦品种(系)采用与RIL相同的实验方法进行株高和2种抗赤霉病类型的调查。
1.2 统计分析
利用Microsoft Excel 2019对数据进行基本处理和统计分析; 利用SPSS的Tukey进行等位变异之间PIS和PDS的T测验。株高的广义遗传力(2)计算公式为22G/(2G+2G×E/E+2eER)[41-42], 其中E和R分别为环境次数和重复数,2G为基因型方差,2G×E为环境对基因型的互作方差,2e为残差。使用IciMapping v4.1计算2和最优线性无偏估计值()[43]。
1.3 基因分型、连锁图谱构建、QTL分析
采用CTAB法[44]从幼苗中提取基因组DNA, 凝胶电泳检测DNA完整性和数量。利用中国金标记(北京)生物技术有限公司的小麦55k单核苷酸多态性(single-nucleotide polymorphism, SNP)芯片对扬麦4号、偃展1号亲本品种和RIL群体进行基因分型, 然后选择、、和等与其他已知基因相关的KASP标记和SSR (simple sequence repeat)对亲本及RIL群体进行基因分型, 检测多态性[11,25,28,45]。标记数据的质控和遗传连锁图谱构建参考Hu等[39]。在构建连锁图谱之前, 对SNP数据进行质控, 然后经过去冗余(删除缺失率大于30%和最小等位基因频率小于5%的SNP)、分群(LOD=8), 最终得到1440个上图SNP, 构建成长度为3574.10 cM的覆盖小麦21条染色体的遗传连锁图谱, 标记之间的平均遗传距离是2.58 cM[39]。利用IciMapping v4.1 (https://www.isbreeding.net/)的完备区间作图法(inclusive composite interval mapping, ICIM)检测与小麦株高显著相关的QTL, LOD阈值设为3.0[43]。图谱绘制使用MapChart 2.3 (https://www.wur.nl/en/ show/Mapchart.htm)。我们利用QTL区间的侧翼SNP的序列信息, 将本研究中鉴定的QTL与先前报道的QTL或基因进行比较(http://202.194.139. 32/blast/blast.html; http://202.194.139.32/genes/), 重叠置信区间内的QTL被视为同一个[46]。
1.4 KASP标记的开发与验证
根据初步QTL作图结果, 参照Li等[47]方法将目标QTL的侧翼标记转化为KASP标记, 追踪相应性状。随后进行KASP测定。使用PHERAstar (BMG LABTECH, 德国)对PCR反应进行荧光检测[45]。使用KlusterCaller软件(LGC Genomics, Beverly, 美国)将开发成功的KASP标记在RIL群体中检测, 与芯片数据进行比较[48]。为了进一步验证主效QTL在不同遗传背景下的效应, 对126个来自国内外的小麦品种(系)进行KASP标记的检测和株高、PIS和PDS的考察。
1.5 候选基因预测
根据主效QTL目标区间侧翼标记的物理位置从小麦参考基因组2.1版本(http://202.194.139.32/ jbrowse-1.12.3-release)中提取物理区间的高置信基因, 利用Triticeae-GeneTribe (http://wheat.cau.edu. cn/TGT/)发掘基因在拟南芥和水稻的同源基因并分析其参与的生物进程和分子功能[49-50]。
2 结果与分析
2.1 亲本和RIL群体株高表现
统计分析显示(表1), 两个亲本的株高具有极显著差异(<0.01)。(最优线性无偏估计值)下亲本扬麦4号和偃展1号的株高分别为98.40 cm和75.21 cm, RIL群体株高变异范围为65.62~119.98 cm, 平均值为97.14 cm。株高性状在6个环境下的广义遗传力(2)为0.81 (表1)。

表1 不同环境下亲本及扬麦4号/偃展1号的株高表型变异及遗传力
E1、E2、E3、E4、E5和E6分别表示2019JZ、2019YZW、2019YZE、2020YZW、2020YZE和2021YZE的环境, BLUE表示最优线性无偏估计值。**表示两亲本间的株高差异显著(< 0.01)。
E1, E2, E3, E4, E5, and E6 indicate the environment of 2019JZ, 2019YZW, 2019YZE, 2020YZW, 2020YZE, and 2021YZE, respectively. BLUE represents the best linear unbiased prediction.**indicates significant difference in plant height between the two parents at< 0.01.
2.2 株高QTL定位分析
共检测到7个株高相关QTL, 分别位于染色体2D、4B、4D、5A和7D上(图1)。、和加性效应为负, 说明矮秆效应来源于扬麦4号, 其余的4个QTL加性效应均为正, 说明矮秆效应来源于偃展1号(表2)。和可同时在6个环境下被检测到, 株高贡献率范围分别为19.48%~44.11% 和10.48%~13.71%, LOD值范围分别是15.50~35.95和8.31~12.70 (表2)。的紧密连锁标记是Rht-D1_SNP, 位于矮秆基因的物理区间。在5个环境下被检测到, 表型贡献率范围为4.13%~11.09%。和在4个环境下被检测到, 表型贡献率范围分别为3.16%~10.76%和3.60%~9.75% (表2)。和在2个环境下被检测到, 株高贡献率范围分别为3.72%~7.06%和8.04%~8.64% (表2)。
连锁群右边是标记名称, 左边是遗传位置(cM), 连锁群中的黑色矩形代表QTL区域。E1、E2、E3、E4、E5和E6分别表示2019JZ、2019YZW、2019YZE、2020YZW、2020YZE和2021YZE的环境,表示最优线性无偏估计值。
The marker name is on the right of the linkage group, the genetic position (cM) is on the left, and the black rectangle in the linkage group represents the QTL region. E1, E2, E3, E4, E5, and E6 indicate the environment of 2019JZ, 2019YZW, 2019YZE, 2020YZW, 2020YZE, and 2021YZE, respectively.represents the best linear unbiased prediction.
