The circular RNA Rap1bpromotes Hoxa5transcription by recruiting Kat7 and leading to increased Fam3aexpression, which inhibits neuronal apoptosis in acute ischemic stroke
Fang-Fang Zhang, Liang Zhang*,, Lin Zhao, Yu Lu, Xin Dong, Yan-Qi Liu, Yu Li, Shuang Guo, Si-Yuan Zheng, Ying Xiao, Yu-Zhu Jiang
Abstract
Key Words:acetylation; apoptosis; circRap1b; circRNAs; epigenetics; histone modification; hypoxia; ischemia; neurons; stroke
Introduction
Stroke is a common neurological disease and a major cause of mortality and disability worldwide (Spescha et al., 2013; Knowland et al., 2014).The two main types of stroke are acute ischemic stroke (AIS) and hemorrhagic stroke.The current US Food and Drug Administration approved treatment for AIS is intravenous injection of recombinant human tissue plasminogen activator within 4.5 hours after symptom onset (Collen and Lijnen, 2009).However,because of the narrow time window and the risk of intracranial hemorrhage and angioedema, only about 7% of patients are eligible for this treatment,and its efficacy is limited (Schwamm et al., 2013; Denorme et al., 2016; Snow,2016; Alberts, 2017).In human patients and in animal models of stroke, acute ischemic primary neurons with blocked arterial blood supply die rapidly,while core and surrounding acute ischemic primary neurons that are infused by other arteries undergo delayed apoptosis to some extent (Broughton et al., 2009; Fann et al., 2013).Although many drugs have shown remarkable neuroprotective effects in animal models, these results have not been confirmed in human clinical trials (Wei et al., 2017).Given the complexity of ischemic brain injury and its systemic consequences, cell therapy is more effective than monotherapy, because it targets multiple mechanisms (Hess and Borlongan, 2008; Vieira et al., 2018).Circular RNAs (circRNAs) are a newly discovered class of non-coding RNAs.They are single-stranded RNAs that form a stable covalent bond between the 3’and 5’ends, generating a circular structure that does not include a polyadenylated tail and is not affected by RNA exonuclease (Memczak et al.,2013; Vicens and Westhof, 2014; Li et al., 2018).Recent studies have shown that circRNAs are highly conserved and abundantly expressed in human cells(Jeck et al., 2013; Memczak et al., 2013; Rybak-Wolf et al., 2015).Importantly,it has been found that circRNA expression is tissue-specific and regulated by different biological processes, such as stroke, neuroinflammation and aging, cardiac hypertrophy and heart failure, and cell growth (Boeckel et al.,2015; Wang et al., 2016, 2021; Du et al., 2017; Huang et al., 2017; Han et al.,2018; Yang et al., 2018b; Chen et al., 2022; Cheng et al., 2022).circRNAs can regulate gene expression at the transcriptional and post-transcriptional levels through miRNA interactions (Memczak et al., 2013).Heart-related circRNAs can act as endogenous sponges formiR-223(Wang et al., 2016), thereby preventing heart failure and cardiac hypertrophy.The circRNAciRS-7regulates the pathophysiology of Parkinson’s disease through adsorption ofmiRNA-7(Salzman, 2016).Recent studies have shown that circRNA expression in neurons (HT22 cells) subjected to transient focal ischemiain vitroand mouse brains subjected to hypoxia/reoxygenation is remarkably altered (Lin et al.,2016; Liu et al., 2017; Mehta et al., 2017).Ras-associated protein 1 (Rap1) is a small GTPase belonging to the Ras family that regulates a variety of cellular processes, including cell adhesion,proliferation, growth, differentiation, polarity, and apoptosis.Rap1aandRap1b, two variants of Rap1, are encoded by different genes but share 95%amino acid homology (Yan et al., 2008; Zhang et al., 2019).It has been shown that miRNAs targeting Rap1b inhibit cell migration, invasion, and metastasis in various tumor types (Zhang et al., 2012, 2019; Bischoff et al., 2014;Peng et al., 2014; Lin et al., 2015); however, the functional role of Rap1b in neurological diseases remains unclear.
Overexpression ofcircRap1bleads to a marked increase in expression of the transcription factorHoxa5.Transcription factors encoded by the hox gene family help determine the fate of neurons (Du et al., 2008).Specifically,Hoxa5was recently shown to be involved in the differentiation of neurons in the mouse brain into autonomic functional brain nuclei (Lizen et al., 2017) and the axonal growth of posterior vagus motor neurons (Barsh et al., 2017).Hoxa5is a crucial regulator of the neuronal inflammation caused by sevoflurane and Gm5106 (Zhu and Ma, 2021).Hoxa5also promotes neuroinflammation by activating the transforming growth factor signaling pathway (Nataf et al., 2019).Hernández et al.(2021) identifiedHoxa5as being important in the neuronal differentiation of human adipose-derived stem cells, and thus a potential candidate gene for the development of cell therapy strategies in neurological diseases.Apart from these studies, however, little is known about the role ofHoxa5in neurological injury, disease, or stroke, so we chose this as a focus for the current study.
Lysine acetyltransferase 7 (KAT7), a member of the MYST domain family,plays a crucial role in transcription and DNA replication by acetylating H3 lysine 14 (H3K14ac) and H4 lysines 5, 8, and 12 (H4K5ac, H4K8ac, H4K12ac)(Sapountzi and Côté, 2011; Taniue et al., 2020).KAT7 is also a driver of cellular senescence (Wang et al., 2021) and is essential for adult hematopoietic stem cell maintenance and self-renewal (Yang et al., 2022).Furthermore,KAT7 promotes efficient germination of tip cells during angiogenesis (Grant et al., 2021), and KAT7 overexpression attenuated cigarette extract-induced emphysema changes and lung cell apoptosis in mice with chronic obstructive pulmonary disease (Chen et al., 2020).
