Uncovering the potential pathological mechanism of acute pancreatitis in patients with COVID-19 by bioinformatics methods
Zhaodi Wang ,Ping Wang ,Xuan Lu ,Congying Song ,Shuai Jiang ,Li Li ,Yuanqiang Lu
1 Department of Nursing, the First Affiliated Hospital of Zhejiang University School of Medicine, Hangzhou 310003, China
2 Department of Geriatric and Emergency medicine, the First Affiliated Hospital of Zhejiang University School of Medicine,Hangzhou 310003, China
3 Key Laboratory for Diagnosis and Treatment of Aging and Physic-chemical Injury Diseases of Zhejiang Province,Hangzhou 310003, China
Severe acute respiratory syndrome coronavirus 2(SARS-CoV-2) has posed a serious challenge to emergency departments that usually encounter emergencies and severe diseases.[1-5]Angiotensin-converting enzyme 2 (ACE2),a protein that interacts with the viral spike protein(s),allows SARS-CoV-2 to penetrate epithelial cells.[6]There is mounting evidence that suggests that the digestive system may also be affected.[7]An observational study described potential patterns of pancreatic injury (elevated amylase and lipase) in patients with coronavirus disease 2019 (COVID-19).[8]Additionally,sporadic case reports have described secondary pancreatitis in patients with SARS-CoV-2 infection and presented imaging evidence.[9]Moreover,a previous study showed that COVID-19 patients not only have a higher risk of developing acute pancreatitis(AP),but also have a significantly higher mortality than those without COVID-19.[10]Using bioinformatics analysis,it may be possible to reveal how COVID-19 and AP are related.In this study,two RNA-seq datasets of SARS-CoV-2 and AP were selected for analysis.
METHODS
The microarray datasets of COVID-19 (GSE152641)and AP (GSE194331) samples were both obtained from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/)to detect mutual differentially expressed genes ( DEGs) (|log2FoldChange| ≥1.5 and adjustedP-value<0.05).A thorough investigation of the gene enrichment analysis was carried out to identify the functional enrichment of common DEGs.Then,a proteinprotein interaction (PPI) network was constructed to extract the hub genes.In addition,the inf iltration of various immune cells in the two diseases was analyzed based on the machine learning tool,Cell-type Identification By Estimating Relative Subsets of RNA Transcripts x (CIBERSORTx).The lncRNA-miRNA-mRNA network was constructed through the Encyclopedia of RNA Interactomes (ENCORI) database(http://starbase.sysu.edu.cn/).
RESULTS
Identification of common DEGs between COVID-19 and AP
Through analysis of GSE152641,we identif ied 1,216 DEGs and confirmed 399 DEGs from GSE194331.Volcano plots displayed dysregulated COVID-19 and AP genes,while comparison revealed 95 common DEGs(supplementary Figure 1).
Enrichment analysis of DEGs
The top 10 items in the categories of molecular function (MF),biological process (BP),and cellular component (CC) of gene ontology (GO) are summarized in Figure 1A. In the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis,the common DEGs were strongly involved in “Staphylococcus aureusinfection”,“transcriptional misregulation in cancer” and“interleukin-17 (IL-17) signaling pathway” (Figure 1B).Similarly,gene set enrichment analysis (GSEA) revealed significant enrichment of immune-related pathways in both diseases (Figures 1 C and D).These results further suggest that similar immune responses may occur during the course of COVID-19 and AP.
PPI network construction and hub gene identification
The PPI network of common DEGs was constructed by string (supplementary Figure 2).The 20 hub genes with the highest scores were then determined using five algorithms based on Cytoscape I.Afterward,we confirmed 14 hub genes,which were all highly expressed in both diseases.Finally,a total of 20 predicted co-expressed genes were included in this co-expression pattern based on the multiple properties of the relationship.Among the 20 co-expressed genes,cathelicidin antimicrobial peptide (CAMP),folate receptor 3 (FOLR3),and interleukin-1 alpha (IL-1α) were included.Thus,they may be involved in the coordinated pathways that cause AP in response to COVID-19.