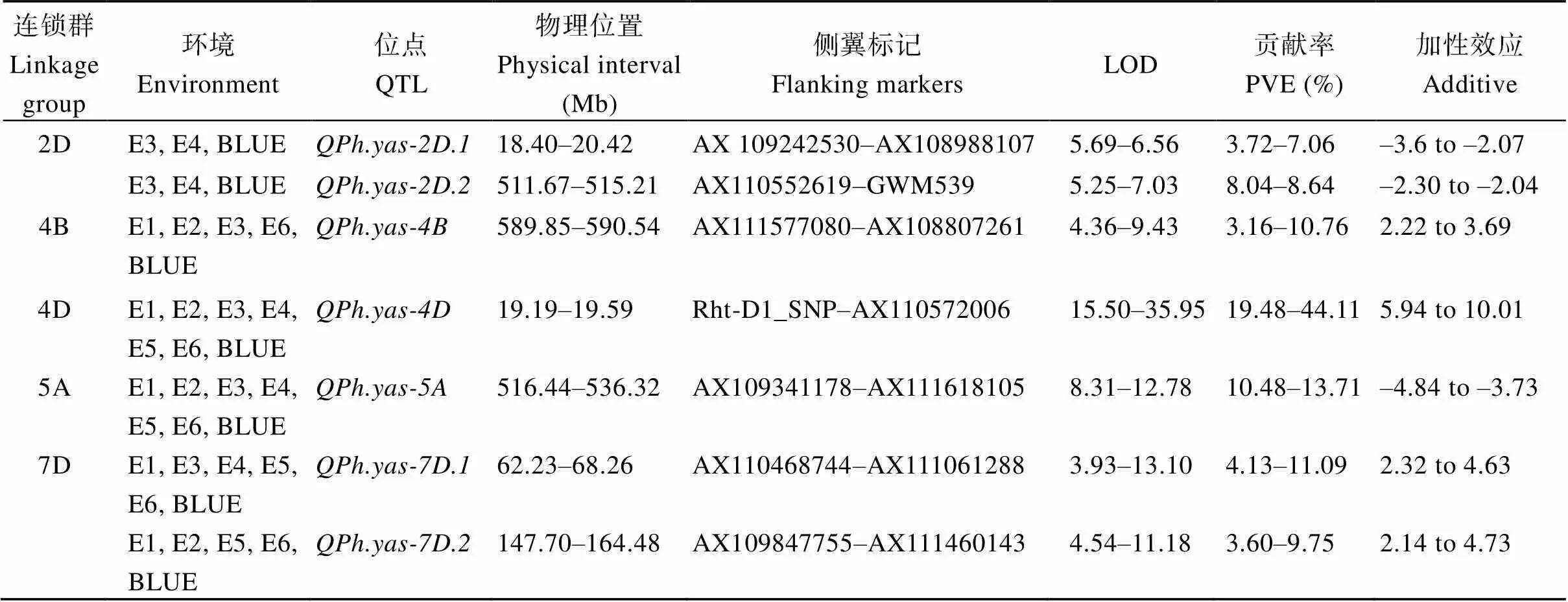
表2 扬麦4/偃展1号群体株高的QTL分析
E1、E2、E3、E4、E5和E6分别表示2019JZ、2019YZW、2019YZE、2020YZW、2020YZE和2021YZE的环境, BLUE表示最优线性无偏估计值。b加性效应为正说明矮秆效应来源于偃展1号, 加性效应为负说明矮秆效应来源于扬麦4号。
E1, E2, E3, E4, E5, and E6 indicate the environment of 2019JZ, 2019YZW, 2019YZE, 2020YZW, 2020YZE, and 2021YZE, respectively. BLUE represents the best linear unbiased prediction.bPositive additive effect indicated that the dwarf effect originates from Yanzhan 1, and the negative additive effect indicates that the dwarf effect originates from Yangmai 4.
2.3 QTL位点对抗赤霉病的效应
分析本研究中定位到的株高相关QTL对赤霉病侵染和扩展严重度的影响, 结果表明,和对赤霉病侵染严重度(PIS值)和赤霉病扩展严重度(PDS值)均无显著效应(图2-A, C, F)。对PIS值无显著效应, 对PDS值具有极显著效应, 携带矮秆等位基因型(YM4等位变异)的家系比携带高秆等位基因型(YZ1等位变异)的家系的PDS值降低24.73% (<0.01) (图2-B);对PIS值无显著效应, 对PDS值具有极显著效应, 携带矮秆等位基因型(YM4等位变异)的家系比携带高秆等位基因型(YZ1等位变异)的家系的PDS值降低14.56% (<0.01) (图2-E)。和对PIS值均具有极显著效应, 携带高秆等位基因型(YM4等位变异)的家系比携带矮秆等位基因型(YZ1等位变异)的家系的PIS值分别降低34.97% (<0.01)和19.09% (<0.01) (图2-D, G)。
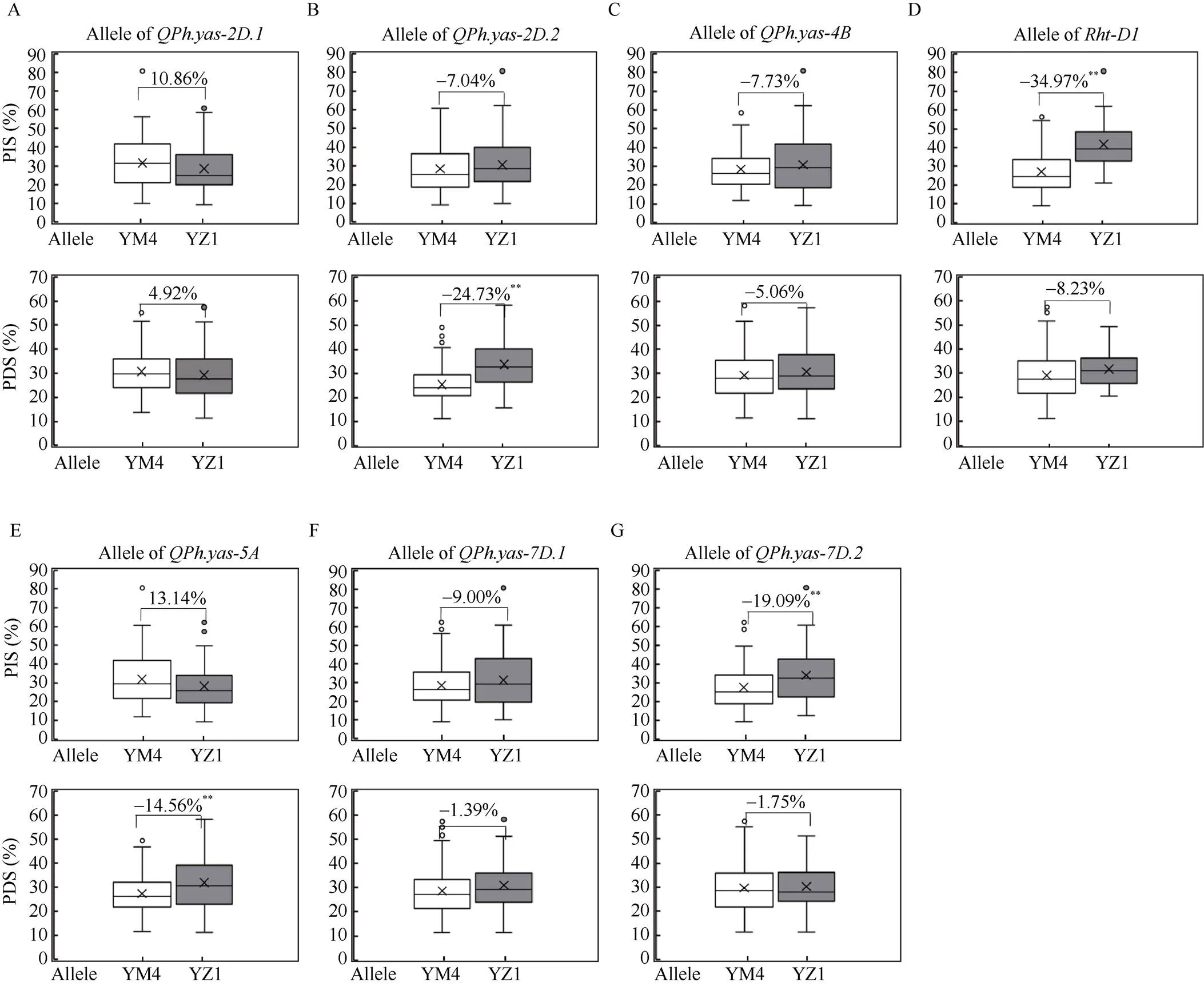
图2 扬麦4号/偃展1号RIL群体株高QTL对赤霉病抗性的效应
PIS: 侵染型平均病小穗率, 土壤表面赤霉病麦粒抛撒法对赤霉病抗性的测定; PDS: 扩展型平均病小穗率, 单花滴注法对赤霉病抗性的测定。‘×’在箱形图中为平均值标记; 箱形图中的点表示离群点; 数据框中的水平线表示中位数。*和**分别代表与偃展1号相比<0.05和<0.01。
PIS: the average percentage of infected spikelets, the measure of FHB resistance in soil surface inoculation; PDS: the average percentage of diseased spikelets, the measure of FHB resistance in point inoculation. ‘×’ in the data box