Family with sequence similarity 3A (FAM3A) is a mitochondrial protein that plays an important role in cellular stress adaptation and survival (Yan et al.,2019) and is involved in the regulation of neuronal apoptosis (Liu et al., 2019).FAM3Ahas been reported to inhibit interleukin-1β-induced chondrocyte apoptosis by activating the pro-survival PI3K/Akt/mTOR pathway (Yan et al., 2019).In addition,FAM3Acan improve hypoxic-ischemic brain injury in neonatal rats (Song et al., 2021).
We hypothesized thatin vitroinhibition of neuronal apoptosis bycircRap1b/Hoxa5might be mediated by Kat7-induced H3K14ac modification in theHoxa5promoter region.The findings from this study were expected to identify novel therapeutic targets for stroke.
Methods
HT22 cell culture and acute ischemic stroke treatment
The mouse hippocampal neuronal cell line HT22 (Cat# PNS-MS-83, RRID:CVCL_0321) was purchased from Procell Life Science & Technology Co., Ltd.(Wuhan, China), and its identify was confirmed by the short tandem repeat method.The cells were cultured in Dulbecco’s modified Eagle medium(DMEM, HyClone, Logan, UT, USA) supplemented with 10% fetal bovine serum(Gibco, Uruguay) in a humidified Calorstat incubator set at 37°C, with 5% CO2and 95% air.The HT22 cells were treated as oxygen-glucose deprivation to establish anin vitromodel of AIS.In brief, the culture medium was replaced by glucose-free medium without fetal bovine serum, and cells were cultured for 24 hours in a humidified tri-gas incubator (Sanyo, Osaka, Japan) at 37°C,with 3% O2and 5% CO2.Then, the previous culture medium was replaced by fresh DMEM supplemented with 10% fetal bovine serum and the cells were re-incubated for 24 hours in a humidified incubator at 37°C, with 5% CO2and 95% air.
Cell transfection
AcircRap1boverexpression (circRap1b(+)) plasmid and corresponding negative control plasmids were constructed based on the pLO5-ciRPuro vector (Geneseed Biotech, Guangzhou, China).Hoxa5andFam3aoverexpression plasmids (Hoxa5(+), concentration: 1150 μg/μL andFam3a(+),concentration: 1232 μg/μL) were constructed (GenePharma, Shanghai,China).Plasmids encoding siRNAs againstHoxa5,Fam3a, andKat7(Hoxa5(–),concentration: 1040 μg/μL;Fam3a(–), concentration: 1182 μg/μL andsiKat7,concentration: 1315 μg/μL) and a corresponding negative control (NC)plasmid were constructed by GeneChem (Shanghai, China).HT22-AIS cells were plated in 24-well plates.When the cell confluency reached 70–80%, the cells were transfected with the above plasmids using the transfection reagent Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) and then screened for successful transfection by selection with G428 or puromycin.The transfection efficiency was determined by quantitative real-time polymerase chain reaction and western blot assay.
Experimental design
Adult male C57BL/6J mice aged 12–14 weeks (n= 60, weighing 24–30 g)were purchased from Spafu (Beijing) Biotechnology Co., Ltd.(Beijing, China,license No.SCXK (Jing) 2019-0010).Animals exhibiting obvious neurological complications were excluded from the experiments.All of the animal experiments were approved by the Ethics Committee of China Medical University (approval No.CMU2021702) on October 8, 2021.The animal experiment procedures followed the Animal Research: Reporting ofIn VivoExperiments (ARRIVE) guidelines (Percie du Sert et al., 2020).Mice were bred in an environment with controlled temperature (24–26°C) and humidity(45–50%) conditions and a 12/12-hour light/dark cycle, as well as standard mouse diet and water.To investigate the effect of AIS on hippocampal neurons in mice, the mice were randomly divided into two groups: the Sham and AIS groups (n= 5/group).To investigate the effect ofcircRap1bandHoxa5on hippocampal neuronal apoptosis, the mice in the AIS group were randomly assigned using a random number list to one of five groups: AIS,AIS +circRap1b(+)NC +Hoxa5(+)NC, AIS +circRap1b(+) +Hoxa5(+)NC, AIS +circRap1b(+)NC +Hoxa5(+), and AIS +circRap1b(+) +Hoxa5(+) (n= 11/group).Permanent middle cerebral artery occlusion (pMCAO) was performed as described previously (Longa et al., 1989).In brief, mice were anesthetized with 1.5% isoflurane (Shengqi pharmaceutical Co, Ltd., Jinan, Shandong,China) in a mixture of 30% O2/70% N2O.After anesthesia, mice were placed on the operation table and disinfected.Then, a midline incision was made to the skin of the neck.The muscle and fascia were bluntly separated.Caution was taken not to injure the vagus nerves during separation of the carotid artery because injury to the vagus nerves may affect breathing and result in death.The common, external, and internal carotid arteries were separated.The part of common carotid artery distal to the heart was clamped at the bifurcation of the internal carotid artery with an arterial clamp.A small cut was made in the common carotid artery, and a nylon wire was inserted into the cut at an angle of 45°.The wire head was anchored into the artery bifurcation, and the prepared wire was then ligated tightly to ensure its insertion into the artery.The artery clamp at the bifurcation was then removed.The wire was generally inserted 8–10 mm, until resistance was felt.The wire was not too long, to avoid it being pulled out when the mice woke up.After closure of the skin wound, the mice were put back in their cages for feeding.Mice in which successful pMCAO was detected within 24 hours were defined as acute ischemic stroke models and used in the subsequent experiments.Plasmids encodingcircRap1b(pLO5-ciR-circRap1b-Puro, Geneseed Biotech, Guangzhou, China) andHoxa5(LV11-Hoxa5-Neo,GenePharma, Shanghai, China) (Additional Figure 1A and B) and plasmid vectors were intracerebroventricularly injected to the mice with induced AIS using anin vivoDNA/RNA transfection reagent (Engreen Biosystem Co., Ltd.,Beijing, China).Each mouse was intracerebroventricularly injected with 2.0 μg/10 μL of plasmid every 12 hours after the pMCAO operation to maintainin vivotransfection efficacy.Figure 1A shows the experimental timeline used to investigate the effects ofcircRap1bandHoxa5on AIS-induced apoptosisin vivo.