Evaluation and analysis of immune cell inf iltration
Considering that the enrichment analysis identified multiple immune-related pathways,we evaluated immune cell infiltration in both diseases (Figures 2 A and B).Among them,only memory-resting CD4 T cells and memory-activated CD4 T cells were significantly different in COVID-19 and AP at the same time,and the trend of difference between the two diseases was consistent.We analyzed the correlation between 14 hub genes and common infiltrating immune cells.Figures 2 C and D show that almost 14 hub genes were negatively correlated with memory-resting CD4 T cells and positively correlated with memory-activated CD4 T cells in both diseases.
Construction of transcriptional regulatory networks
We aimed to detect substantial changes occurring at the transcriptional level and gain insights into the hub genes (supplementary Figure 3).The results showed that 49 transcription factor (TF) targets were predicted from the Jellyfish-based Assembly Sequence Polisher for Error Reduction (JASPER) database to generate the co-regulatory network (63 nodes and 101 edges).Two critical genes (PPARGandS100A8) had a higher degree.In addition,a miRNA-gene interaction network was built.The hsa-miR-27a-3p was highly enriched.
LncRNA-miRNA-mRNA ceRNA regulatory network
The lncRNA-miRNA-mRNA ceRNA regulatory network can lead to a much better understanding of gene function and regulation.The ceRNA regulatory network was visualized using Cytoscape and included 64 lncRNAs,77 miRNAs and 14 mRNAs (supplementary Figure 4).
DISCUSSION
The exact relationship between COVID-19 and AP,however,is still controversial and difficult to understand.The following is a summary of the explanations that could explain the relationship between AP and COVID-19: (1) Since ACE2 receptor proteins are present in the pancreatic ductal,acinar,and islet cells,infection of the pancreas with SARS-CoV-2 is undoubtedly possible.[11](2) Alterations in inflammatory responses induced by SARS-CoV-2 infection may result in pancreatic damage .[12,13](3) Additionally,medications used to treat COVID-19 may have a negative impact on the development of AP.To develop new tactics,it is vital to reveal the exact relationship between COVID-19 and AP.In this study,we used bioinformatics-related methods to investigate the potential interaction between COVID-19 and AP.
Inflammation may play a substantial role in the mechanism of COVID-19-induced AP according to the findings of BP term GO annotation analysis,which showed that the DEGs were considerably enriched in leukocyte migration and controlled inflammatory response .The notable GO-CC terms,such as tertiary granule,special granule,and secretory granule lumen,are related to neutrophil degranulation. Simultaneously,according to the enrichment pathway results,the common DEGs primarily focused on neutrophil degranulation and the IL-17 signaling pathway.The activated neutrophils play a critical role in the pathophysiology and progression of AP by initiating chemokine and cytokine cascades.