indicates the mean value; the dots in the boxplots are the outliers; the horizontal line in the data box indicates the median. * and ** represent significant difference at< 0.05 and< 0.01 compared with Yanzhan 1, respectively.
2.4 KASP标记开发及验证
定位到的株高QTL中,和是效应值较大且稳定的位点,位于矮秆基因区间[11]。根据另一主效位点目标区间两侧的分子标记侧翼序列, 最终成功将位于峰值区间紧密连锁的SNP标记AX109341178转化为KASP标记并命名为KASP-5A[51]。该SNP碱基突变是C/G (扬麦4号是C, 偃展1号是G), 共用引物序列是5'- TCTGCGGGCACATCAGTTAG-3', 特异性引物1序列是5′-GAAGGTGACCAAGTTCATGCTTGGACA CCGAAGTAGTTCCC-3′, 尾部添加能够与FAM荧光结合的特异性序列; 特异性引物2序列是5′- GAAGGTCGGAGTCAACGGATTTGGACACCGAAGTAGTTCCT-3′, 尾部添加能够与HEX荧光结合的特异性序列。
2.5 QPh.yas-5A在自然群体中的效应分析
利用126份自然群体试验材料验证位点对株高和赤霉病抗性的效应(附表1)。研究发现位点上的矮秆等位变异(YM4等位变异)仍可显著降低株高和赤霉病扩展严重度, 株高和PDS值分别降低2.38% (<0.05)和28.96% (<0.01) (图3-A, C)。其对赤霉病侵染严重度无显著影响, 与RIL群体结果一致(图2-E和图3-B)。
PH: 株高; PIS: 侵染型平均病小穗率, 土壤表面赤霉病麦粒抛撒法对赤霉病抗性的测定; PDS: 扩展型平均病小穗率, 单花滴注法对赤霉病抗性的测定。‘×’在箱形图中为平均值标记; 箱形图中的点表示离群点; 数据框中的水平线表示中位数。*和**分别代表与偃展1号相比< 0.05和<0.01。
PH: plant height; PIS: the average percentage of infected spikelets, the measure of FHB resistance in soil surface inoculation; PDS: the average percentage of diseased spikelets, the measure of FHB resistance in point inoculation. ‘×’ in the data box indicates the mean value; the dots in the boxplots are the outliers; the horizontal line in the data box indicates the median. * and ** represent significant difference at< 0.05 and< 0.01 compared with Yanzhan 1, respectively.