Quantitative polymerase chain reaction
Total RNA was extracted from HT22 cells using Trizol (Invitrogen).The purity and concentration of the extracted RNA were determined using a NanoDrop 2000 ultra-micro spectrophotometer (Thermo Fisher Scientific, Waltham,MA, USA).Hoxa5andFam3amRNA expression levels were determined using a one-step Prime Script TM RT-PCR Kit (RR064A, Takara, Tokyo, Japan).β-Actin was used as the internal reference.Three micrograms of total RNA were reverse transcribed to complementary DNA using the RiboTMReverse Transcription Kit (RiboBio, Guangzhou, China).The expression levels of lin-Rap1b andcircRap1bwere quantified using SYBR Premix Ex Taq II (Takara),random primers, and oligo dT primers, withβ-actinas the internal reference.RNase R (Beyotime, Shanghai, China) was used to digest linear RNAs but not circular RNAs, to eliminate the potential influence of lin-Rap1b, and to confirm the presence ofcircRap1b.The SYBR Premix Ex Taq II kit was used to detect precipitation of theHoxa5promoter region by H3K14ac, and the amount pulled down by H3K14ac was calculated as a percentage of the input.The relative expression levels of lin-Rap1b,CircRap1b,Hoxa5, andFam3amRNA and theHoxa5promoter region were calculated using the 2–∆∆Ctmethod (Zhang et al., 2020; Dong et al., 2021).The primers are listed in Additional Table 1.
Western blot assay
Firstly, a protein lysate solution was prepared using a 1:100 ratio of phenylmethylsulfonyl fluoride to radioimmunoprecipitation assay buffer.An equal volume of lysate solution was added to HT22 cells to extract protein.The protein concentration was detected using a bicinchoninic acid (BCA) kit (Beyotime), and the volume needed to load 40 mg protein on a gel was calculated.Secondly, an 8–10% separating gel and stacking gel were prepared, based on the molecular size of the target protein.The gel was soaked in electrophoresis solution, the denatured proteins were loaded into the well using the calculated loading volumes, and the gel was subjected to a constant current of 80 V for electrophoretic separation.Then,the proteins were transferred to a membrane at a constant current of 120 mA, with the transfer time depending on the size of the proteins.Next,the membrane was blocked with 5% skim milk for 2 hours, after which the blocking solution was washed off with Tris-buffered saline containing 0.1%Tween-20.The primary antibodies were diluted with the primary antibody dilution solution to the dilution ratio recommended by the antibody manuals.The membrane was sealed in a plastic sleeve containing the appropriate diluted primary antibody and incubated overnight at 4°C.The antibodies used were as follows: Hoxa5 (rabbit, 1:800, Affinity Bioscience, Beijing,China, Cat# DF4123, RRID: AB_2836488), Fam3a (rabbit, 1:800, Proteintech,Chicago, IL, USA, Cat# 20588-1-AP, RRID: AB_2878704), Kat7 (rabbit, 2 μg/mL,Abcam, Cambridge, UK, Cat# ab70183, RRID: AB_1269226), and β-actin(rabbit, 1:1000, Proteintech, SRRID: AB_10700003).The membranes were then washed with Tris-buffered saline containing 0.1% Tween-20 and incubated in goat anti-rabbit secondary antibody (1:10,000, Proteintech,Cat# SA00001-2, RRID: AB_2722564) for 2 hours at 37°C on a shaker.Lastly,the chemiluminescent solution was prepared.Enhanced chemiluminescence(ECL), photographing, and integrated density values (IDVs) analysis were performed using Gelpro 32 software (Media Cybernetics, Bethesda, MD, USA),using β-actin as an endogenous control.The Hoxa5 and Fam3a expression levels were calculated based on band density (Additional Figure 2).
Fluorescence in situhybridization
To investigatecircRap1blocalization in HT22 cells, fluorescencein situhybridization (FISH) assays were carried out using acircRap1bprobe, as previously described (Zhang et al., 2020).Briefly, HT22 cells were fixed on slides in 4% formaldehyde (Sigma-Aldrich, St.Louis, MO, USA) for 15 minutes and then washed with phosphate-buffered saline (containing 1% diethyl pyrocarbonate [DEPC], Dingguo, Guangzhou, China).After digestion with PCR-grade proteinase-K (Roche Diagnostics, Mannheim,Germany), the hybridization mix was prepared withcircRap1bprobe (greenlabeled, Servicebio, Wuhan, China) (5′-CCT TGA ACA AAC TGT ACA GT-3′)in hybridization solution (Invitrogen).After washing with washing buffer(containing 1% DEPC), the sections were stained with anti-digoxin rhodamine conjugate (1:100, Exon Biotech, Guangzhou, China) at 37°C for 1 hour in the dark.The sections were stained with 4′,6-diamidino-2-phenylindole (DAPI;Beyotime) for nuclear staining.All fluorescence images were captured using an Olympus BX51 fluorescence microscope (Olympus, Tokyo, Japan).The results from the FISH assays showed thatcircRap1bwas mainly present in the nucleus.