[14]In addition,the IL-17 level and neutrophilia have been identified as markers associated with poor prognosis and severe respiratory symptoms in patients with COVID-19.Taken together,monitoring neutrophil activity may help to identify AP in COVID-19. In addition,MF analysis of GO terms showed that the DEGs were highly enriched for receptor for advanced glycation endproducts(RAGE) binding,calcium-dependent protein binding,and long-chain fatty acids binding. Leukocyte calciumdependent binding proteins are essential for inflammatory cell transmigration into epithelial tissue.Previous research has demonstrated that calcium-binding protein increases the severity of pancreatitis and intra-acinar events.[15]On the other hand,RAGE may not only contribute to the tissue damage induced by SARS-CoV-2 infection,[16]but also contribute to the development of pancreatitis by mediating nucleosome-induced absent in melanoma 2 (AIM2) inflammasome activation and the release of proinf lammatory mediators from macrophages.[17]
Both COVID-19 and AP have a pathophysiology that is strongly inf luenced by immune inf lammation.[18]According to the results of immune infiltration,COVID-19 patients and AP patients showed decreased memory-resting CD4 T cells and increased memory-activated CD4 T cells. T-cell immunity plays an indispensable role in the control of primary SARS-CoV-2 infection.[19]CD4 T cells have the ability to differentiate into several different cell types with the capacity to have direct antiviral activities.Among them,T helper type 1 (Th1) cells provide direct antiviral functions via the production of interferon-gamma (IFN-γ).[20]In animal studies,IFN- γ has been shown to increase serum interleukin(IL)-18 and reduce serum IL-27 to aggravate inf lammation in AP.[21]Therefore,the activation of memory CD4 T cells may be the cause of COVID-19-induced AP,which needs further verification and support by more experiments.We further deduced that hub genes highly expressed in both diseases,in particularARG1,FCGR1A,LCN2,andMCEMP1,may cause the emergence of AP in patients with COVID-19.Accordingly,ARG1has been demonstrated to be significantly upregulated in the whole blood of COVID-19 patients,suggesting that it may be a useful diagnostic biomarker.[22]
Non-coding RNAs (ncRNAs),including miRNAs and lncRNAs,cannot encode proteins,but they can perform their functions by regulating coding RNAs.The application of ncRNAs as biomarkers or intervention targets can provide new insights into the diagnosis and treatment of diseases.A coding transcriptional map of the peripheral immune response in COVID-19 patients revealed that hsa-miR-146a-5p is a crucial player in the etiology of the disease and acts as a hub regulator of the host immune response,[23]which is consistent with our findings. The hsa-miR-27a-3p,which was the most highly expressed miRNA in our study,has been identif ied as a crucial factor in regulating protein ubiquitination of SARS-CoV-2 infection.[24]We demonstrated the potential correlation between hsa-miR-34a-5p and AP,although further studies are needed.Our findings provide further details on the possible pathogenic mechanism behind COVID-19-induced AP and lay the groundwork for further research.