2.6 QPh.yas-5A区间的候选基因发掘
进一步利用小麦中国春参考基因组2.1版本的信息, 在的定位区间挖掘到191个高置信基因, 其中有146个基因有注释功能, 主要涉及合成细胞色素P450 (水稻的同源基因是、和, 拟南芥的同源基因是)、脱水反应元件结合蛋白(水稻的同源基因是、和, 拟南芥的同源基因是)、乙烯响应转录因子(水稻的同源基因是和, 拟南芥的同源基因是和)、转录因子MYC2 (水稻的同源基因是)和细胞壁受体相关激酶等(水稻的同源基因是和) (附表2)。
3 讨论
3.1 QTL的比较分析
本研究共检测到7个株高QTL。由表2可得,的物理位置区间是18.40~20.42 Mb, 位于的置信区间附近,在染色体2D上511.67~515.21 Mb区间, 与以往研究中检测到的株高 QTL/基因存在差异, 推测是一个新的株高位点[22,52]。位于4B染色体上589.85 Mb到590.54 Mb之间, 与之前报道的[53]、[54]、[55]和[56]具有相近的置信区间, 这一区间与(544.65 Mb)的QTL相差45.20 Mb[57]。的物理区间在62.23~68.26 Mb, 与此前在7D染色体上检测到的株高QTL或基因的定位区间不同, 但是与春化基因在相同物理区间[57-59],是在7D染色体上检测到的另一个与株高有关的QTL, 其物理位置区间为147.70~395.89 Mb, 与控制抽穗期和株高相关基因(237.61 Mb)在相同的置信区间[60]。的物理位置区间是19.19 Mb至19.59 Mb, 与矮秆基因重叠[61]。经过检测, 发现偃展1号和扬麦4号分别携带的矮秆等位变异和高秆等位变异。的物理位置区间为516.44~536.32 Mb, 距离Yan等[62]在扬麦158/偃展1号 RIL群体中挖掘到的(506.00 Mb) 10 Mb和Zhang等[63]挖掘到的矮秆效应来源于KN9204的(479.12~487.31 Mb) 29 Mb, 与Schnurbusch等[64]在‘Arina×Forno’ F5:7杂交群体中挖掘出来的(530.98 Mb)和胡文静等[51]在人工合成小麦C615中挖掘到的株高位点(519.89 Mb)位置重叠。此外,与位于5A染色体上的矮秆基因(698.89 Mb)相距较远[16]。
3.2 株高位点的抗赤霉病遗传效应分析
以往研究中发现,,对赤霉病具有显著效应, 其中增加株高的基因型,和对赤霉病抗性具有显著的增效作用[36-38]。本研究结果表明和的矮秆等位变异可显著增加PIS值, 而与矮秆等位变异可显著降低PDS值。其中对株高的效应值仅次于, 且在所有环境中均能检测到, 在育种中可加以利用。Hu等[46]利用同一RIL群体进行了抗扩展型赤霉病QTL定位, 共挖掘到了5个抗赤霉病QTL, 其在2D和5A染色体上分别定位到了(528.39~531.66 Mb)和(547.10~548.10 Mb), 与本研究定位到的2D和5A染色体上株高QTL虽然不在同一区间, 但是相距较近:与相距13.18 Mb,与相距10.78 Mb。因此, 我们推测在2D和5A的这两个相近区间上存在株高和抗赤霉病的基因, 并且具有连锁关系, 还需通过精细定位和克隆进行验证。
3.3 QPh.yas-5A 的矮秆效应来源和育种价值分析
矮秆效应来源于扬麦4号, 扬麦4号是江苏里下河地区农业科学研究所用南大2419×胜利麦杂交后的F5代, 再与阿夫杂交育成[26], 利用KASP-5A对南大2419、胜利麦和阿夫进行检测, 结果表明仅阿夫在该位点呈现与扬麦4号相同的等位变异, 说明的矮秆等位变异来源于阿夫。利用的功能标记KASP-5A在126份小麦品种(系)中进行分子检测, 再与自然群体的株高、PIS和PDS表型数据结合进行分析, 结果表明对赤霉病抗扩展和对株高的效应与RIL群体中的结果一致。位点的矮秆等位变异不仅显著降低小麦株高还显著增加赤霉病抗扩展能力, 因此具有较大的育种应用潜力。中国早期的地方品种望水白、白三月黄等和早期栽培品种例如苏麦3号、扬麦4 号等均携有、位点的高秆等位变异, 其中苏麦3号和扬麦4号携有的矮秆等位变异, 望水白、白三月黄和苏麦3号的常年株高是125~140 cm, 但是扬麦4号的株高常年是92~100 cm, 经过检测发现望水白、白三月黄和苏麦3号均不携有的矮秆等位变异, 说明对扬麦4号的降秆起到重要作用。一般小麦品种(系)的PDS值小于25%可以界定为赤霉病抗扩展达到抗-中抗的水平, 株高小于85 cm可以界定为矮秆, 我们以此为依据结合的分子标记辅助选择发掘RIL群体和自然群体中矮秆且抗性好的材料, 发现RIL群体中家系RIL-31、RIL-32、RIL-50、RIL-136、RIL-146和RIL-147携有矮秆等位变异, 株高小于85 cm, 且PDS值小于25%, 自然群体中鄂麦174、宁1616、宁17342、宁麦24、宁麦15318、苏麦5号、扬辐麦3048、扬辐麦4188、扬辐麦9、扬麦20、扬麦21和镇麦11这12个品种携有矮秆等位变异, 株高小于85 cm, 且PDS值小于25%, 这些筛选出的品种(系)可以作为抗赤霉病遗传改良的优异抗源。
3.4 QPh.yas-5A区间的候选基因分析
的定位区间挖掘到的高置信基因主要涉及合成细胞色素P450、脱水反应元件结合蛋白、乙烯响应转录因子、转录因子MYC2和细胞壁受体相关激酶等。前人报道中证实小麦对赤霉病病原体和真菌毒素的应答与细胞色素P450相关基因的表达显著相关[65], 靶向沉默宿主诱导的细胞色素P450的CYP51 (lanosterol C-14α-demethylase)基因是一种控制真菌病害的方法[66]。乙烯响应转录因子(ethylene-responsive transcription factor)作为转录激活剂, 与GCC-box致病相关启动子元件结合, 在植物发育过程中参与基因表达的调控, 或受胁迫因子和胁迫信号转导通路组分的介导[67-68]。细胞壁受体相关激酶(wall-associated receptor kinase 5)如丝氨酸/苏氨酸蛋白激酶可能作为细胞外基质成分的信号受体,与果胶的结合可能在控制细胞扩张、形态发生和发育方面具有重要意义[69]。综合上述分析结果表明区间不仅与植物生长发育相关, 与抗病相关的机制也具有一定的联系, 因此精细定位和克隆该基因对小麦产量与抗病育种具有一定的价值。
4 结论
定位到7个株高的QTL, 其中和在6个环境中均能检测到, 表型贡献率范围分别为19.48%~44.11%和10.48%~13.71%,与已知矮秆基因一致。和位点的矮秆等位变异可以显著降低株高和赤霉病扩展严重度。分析目标区间的候选基因发现该区间与植物生长发育和对病害的响应有关。进一步开发位点的分子标记KASP-5A, 可促进其应用于小麦株高性状的遗传改良。
附表 请见网络版: 1) 本刊网站http://zwxb. chinacrops.org/; 2) 中国知网http://www.cnki.net/; 3) 万方数据http://c.wanfangdata.com.cn/Periodical- zuowxb.aspx。
[1] Liu T, Wu L, Gan X, Chen W, Liu B, Fedak G, Cao W, Chi D, Liu D, Zhang H, Zhang B. Mapping quantitative trait loci for 1000-grain weight in a double haploid population of common wheat., 2020, 21: 3960.