RNA pull-down
The interaction betweencircRap1band Kat7 (or H3K14ac) was detected using a Pierce magnetic RNA-protein pull-down kit (Thermo Fisher Scientific)according to the manufacturer’s instructions.The assay was performed as previously described (Zhang et al., 2020).Anti-Kat7 (rabbit, 5 μg/mL, Abcam,Cat# ab70183, RRID: AB_1269226) and H3K14ac antibodies (rabbit, 5 μg/mL,Abcam, Cat# ab82501, RRID: AB_10865341) were used for the assay.
RNA immunoprecipitation assay
An EZ-Magna RNA-Binding Protein Immunoprecipitation Kit (Millipore,Bedford, MA, USA) was utilized to carry out RNA immunoprecipitation (RIP)according to the manufacturer’s protocol.The assay was performed as previously described (Zhang et al., 2020).The following antibodies were used:anti-Kat7 (rabbit, 5 μg/mL, Abcam, Cat# ab70183, RRID: AB_1269226), anti-IgG (Millipore), and anti-SNRNP70 (Millipore).
Flow cytometry
HT22 cell apoptosis was assessed using an ApoScreen Annexin 7AAD/PE kit (Southern Biotech, Birmingham, AL, USA).After the cells to be tested were trypsinized, washed with phosphate-buffered saline, and centrifuged twice, refrigerated 10× Annexin V binding buffer solution was diluted to a 1×concentration and placed on ice, and the cells were resuspended in 120 μL of cold 1× Annexin V to obtain a 1 × 106cells/mL cell suspension.According to the manufacturer’s instructions, after the addition of 10 μL of conjugated Annexin V-PE, the cells were mixed gently and incubated at 2–8°C for 15 minutes in the dark.Subsequently, 10 μL of 7-AAD stain followed by 380 μL of cold 1× Annexin V binding buffer was added.Lastly, the cells were analyzed by FACScan flow cytometry (BD Biosciences, New York, NY, USA) to determine the rate of apoptosis.
Hematoxylin-eosin staining
Whole brains were fixed in 4% formaldehyde (pH 7.0) (Biosharp, Beijing,China) for up to 24 hours, then embedded in high-efficiency sectioning paraffin using routine procedures (Shanghai Huayong Paraffin Wax Co.LTD., Shanghai, China).The hippocampal CA1 region was cut to 4 μm-thick sections using a LEICA RM2245 microtome (LEICA, Heidelberg, Baden-Wurttemberg, Germany), and the sections were mounted on glass slides using Neutral Balsam FMP (HUSHI, Shanghai, China).The slides were stained with hematoxylin and eosin.Neuronal atrophy and nuclear pyknosis in the hippocampal CA1 region were observed using a VENTANA HE 600 staining system (Roche, Innovation Park Drive Tucson, AZ, USA).
Terminal deoxynucleotidyl transferase dUTP nick-end labeling assay
The brain tissues were embedded in paraffin using routine procedures, the hippocampal CA1 region was cut into 4 μm-thick sections, and the sections were mounted on glass slides.The slides were then subjected to terminal deoxynucleotide transferase dUTP nick-end labeling (TUNEL) using a One Step TUNEL Apoptosis Assay Kit (Beyotime) according to the manufacturer’s protocol, as previously described (Liu et al., 2019).The cell apoptosis rate was calculated using the following formula: apoptosis rate = the number of TUNEL-positive cells in the CA1 area/total number of cells in the CA1 area.All fluorescence images were captured using an Olympus BX51 fluorescence microscope.
Reporter vector construction and luciferase reporter assay
As previously described (He et al., 2020), theFam3apromoter region (–3000 to 0 bp) was PCR-amplified and cloned into the pmirGLO of Dual Luciferase Expression Vector (Promega, Madison, WI, USA) to construct wild-type and mutated luciferase reporter vectors (Generay Biotech Co., Shanghai, China).Then, HEK293T cells (Chinese Academy of Sciences, Shanghai, China) were co-transfected with the reporter vectors and a plasmid encoding full-length mouseHoxa5or empty vector (GeneChem) using Lipofectamine 3000.The relative luciferase activities were measured 48 hours after co-transfection using a luciferase reporter gene assay kit (YPH, Beijing, China), following the manufacturer’s protocols.