CONCLUSIONS
Our results provide evidence that the altered inf lammatory responses induced by SARS-CoV-2 infection may be the cause of COVID-19-induced AP.The activation of memory CD4 T cells may be the main cause of COVID-19-induced AP.These findings provide information for those trying to understand the underlying relationships between COVID-19 and AP.
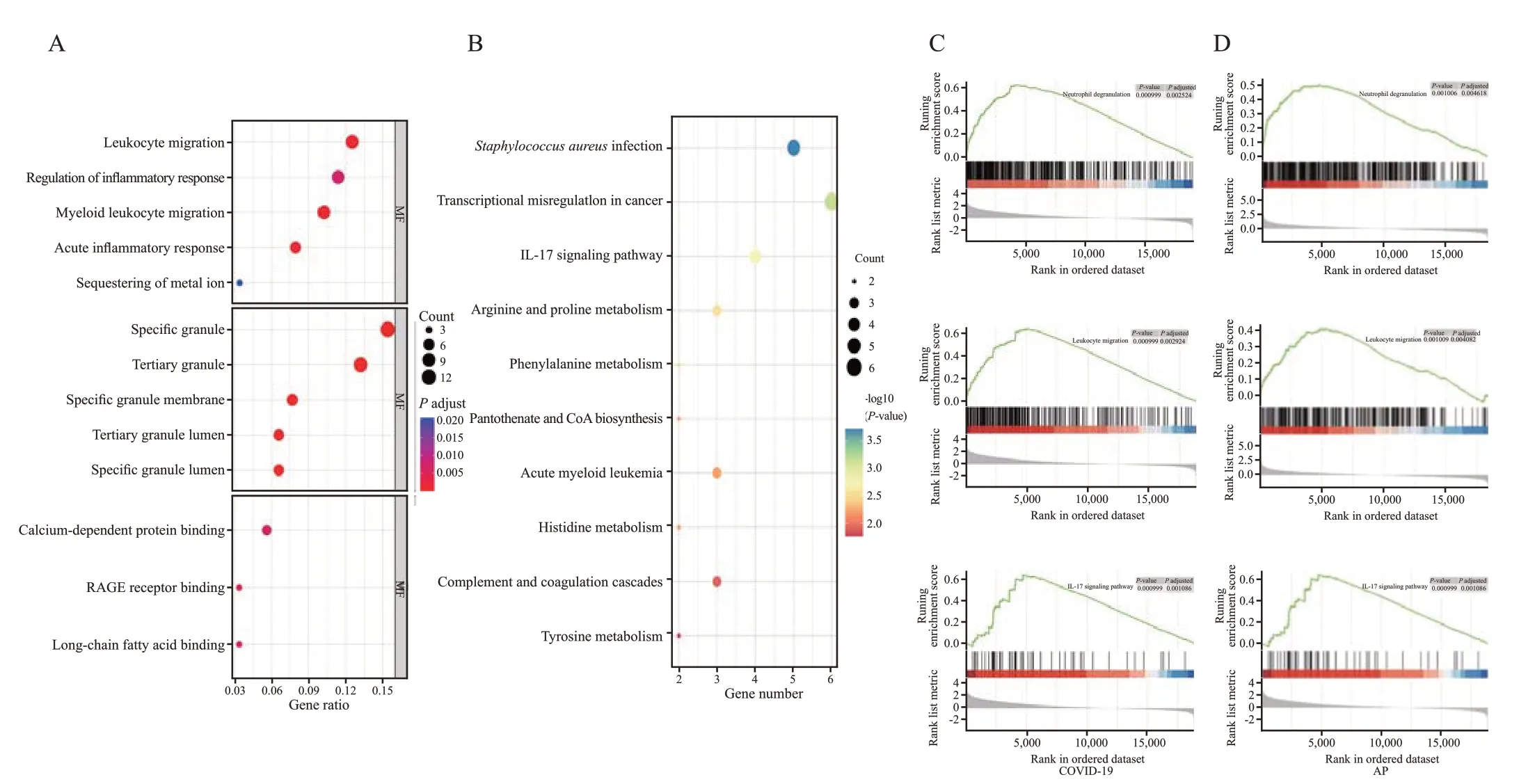
Figure 1.Enrichment analyses of DEGs.A and B: results of GO and KEGG enrichment analysis;C and D: results of GSEA in COVID-19 and AP.RAGE: receptor for advanced glycation endproducts;GSEA: gene set enrichment analysis;DEGs: differentially expressed genes;GO: gene ontology;KEGG: Kyoto Encyclopedia of Genes and Genomes;AP: acute pancreatitis.
ACKNOWLEDGMENTS
We thank all the patients who participated in this study and donated samples,as well as the GEO database for providing their platform.
Funding:This study was sponsored by the Key Research and Development Program of Zhejiang Province (2019C03076).The funders had no role in study design,data collection and analysis,decision to publish,or preparation of the manuscript.
Ethical approval:Not applicable.
Conf licts of interest:The authors declare that they have no conf licts of interest.
Contributors:ZDW and PW contributed equally to this work and share first authorship.ZDW and PW conceived,designed this research and drafted the manuscript.ZDW carried out the data analysis and data interpretation.PW is responsible for literature searching on the background and the image processing.XL and CYS annotated the picture and wrote the conclusion.JS and LL searched on the background of diseases and proofread the references.YQL reviewed and revised the manuscript.All authors contributed to the article and approved the submitted version.
All the supplementary files in this paper are available at http://wjem.com.cn.
 World journal of emergency medicine2023年5期
World journal of emergency medicine2023年5期
- World journal of emergency medicine的其它文章
- Emergency department approach to monkeypox
- The neuro-prognostic value of the ion shift index in cardiac arrest patients following extracorporeal cardiopulmonary resuscitation
- A prospective cohort study on serum A20 as a prognostic biomarker of aneurysmal subarachnoid hemorrhage
- Mendelian randomization study to investigate the causal relationship between plasma homocysteine and chronic obstructive pulmonary disease
- Cardiopulmonary prognosis of prophylactic endotracheal intubation in patients with upper gastrointestinal bleeding undergoing endoscopy
- Effects of mesencephalic astrocyte-derived neurotrophic factor on sepsis-associated acute kidney injury