[2] Su Z, Jin S, Lu Y, Zhang G R, Chao S M, Bai G H. Single nucleotide polymorphism tightly linked to a major QTL on chromosome 7A for both kernel length and kernel weight in wheat., 2016, 36: 1–11.
[3] Sakamoto T, Matsuoka M. Generating high-yielding varieties by genetic manipulation of plant architecture., 2004, 15: 144–147.
[4] Peng J R, Richards D E, Hartley N M, Murphy G P, Devos K M, Flintham J E, Beales J, Fish L J, Worland A J, Pelica F, Sudhakar D, Christou P, Snape J W, Gale M D, Harberd N P. ‘Green revolution’ genes encode mutant gibberellin response modulators., 1999, 400: 256–261.
[5] Bellucci A, Torp A M, Bruun S, Magid J, Andersen S B, Rasmussen S K. Association mapping in Scandinavian winter wheat for yield, plant height, and traits important for second-generation bioethanol production., 2015, 6: 1046.
[6] Cadalen T, Sourdille P, Charmet G, Tixier M H, Gay G, Boeuf C, Bernard S, Leroy P, Bernard M. Molecular markers linked to genes affecting plant height in wheat using a doubled-haploid population., 1998, 96: 933–940.
[7] Börner A, Schumann E, Fürste A, Cöster H, Leithold B, Röder S, Weber E. Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (L.)., 2002, 105: 921–936.
[8] Borrell A K, Incoll L D, Dalling M J. The influence of theandalleles on the growth of wheat stems and ears., 1991, 67: 103–110.
[9] Tang N, Jiang Y, He B R, Hu Y G. The effects of dwarfing genes (,, and) with different sensitivity to GA (3) on the coleoptile length and plant height of wheat., 2009, 8: 1028–1038.
[10] Akman H, Bruckner P. Marker assisted selection foranddwarfing genes in winter wheat breeding program., 2012, 29: 139.
[11] Rasheed A, Wen W E, Gao F M, Zhai S N, Jin H, Liu J D, Guo Q, Zhang Y J, Dreisigacker S, Xia X C, He Z H. Development and validation of KASP assays for genes underpinning key economic traits in bread wheat., 2016, 129: 1843–1860.
[12] Korzun V, Roder M S, Ganal M W, Worland A J, Law C N. Genetic analysis of the dwarfing gene () in wheat. Part I: Molecular mapping ofon the short arm of chromosome 2D of bread wheat (L.)., 1998, 96: 1104–1109.
[13] Chai L, Xin M, Dong C, Chen Z, Zhai H, Zhuang J, Cheng X, Wang N, Geng J, Wang X, Bian R, Yao Y, Guo W, Hu Z, Peng H, Bai G, Sun Q, Su Z, Liu J, Ni Z. A natural variation in Ribonuclease H-like gene underliesto confer ‘Green Revolution’ trait in wheat., 2022, 15: 377–380.
[14] Xiong H C, Zhou C Y, Fu M Y, Guo H J, Xie Y D, Zhao L S, Gu J Y, Zhao S R, Ding Y P, Li Y T, Zhang J Z, Wang K, Li X J, Liu L X. Cloning and functional characterization of, a ‘Green Revolution’ replacement gene in wheat., 2022, 15: 373–376.
[15] 徐相波, 张爱民, 李新华, 孙永堂. 小麦矮源的利用和矮秆基因的研究进展. 核农学报, 2001, 15: 188–192. Xu X B, Zhang A M, Li X H, Sun Y T. Utilization of dwarf source and research progress of dwarf gene in wheat., 2001, 15: 188–192 (in Chinese with English abstract).
[16] Sun L, Yang W, Li Y, Shan Q, Ye X, Wang D, Yu K, Lu W, Xin P, Pei Z, Guo X, Liu D, Sun J, Zhan K, Chu J, Zhang A. A wheat dominant dwarfing line with, which reduces stem cell length and affects gibberellic acid synthesis, is a 5AL terminal deletion line., 2019, 97: 887–900.
[17] Ford B A, Foo E, Sharwood R, Karafiatova M, Vrána J, MacMillan C, Nichols D S, Steuernagel B, Uauy C, Doležel J, Chandler P M, Spielmeyer W.semidwarfism in wheat is due to increasedexpression and reduced GA content., 2018, 177: 168–180.
[18] Peng Z S, Li X, Yang Z J, Liao M L. A new reduced height gene found in the tetraploid semi-dwarf wheat landrace Aiganfanmai., 2011, 10: 2349–2357.
[19] Tian X, Xia X, Xu D, Liu Y, Xie L, Hassan M A, Song J, Li F, Wang D, Zhang Y, Hao Y, Li G, Chu C, He Z, Cao S., an ancient variation of, reduces plant height without yield penalty in wheat., 2022, 233: 738–750.
[20] Borrill P, Mago R, Xu T, Ford B, Williams S J, Derkx A, Bovill W D, Hyles J, Bhatt D, Xia X, MacMillan C, White R, Buss W, Molnár I, Walkowiak S, Olsen O A, Doležel J, Pozniak C J, Spielmeyer W. An autoactivegene causesdwarfism in wheat., 2022, 119: e2209875119.
[21] Chen L, Yang Y, Cui C G, Lu S, Lu Q M, Du Y Y, Su R, Chai Y M, Li H J, Chen F Z, Yu F, Hu Y G. Effects ofandon developmental and agronomic traits indwarf plants of bread wheat., 2018, 219: 24–32.