JASPAR analysis
JASPAR is a database of transcription factor binding site motifs that was used to predict binding sites ofHoxa5(or HOXA5) in theFam3a(or FAM3A)promoters.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation (ChIP) assays were conducted using a Simple-ChIP enzymatic ChIP kit (Cell Signaling Technology, Boston, MA,USA).In brief, HT22 cells in the logarithmic growth phase were incubated with 1% formaldehyde in a 10-cm cell culture dish for 10 minutes at room temperature (about 20°C), then treated with glycine for 5 minutes.Next, the cells were lysed with PMSF-containing lysis buffer, sonicated, and incubated with nuclease for 20 minutes to digest the chromosomal DNA.Appropriate DNA digestion confirmed by agarose gel electrophoresis, and a sample of the digested DNA was set aside for use as the 2% input positive control.The remaining DNA was incubated with an anti-Hoxa5antibody (1:100, Affinity Bioscience, Beijing, China, Cat# DF4123), an anti-methylated H3K14ac antibody (5 μg/mL, Abcam, Cat# ab82501), or a negative control anti-IgG antibody overnight at 4°C on a shaker.All DNA bound to the antibodies was then purified using spin columns.The target DNA products were PCRamplified using sequence-specific primers, and images were collected after agarose gel electrophoresis.The primers used for each PCR reaction are provided in Additional Table 2.For each PCR reaction, the corresponding input DNA was included in parallel as a positive control for PCR validation.Then, quantitative real-time PCR was performed using SYBR Premix Ex Taq II (Tli RNase H Plus) (Takara) to quantitate the DNA immunoprecipitated by the anti-H3K14ac antibody; the primers used for this reaction are provided in Additional Table 3.The results were normalized to input and are displayed as enrichment relative to the IgG control.
Chromatin isolation by RNA purification
Firstly, we designed an antisense DNA oligonucleotide probe specific for fulllengthcircRap1b.For chromatin preparation, cell pellets were rapidly thawed in a 37°C water bath and resuspended in buffer (0.1 M Tris pH 7.0, 10 mM potassium acetate (KOAc), 15 mM magnesium acetate (MgOAc)).The cells were then treated with a solution containing 1% NP-40, 1 mM dithiothreitol(DTT), 1 mM PMSF, complete protease inhibitor, and 0.1 U/μL superoxide on ice for 10 minutes.Next, the cell suspension was centrifuged at 2500 ×gfor 5 minutes, followed by chemical cross-linking performed by fixing cells in an appropriate amount of 1% formaldehyde in phosphate-buffered saline for 10 minutes at room temperature.Cross-linking reactions were quenched with 0.125 M glycine for 5 minutes.Nuclei were lysed in nucleolysis buffer on ice for 10 minutes followed by sonication until most of the chromatin was lysed.The sizes of the resulting DNA fragments ranged from 100–500 bp.
Next, 100 pmol probe was added to 3 mL of diluted chromatin, and the solution was mixed for 4 hours at 37°C with end-to-end rotation.Streptavidin magnetic beads were washed three times in nuclear lysis buffer, blocked with 500 ng/μL total yeast RNA and 1 mg/mL bovine albumin (BSA) for 1 hour at room temperature, washed three times in nuclear lysis buffer, and then restored to their original volume.Then, 100 μL of the cleaned streptavidin magnetic beads per 100 pmol of probe were added, and the entire reaction was mixed at 37°C for 30 minutes.The bead-biotin probe-RNA-chromatin complex was captured with a magnet and washed five times with a 40×volume of washing solution.Finally, the washing buffer was carefully aspirated with a pipette, and the RNA was extracted with Trizol and chloroform.The extracted RNA was then subjected to quantitative reverse transcription PCR to detect enriched transcripts.
Beads were resuspended with three times the original volume of DNA elution buffer.DNA was eluted with a mixture of 100 μg/mL RNase A and 0.1 U/μL RNase H.RNase elution was performed twice at 37°C, and the two eluates were mixed to combine.DNA was extracted with an equal volume of phenolchloroform-isoamyl ester and then precipitated with ethanol overnight at –80°C.The precipitated DNA was subjected to quantitative PCR.The sequences of the probes are provided in Additional Table 4.
Statistical analysis
All data in this study were analyzed using GraphPad Prism version 5.0.1 for Windows (GraphPad Software, San Diego, CA, USA, www.graphpad.com) and are expressed as the mean ± standard deviation (SD).The Student’st-test or one-way analysis of variance followed by Bonferroni correction was used to assessed differences between groups.P< 0.05 was considered statistically significant.
Results
circRap1bexpression decreases after AIS in vivo and inhibits neuronal apoptosis in vitro
The acute ischemia models (mouse AIS and HT22-AIS) were constructed using C57BL/6J mice and hippocampal neuron cells HT22, respectively, forin vivoandin vitrostudies.Hippocampal CA1 neuron apoptosis in the sham and AIS groups was detected by hematoxylin-eosin (HE) staining and TUNEL assay(Figure 1B and C).HE staining showed that neurons in the hippocampal CA1 area of the AIS mice appeared to be atrophic and exhibited nuclear pyknosis(Figure 1A).TUNEL analysis showed that the apoptosis rate was markedly higher in the AIS group than in the sham group (Figure 1CandD).circRap1bexpression was very low in mouse AIS hippocampus samples and HT22-AIS cells, as detected by quantitative polymerase chain reaction (qPCR) (Figure1E–H).Flow cytometry analysis showed that the apoptosis rate of HT22-AIS cells was markedly higher than that of normal HT22 cells (Figure 1IandJ).Furthermore,circRap1binhibited HT22-AIS cell apoptosis (Figure 1KandL).