[22] Chai L, Chen Z, Bian R, Zhai H, Cheng X, Peng H, Yao Y, Hu Z, Xin M, Guo W, Sun Q, Zhao A N. Dissection of two quantitative trait loci with pleiotropic effects on plant height and spike length linked in coupling phase on the short arm of chromosome 2D of common wheat (L.)., 2018, 131: 1815–1831.
[23] Li T, Deng G B, Su Y, Yang Z, Tang Y Y, Wang J H, Qiu X B, Pu X, Li J, Liu Z H, Zhang H L, Liang J J, Yang W Y, Yu M Q, Wei Y M, Long H. Identification and validation of two major QTL for spike compactness and length in bread wheat (L.) showing pleiotropic effects on yield-related traits., 2021, 134: 3625–3641.
[24] Zhang L, Luo P G, Ren Z L, Zhang H Y. ControllingFusarium head blight of wheat (L.) with genetics., 2011, 2: 263–270.
[25] Zhang K P, Wang J J, Qin H J, Wei Z Y, Hang L B, Zhang P W, Reynolds M, Wang D W. Assessment of the individual and combined effects ofandon plant height, time to heading and yield traits in common wheat., 2019, 7: 845–856.
[26] Zhu Z W, Hao Y F, Mergoum M, Bai G H, Humphreys G, Cloutier S, Xia X C, He Z H. Breeding wheat for resistance to Fusarium head blight in the Global North: China, USA, and Canada., 2019, 7: 730–738.
[27] 陈云, 王建强, 杨荣明, 马忠华. 小麦赤霉病发生危害形势及防控对策. 植物保护, 2017, 43: 11–17. Chen Y, Wang J Q, Yang R M, Ma Z H. Current situation and management strategies of Fusarium head blight in China., 2017, 43: 11–17 (in Chinese with English abstract).
[28] Zhu Z W, Xu X T, Fu L P, Wang F J, Dong Y C, Fang Z W, Wang W, Chen Y P, Gao C B, He Z H, Xia X C, Hao Y F. Molecular mapping of quantitative trait loci for fusarium head blight resistance in a doubled haploid population of Chinese bread wheat., 2021, 105: 1339–1345.
[29] Schroeder H W, Christensen J J. Factors affecting resistance of wheat to scab caused by., 1963, 53: 831–838.
[30] 胡文静, 张勇, 陆成彬, 王凤菊, 刘金栋, 蒋正宁, 王金平, 朱展望, 徐小婷, 郝元峰, 何中虎, 高德荣. 小麦品种扬麦16赤霉病抗扩展QTL定位及分析. 作物学报, 2020, 46: 157–165. Hu W J, Zhang Y, Lu C B, Wang F J, Liu J D, Jiang Z N, Wang J P, Zhu Z W, Xu X T, Hao Y F, He Z H, Gao D R. Mapping and genetic analysis of QTLs for Fusarium head blight resistance to disease spread in Yangmai 16., 2020, 46: 157–165 (in Chinese with English abstract).
[31] 陆成彬, 范金平, 印娟, 王朝顺, 褚正虎. 小麦主要农艺性状对赤霉病抗性的影响. 安徽农业科学, 2013, 41: 1091–1092. Lu C B, Fang J P, Yin J, Wang C S, Chu Z H. Effects of main agronomic traits of wheat on the resistance of Fusarium head blight., 2013, 41: 1091–1092 (in Chinese with English abstract).
[32] 陆成彬, 张伯桥, 范金平, 吴荣林, 王朝顺, 褚正虎. 2个重组自交系群体的小麦赤霉病抗性与表型性状相关性. 江苏农业科学, 2012, 40: 99–101. Lu C B, Zhang B Q, Fan J P, Wu R L, Wang C S, Chu Z H. Correlation between resistance to Fusarium head blight and phenotypic traits in two recombinant inbred lines., 2012, 40: 99–101 (in Chinese with English abstract).
[33] 陈士强, 陈秀兰, 张容, 王建华, 王锦荣, 黄向明, 何震天. 小麦赤霉病抗性与株高的相关性研究. 江苏农业科学, 2015, 43: 144–147. Chen S Q, Chen X L, Zhang R, Wang J R, Huang X M, He Z T. Study on correlation between resistance toFusariumhead blight and plant height in wheat., 2015, 43: 144–147 (in Chinese with English abstract).
[34] Draeger R, Gosman N, Steed A, Chandler E, Thomsett M, Srinivasachary, Schondelmaier J, Buerstmayr H, Lemmens M, Schmolke M, Mesterhazy A, Nicholson P. Identification of QTLs for resistance to Fusarium head blight, DON accumulation and associated traits in the winter wheat variety Arina., 2007, 115: 617–625.
[35] Mao S L, Wei Y M, Cao W. Confirmation of the relationship between plant height and Fusarium head blight resistance in wheat (L.) by QTL meta-analysis., 2010, 174: 343–356.
[36] Srinivasachary Gosman N, Steed A, Simmonds J, Leverington-Waite M, Wang Y, Snape J, Nicholson P. Susceptibility to Fusarium head blight is associated with thesemi-dwarfing allele in wheat., 2008, 116: 1145–1153.
[37] Srinivasachary Gosman N, Steed A, Hollins T W, Bayles R, Jennings P, Nicholson P. Semi-dwarfingandloci of wheat differ significantly in their influence on resistance to Fusarium head blight., 2009, 118: 695–702.
[38] Hu W J, Wu H Y, Lu C B, Zheng X, Jia J, Xu W G. Genetic dissection of quantitative trait loci for spikelets compactness in two Yanzhan 1-derived recombinant inbred line wheat populations., 2022, 141: 719–732.
[39] Hu W J, Zhu D M, Zhang Y, Liu J, Zhao D, Liao S, Jia J Z, Xu W G. Quantitative trait loci mapping for heading date and spikelet number in wheat (L.) based on two recombinant inbred line populations., 2023, 70: 1179–1195.
[40] Hu W J, Liao S, Zhao D, Jia J Z, Xu W G, Cheng S H. Identification and validation of quantitative trait loci for grain size in bread wheat (L.)., 2022,12: 822.
[41] Nyquist W E, Baker R J. Estimation of heritability and prediction of selection response in plant populations., 1991, 10: 235–322.