Hoxa5expression is low in the hippocampus of a mouse model of acute
ischemia and in HT22-AIS cells, and inhibits HT22-AIS cell apoptosisTo further explore the molecular mechanism by whichcircRap1binhibits HT22-AIS cell apoptosis, we overexpressedcircRap1bin HT22-AIS cells and detected the mRNA expression profile by qPCR.The results showed that,among the proteins that were differentially expressed between HT22 cells and HT22-AIS cells, the transcription factorHoxa5was the most markedly upregulated (Additional Figure 3A).As shown inFigure 2A–E,Hoxa5mRNA and protein expression levels were markedly lower in mouse AIS hippocampustissues than in the Sham group (Additional Figure 3B) and in HT22-AIS cells than in normal HT22 cells, as detected by qPCR and western blot assay.Flow cytometry analysis showed that the apoptosis rate of HT22-AIS cells was markedly decreased byHoxa5overexpression and markedly increased byHoxa5knockdown (Figure 2FandG).These results indicate thatcircRap1bmost likely inhibits HT22-AIS cell apoptosis by regulatingHoxa5expression.
CircRap1binduces H3K14ac modification in the Hoxa5 promoter region by recruiting the acetyltransferase Kat7, thereby promotingHoxa5
transcriptionWhencircRap1bwas overexpressed in HT22-AIS cells,Hoxa5mRNA expression increased (Figure 3A).To further investigate the mechanism by whichcircRap1bregulatesHoxa5transcription, we first performed FISH assays to determine the localization ofcircRap1bin HT22 cells.The results showed thatcircRap1bwas mainly present in the nucleus (Figure 3B).Kat7 is often involved in transcriptional regulation, so we hypothesized that Kat7 participates in thecircRap1b-mediated regulation ofHoxa5transcription.To test this, we performed an RNA pull-down assay and found that Kat7 was bound tocircRap1b(Figure 3C).RIP assay showed that U1 enrichment in the anti-SNRNP70 group was markedly higher than anti-normal IgG group (Figure3D), demonstrating that the pull-down assay was performed successfully.circRap1benrichment was markedly greater in the anti-Kat7 group than in the negative control IgG group and the negative RNA control GAPDH group (Figure 3E), which further verifiedcircRap1bbinding to Kat7.Next,we divided theHoxa5promoter region (–3000 to 0 bp) into six segments(P1–P6), designed primers specific to these segments (Additional Table 5),and performed chromatin immunoprecipitation (ChIP) using anti-H3K14ac,-H4K5ac, and -H4K12ac antibodies.The H3K14ac modification was present in promoter region –1000 to 0 bp (P1 and P2) (Figure 3F), but the H4K5ac and H4K12ac modifications were not detected (Additional Figure 4AandB).Next we used qPCR to amplify the immunoprecipitated DNA fragment and found that H3K14ac was enriched in the P1 and P2 regions, as shown inFigure 3G, compared with the IgG negative control.The H3K14ac enrichment was markedly higher in the P1 region than in the P2 region.To further prove thatcircRap1band Kat7 jointly regulate H3K14ac modification in theHoxa5promoter region, we transiently transfected HT22-AIS cells with a plasmid encodingcircRap1bor a negative control plasmid (circ-NC) and with a plasmid encoding a small interfering RNA specific for siKat7 plasmids or a negative control plasmid (siNC), yielding the following HT22-AIS cell lines: circ-NC +siNC,circRap1b+ siNC, andcircRap1b+ siKat7.H3K14ac enrichment in each group compared with the circ-NC + siNC group was then detected by ChIPPCR, as shown inFigure 3HandI.H3K14ac enrichment in the circ + siNC group was markedly higher than that in the circ-NC + siNC group, while H3K14ac enrichment in the circ + siKat7 group was markedly lower than that in the circ + siNC group and the H3K14ac enrichment in circ + siKat7 group was reversed to the level of circ-NC + siNC group.These results indicate that Kat7 knockdown attenuated thecircRap1b-mediated increase in H3K14ac enrichment in theHoxa5promoter region.TheHoxa5mRNA and protein expression levels changed accordingly (Figure 3J–L).We further demonstrated the interaction between H3K14ac andcircRap1bby RNA pull-down (Figure3M).Next, we performed RNA purification isolated chromatin (ChIRP) assays.In total, 20–40% ofcircRap1binput was retrieved from the RNA fraction usingtiling probes.The –1000 to –500 bp region of theHoxa5promoter exhibited 50% retrieval, and the –500 to 0 bp region exhibited 20% retrieval from the DNA fraction (Figure 3NandO).The results described above indicate thatcircRap1bpromotesHoxa5transcription by recruiting the acetyltransferase Kat7 to induce H3K14ac modification in theHoxa5promoter region.
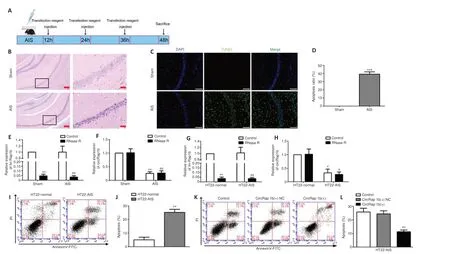
Figure 1|CircRap1boverexpression inhibits HT22-AIS cell apoptosis.
Hoxa5regulates neuronal apoptosis by activating Fam3atranscriptionFam3ais a neuronal apoptosis-related protein (Liu et al., 2019), as shown inFigure 4A–E.We found thatFam3amRNA and protein expression levels were markedly decreased in mouse AIS hippocampus tissues (Additional Figure 5A) and HT22-AIS cells.Flow cytometry analysis showed that the rate of HT22-AIS apoptosis decreased markedly in response toFam3aoverexpression and increased markedly in response toFam3aknockdown (Figure 4FandG).Importantly, theFam3amRNA and protein expression levels were upregulated in response toHoxa5overexpression and down-regulated in response toHoxa5knockdown (Figure 4H–J).To investigate the mechanism by whichHoxa5regulatesFam3atranscription, we used the bioinformatics database JASPAR to predictHoxa5binding sites in theFam3apromoter region (–3000 to 0 bp).Four putative binding sites were identified, three of which were in the antisense strand and one of which was in the sense strand.Next, ChIP and luciferase reporter assays were performed to confirm these predicted sites.The results showed thatHoxa5bound to the site in the –1000 to –500 bp region of theFam3apromoter, and that this binding interaction activatedFam3atranscription (Figure 4K–M), resulting in an increase inFam3aexpression.To conclude, these findings suggest thatHoxa5regulates neuronal apoptosis by activatingFam3atranscription.