[42] Holland J B, Nyquist W E, Cervantes-Martínez C T. Estimating and interpreting heritability for plant breeding: an update., 2003, 22: 9–112.
[43] Meng L, Li H H, Zhang L Y, Wang J K. QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations., 2015, 3: 269–283.
[44] Ma Z Q, Sorrells M E. Genetic analysis of fertility restoration in wheat using restriction fragment length polymorphisms., 1995, 35: 1137–1143.
[45] Xu X T, Zhu Z W, Jia A L, Wang F J, Wang J P, Zhang Y L, Fu C, Fu L P, Bai G H, Xia X C, Hao Y F, He Z H. Mapping of QTL for partial resistance to powdery mildew in two Chinese common wheat cultivars., 2019, 216: 1.
[46] Hu W J, Gao D R, Zhang Y, Zheng X, Lu C, Wu H, Xu W, Cheng S H, Jia J Z. Mapping quantitative trait loci for type II fusarium head blight resistance in two wheat recombinant inbred line populations derived from Yangmai 4 and Yangmai 5., 2023, 107: 422–430.
[47] Li T, Deng G B, Su Y, Yang Z, Tang Y Y, Wang J H, Qiu X B, Pu X, Li J, Liu Z H, Zhang H L, Liang J J, Yang W Y, Yu M Q, Wei Y M, Long H. Identification and validation of two major QTLs for spike compactness and length in bread wheat (L.) showing pleiotropic effects on yield-related traits., 2021, 134: 3625–3641.
[48] Jiang P, Zhang X, Wu L, He Y, Zhuang W, Cheng X, Ge W, Ma H, Kong L. A novel QTL on chromosome 5AL of Yangmai 158 increases resistance to Fusarium head blight in wheat., 2019, 69: 249–258.
[49] Chen Y, Song W, Xie X, Wang Z, Guan P, Peng H, Jiao Y, Ni Z, Sun Q, Guo W. A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the Triticeae tribe as a pilot practice in the plant pangenomic era., 2020, 13: 1694–1708.
[50] Ma S W, Wang M, Wu J H, Guo W L, Chen Y M, Li G W, Wang Y P, Shi W M, Xia G M, Fu D L, Kang Z S, Ni F. WheatOmics: a platform combing multiple omics data to accelerate functional genomics studies in wheat., 2021, 14: 1965–1968.
[51] 胡文静, 李东升, 裔新, 张春梅, 张勇. 小麦穗部性状和株高的QTL定位及育种标记开发和验证. 作物学报, 2022, 48: 1346–1356. Hu W J, Li D S, Yi X, Zhang C M, Zhang Y. Molecular mapping and validation of quantitative trait loci for spike-related traits and plant height in wheat., 2022, 48: 1346–1356 (in Chinese with English abstract).
[52] Botwright T L, Rebetzke G J, Condon A G, Richards R A. Influence of the gibberellin-sensitivedwarfing gene on leaf epidermal cell dimensions and early vigour in wheat (L.)., 2015, 95: 631–639.
[53] Huang X Q, Cöster H, Ganal M W, Röder M S. Advanced backcross QTL analysis for the identification of quantitative trait loci alleles from wild relatives of wheat (L.)., 2003, 106: 1379–1389.
[54] McCartney C A, Somers D J, Humphreys D G, Lukow O, Ames N, Noll J, Cloutier S, McCallum B D. Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL4452 × ‘AC Domain’., 2005, 48: 870–883.
[55] Hai L, Guo H J, Wagner C, Xiao S H, Friedt W. Genomic regions for yield and yield parameters in Chinese winter wheat (L.) genotypes tested under varying environments correspond to QTL in widely different wheat materials., 2008, 175: 226–232.
[56] Jia H Y, Wan H S, Yang S H, Zhang Z Z, Kong Z X, Xue S L, Zhang L X, Ma Z Q. Genetic dissection of yield-related traits in a recombinant inbred line population created using a key breeding parent in China’s wheat breeding., 2013, 126: 2123–2139.
[57] Yan H, Li G, Shi J, Tian S, Zhang X, Cheng R, Wang X, Yuan Y, Cao S, Zhou J, Kong Z, Jia H, Ma Z. Genetic control of Fusarium head blight resistance in two Yangmai 158-derived recombinant inbred line populations., 2021, 134: 3037–3049.
[58] Gao F, Wen W, Liu J, Rasheed A, Yin G, Xia X, Wu X, He Z. Genome-wide linkage mapping of QTL for yield components, plant height and yield-related physiological traits in the Chinese wheat cross Zhou 8425B/Chinese Spring., 2015, 6: 1099.
[59] Zhang N, Fan X, Cui F, Hao C, Zhang W, Zhao X, Yang L, Pan R, Chen M, Han J, Ji J, Liu D, Zhao Z, Tong Y, Zhang A, Wang T, Li J. Characterization of the temporal and spatial expression of wheat (L.) plant height at the QTL level and their influence on yield-related traits., 2017, 130: 1235–1252.
[60] Zhang L, Zhang H, Qiao L Y, Miao L F, Yan D, Liu P, Zhao G Y, Jia J Z, Gao L F. Wheat MADS-box genenegatively regulates heading date., 2021, 9: 1115–1123.
[61] Tian X, Zhu Z, Xie L, Xu D, Li J, Fu C, Fu C, Chen X, Wang D, Xia X, He Z, Cao S. Preliminary exploration of the source, spread, and distribution ofreducing height in bread wheat., 2019, 59: 19–24.
[62] Yan H, Li G, Shi J, Tian S, Zhang X, Cheng R, Wang X, Yuan Y, Cao S, Zhou J, Kong Z, Jia H, Ma Z. Genetic control of Fusarium head blight resistance in two Yangmai 158-derived recombinant inbred line populations., 2021, 134: 3037–3049
[63] Zhang N, Fan X, Cui F, Zhao C, Zhang W, Zhao X, Yang L, Pan R, Chen M, Han J, Ji J, Liu D, Zhao Z, Tong Y, Zhang A, Wang T, Li J. Characterization of the temporal and spatial expression of wheat (L.) plant height at the QTL level and their influence on yield-related traits., 2017, 130: 1235–1252.
[64] Schnurbusch T, Paillard S, Fossati D, Messmer M, Schachermayr G, Winzeler M, Keller B. Detection of QTLs for Stagonospora glume blotch resistance in Swiss winter wheat., 2003, 107: 1226–1234.
[65] Koch A, Kumar N, Weber L, Keller H, Imani J, Kogel K H. Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species., 2013, 110: 19324–19329.
[66] Li X, Zhang J B, Song B, Li H P, Xu H Q, Qu B, Dang F J, Liao Y C. Resistance to fusarium head blight and seedling blight in wheat is associated with activation of a cytochrome P450 gene., 2010, 100: 183–191.
[67] Oñate-Sánchez L, Singh K B. Identification ofethylene-responsive element binding factors with distinct induction kinetics after pathogen infection., 2002, 128: 1313–1322.
[68] Berrocal-Lobo M, Molina A, Solano R. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 inconfers resistance to several necrotrophic fungi., 2002, 29: 23–32.
[69] Theologis A, Ecker J R, Palm C J, Federspiel N A, Kaul S, White O, Alonso J, Altafi H, Araujo R, Bowman C L, Brooks S Y, Buehler E, Chan A, Chao Q, Chen H, Cheuk R F, Chin C W, Chung M K, Conn L, Conway A B, Conway A R, Creasy T H, Dewar K, Dunn P, Etgu P, Feldblyum T V, Feng J, Fong B, Fujii C Y, Gill J E, Goldsmith A D, Haas B, Hansen N F, Hughes B, Huizar L, Hunter J L, Jenkins J, Johnson-Hopson C, Khan S, Khaykin E, Kim CJ, Koo H L, Kremenetskaia I, Kurtz D B, Kwan A, Lam B, Langin-Hooper S, Lee A, Lee J M, Lenz C A, Li J H, Li Y, Lin X, Liu S X, Liu Z A, Luros J S, Maiti R, Marziali A, Militscher J, Miranda M, Nguyen M, Nierman W C, Osborne B I, Pai G, Peterson J, Pham P K, Rizzo M, Rooney T, Rowley D, Sakano H, Salzberg S L, Schwartz J R, Shinn P, Southwick A M, Sun H, Tallon L J, Tambunga G, Toriumi M J, Town C D, Utterback T, Van Aken S, Vaysberg M, Vysotskaia V S, Walker M, Wu D, Yu G, Fraser C M, Venter J C, Davis R W. Sequence and analysis of chromosome 1 of the plant., 2000, 408: 816–820.
Detection and verification of QTL for plant height in Yangmai 4/Yanzhan 1 recombinant inbred lines population and their genetic effects on Fusarium head blight resistance
ZHAO Die1,2, HU Wen-Jing2,3,*, CHENG Xiao-Ming2, WANG Shu-Ping1, ZHANG Chun-Mei2, LI Dong-Sheng2, and GAO De-Rong2,3,*
1College of Agriculture, Yangtze University, Jingzhou 434025, Hubei, China;2Lixiahe Institute of Agriculture Sciences / Key Laboratory of Wheat Biology and Genetic Improvement for Low and Middle Yangtze Valley, Ministry of Agriculture and Rural Affairs, Yangzhou 225007, Jiangsu, China;3Jiangsu Co-Innovation Center for Modern Production Technology of Grain Crops / Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding, Yangzhou University, Yangzhou 225009, Jiangsu, China
Plant height (PH) is associated with fusarium head blight (FHB) resistance in wheat. In this study, a recombinant inbred lines (RIL) population derived from the cross of Yangmai 4/Yanzhan 1 (YM4/YZ1) was used to mine the quantitative trait loci (QTL) of PH traits using 55K SNP (single-nucleotide polymorphism) chip data, combined with the PH data of RIL population and their parents in six environments for three continuous years.Soil surface inoculation method and single spikelet inoculation were used to identify the FHB resistance to infection (Type I) and spread (Type II), respectively. Seven QTLs related to PH were detected on chromosomes 2D, 4B, 4D, 5A, and 7D, and onlymay be a new QTL of PH after comparing with previous studies. The dwarfing effects of,, andwere derived from YM4, and the dwarfing effects of the other four QTLs were derived from YZ1. Bothandcould be detected in six environments, and phenotypic variation explained (PVE) rates ranged from 19.48%–44.11% and 10.48%–13.71%, respectively. The increasing alleles at theand(YM4 allele) significantly reduced the average percentage of infected spikelets (PIS) by 34.97% and 19.09%, respectively. The dwarfing alleles atand(YM4 allele) significantly reduced the average percentage of diseased spikelets (PDS) by 24.73% and 14.56%, respectively. The dwarfing allele ofwas derived from Funo. Furthermore, we preliminary analyzedthe genes within the physical interval ofusing the reference genome information of wheat version 2.1. A total of 146 high-confidence annotated genes were detected in the target interval, which were mainly involved in the synthesis of cytochrome P450, dehydration response element-binding protein, ethylene response transcription factor, transcription factor MYC2, and cell wall receptor-associated kinases. The SNP marker closely linked towas further converted into kompetitive allele-specific PCR marker KASP-5A, and its effect on plant height and FHB resistance was then verified in 126 wheat cultivars (lines). The results of this study could provide a solid foundation for future fine mapping.
; plant height; fusarium head blight; QTL;; candidate gene
2023-05-24;
2023-06-15.
通信作者(Corresponding author): 胡文静, E-mail: huren2008@126.com; 高德荣, E-mail: gdr@wheat.org.cn
10.3724/SP.J.1006.2023.31005
E-mail: zd2021720791@163.com
2023-01-10;
本研究由国家自然科学基金项目(31901544), 泰州市科技计划项目(TN202117), 江苏现代农业产业单项技术研发(CX(21)3063)和江苏省重点研发项目(BE2021335)资助。
This study was supported by the National Natural Science Foundation of China (31901544), the Taizhou Science and Technology Project (TN202117), the Jiangsu Modern Agricultural Industry Single Technology Research and Development (CX(21)3063), and the National Key Research and Development Program of Jiangsu (BE2021335).
URL: https://kns.cnki.net/kcms2/detail/11.1809.S.20230613.1216.006.html
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).