CircRap1bregulates Fam3aexpression through Hoxa5, thereby regulating neuronal apoptosis
Given thecircRap1bandHoxa5expression levels in the HT22-AIS model and their effects on apoptosis, as described above, we next sought to clarify the mechanism by whichcircRap1bandHoxa5coregulate HT22-AIS apoptosis.To do this, HT22-AIS cells were cotransfected withcircRap1b(+),Hoxa5(-), and their corresponding negative control plasmids, and the rate of cell apoptosis was detected by flow cytometry.The results showed thatcircRap1boverexpression inhibited apoptosis,Hoxa5knockdown promoted apoptosis, andHoxa5knockdown reversed the inhibitory effect ofcircRap1boverexpression alone on cell apoptosis (Figure 5AandB).To investigate howcircRap1bandHoxa5affect cell apoptosis, we determined their effects on the expression of the neuronal apoptosis-related proteinFam3a.circRap1boverexpression resulted in an increase inFam3aexpression (Figure 5CandD) and inHoxa5expression (Figure 5EandF).AsFigure 5GandHshow,Hoxa5knockdown reversed theFam3aupregulation induced bycircRap1boverexpression, suggesting thatcircRap1bregulatedFam3aexpression throughHoxa5.AsAdditional Figure 5BandCshow,circRap1borHoxa5overexpression decreased the rate of cell apoptosis, and combinedcircRap1bandHoxa5overexpression exerted the strongest inhibitory effect on cell apoptosis.Taken together, these results suggest thatcircRap1bregulatesHoxa5transcription and expression, and subsequentlyFam3aexpression,ultimately affecting cell apoptosis.
circRap1band Hoxa5inhibit neuronal apoptosisin vivo
Given thatcircRap1bregulatesHoxa5transcription by histone modification to regulate neuronal apoptosis, we further explored its potential clinical relevance.To do this,circRap1b(+) andHoxa5(+), as well asin vivotransfection reagents, were injected into the lateral ventricle of the AIS mouse model.As shown inFigure 6A, the number of apoptotic cells in the hippocampal CA1 region was markedly decreased in the AIS +circRap1b(+) +Hoxa5(+)NC, AIS+circRap1b(+)NC +Hoxa5(+), and AIS +circRap1b(+) +Hoxa5(+) groups, and the lowest number of apoptotic cells was observed in the AIS +circRap1b(+)+Hoxa5(+) group.The results from the TUNEL assay showed that the cell apoptosis rate was markedly lower in the AIS +circRap1b(+) +Hoxa5(+)NC,AIS +circRap1b(+)NC +Hoxa5(+), and AIS +circRap1b(+) +Hoxa5(+) groups,compared with the AIS +circRap1b(+)NC +Hoxa5(+)NC group (Figure 6B).Furthermore, the cell apoptosis rate was lower in the AIS +circRap1b(+) +Hoxa5(+) group compared with the AIS +circRap1b(+)NC +Hoxa5(+) group and was markedly lower in the AIS +circRap1b(+) +Hoxa5(+) group compared with the AIS +circRap1b(+) +Hoxa5(+)NC group (Figure 6C).Next, we performed qPCR and found thatFam3amRNA expression was higher in the AIS +circRap1b(+) +Hoxa5(+)NC and AIS +circRap1b(+)NC +Hoxa5(+) groups and highest in the AIS +circRap1b(+) +Hoxa5(+) group when compared with the AIS +circRap1b(+)NC +Hoxa5(+)NC group (Additional Figure 5D).Taken together, these findings suggest thatcircRap1bandHoxa5may be novel treatment targets in acute ischemic stroke.
Discussion
Previous studies have confirmed that circRNAs help regulate the occurrence and progression of ischemic cerebrovascular diseases (Lin et al., 2016;Mehta et al., 2017; Han et al., 2018).circDLGAP4regulates HER1 expression by binding to miR-143, thereby affecting endothelial cell dedifferentiation into mesenchymal cells and reducing cerebral infarction size and bloodbrain barrier injury.It has also been shown that circRNAs are involved in the regulation of apoptosis in a variety of cells, including neurons.For example,circ_0000950 negatively regulates neuronal apoptosis via miR-103 during the development of Alzheimer’s disease (Yang et al., 2018a).In addition,circ_0000296is down-regulated in hippocampal tissues and HT22 cells in response to chronic cerebral ischemia/hypoxia (CCI), andcirc_0000296overexpression significantly inhibits CCI-induced neuronal apoptosis (Huang et al., 2021).In the present study, we found thatcircRap1bexpression levels were low in hippocampal tissues from a mouse model of AIS and in HT22-AIS cells, and thatcircRap1boverexpression markedly inhibited AIS-induced neuronal apoptosis.Hoxa5expression was strongly up-regulated in response tocircRap1boverexpression.Transcription factors encoded by the Hox gene family contribute to determination of neuronal fate (Boeckel et al., 2015).Specifically,Hoxa5has recently been shown to be involved in axonal growth of autonomously functioning nuclear cells (Salzman, 2016) and post-vagal motor neurons (Lin et al., 2016) in the mouse brain.Rosa Hernández et al.(2021) suggested thatHoxa5plays an important role in the differentiation of human adipose-derived stem cells and is therefore a potential treatment target that could be explored for the treatment of neurological disorders(Hernández et al., 2021).Our findings confirm thatHoxa5expression is low in hippocampal tissues from the mouse model of AIS and in HT22-AIS cells.Hoxa5overexpression markedly inhibited AIS-induced neuronal apoptosis,andHoxa5knockdown markedly increased the apoptosis rate of HT22-AIS cells.Consistent with our study,HOXA5overexpression in fibroblasts associated with hypertrophic scar or keloid attenuated cell proliferation,migration, and collagen synthesis, while accelerating apoptosis (Liang et al.,2021).Silencing HOXA5 gene expression induced Jurkat cell apoptosis and cell cycle arrest (Huang et al., 2016).HOXA5acts directly downstream of retinoic acid receptors and is involved in retinoic acid-induced apoptosis and growth inhibition (Chen et al., 2007).Hoxa13 is down-regulated in hippocampal tissues and HT22 cells in response to CCI, andHoxa13overexpression attenuated the CCI-induced apoptosis of HT22 hippocampal neurons (Liu et al., 2019).A mouseHoxa13mutant inhibits urinary epithelial cell apoptosis by down-regulating Bmp7 expression (Morgan et al., 2003).
Histone modifications mainly include methylation, acetylation,phosphorylation, ubiquitination, and sumoylation (Dong and Cui, 2019).Histone acetylation is dynamically regulated by the histone acetyl transferase and histone deacetylase enzymes.Previous studies have shown that Kat7 tends to catalyze the acetylation of lysine 5 and lysine 12 of histone H4,and lysine 14 of histone H3 (Miotto and Struhl, 2010; Feng et al., 2016).Furthermore,circFoxo3overexpression inhibits the effects of oxygen-glucose deprivation; induces autophagy, apoptosis, inflammation, and cardiomyocyte injuryin vitro; and inhibits HMGB1 expression by reducing the KAT7,H3K14ac, and RNA Poly II enrichment at the HMGB1 promoter, thus inhibiting autophagy and alleviating myocardial ischemia/reperfusion injury (Sun et al., 2021).Overexpression of CDK11(P58) in mammalian cells enhances the acetyltransferase activity of Kat7 to free histones.CDK11(P58) is a regulatory protein that interacts with Kat7in vivoandin vitro, plays an important role in cell cycle progression, and is closely related to apoptosis (Zong et al.,2005).In this study, we found thatHoxa5mRNA and protein expression levels were markedly elevated in response tocircRap1boverexpression.circRap1blocalized to the nucleus and interacted with Kat7 in theHoxa5promoter region.Furthermore, H3K14ac was enriched in theHoxa5promoter region, mainly enriched from –1000 to 0 bp (regions P1 and P2), while H4K5ac and H4K12ac were not detected in this region.Co-transfection ofcircRap1boverexpression and Kat7 knockdown plasmids into HT22-AIS cells reversed the increase in H3K14ac enrichment andHoxa5mRNA and protein expression induced bycircRap1boverexpression.Therefore, the findings from our study support a model in whichcircRap1brecruits Kat7 to induce H3K14ac modification in theHoxa5promoter region, thereby activatingHoxa5transcription.Similarly,circMRPS35enriches H4K5ac binding to theFOXO1andFOXO3apromoter regions through the recruitment of KAT7,thereby inhibiting the progression of gastric cancer (Jie et al., 2020).In addition,lncRNA-MRCCAT1mediatesNPR3transcription by binding EZH2 to induce H3K27me3 binding to theNPR3promoter region (Li et al., 2017).Furthermore, EZH2 binding to the HIV-1 LTR promoter was abrogated byMALAT1binding to PRC2, resulting in removal of H3K27me3 from the LTR promoter and reversing the epigenetic silencing of the corresponding transcript (Qu et al., 2019).
FAM3Ais a mitochondrial protein that plays an important role in cellular stress adaptation and survival and participates in the regulation of neuronal apoptosis (Liu et al., 2019; Yan et al., 2019).Song et al.(2016) reported thatFAM3Ais an important regulator of ER-stress-induced HT22 cell death.FAM3Aprotects HT22 cells from hydrogen peroxide-induced oxidative stress by activating the PI3K/Akt pathway (Song et al., 2015).FAM3Aexpression is down-regulated in hippocampal tissues and neurons cells in response to CCI, andFAM3Aoverexpression inhibits CCI-induced hippocampal neuron apoptosis (Liu et al., 2019).In this study, we found thatFam3awas markedly down-regulated in hippocampal tissues from a mouse model of AIS and in HT22-AIS cells;Fam3aoverexpression markedly inhibited HT22-AIS cell apoptosis; andFam3aknockdown markedly promoted HT22-AIS cell apoptosis.A previous study reported thatFAM3Ainhibits interleukin 1β-induced chondrocyte apoptosis by activating the pro-survival PI3K/AKT/mTOR pathway (Yan et al., 2019).Furthermore,FAM3Apositively regulates post-ischemia angiogenesis in endothelial cells (Xu et al., 2019).In addition,FAM3Aimproves hypoxic-ischemic brain injury in neonatal mice (Song et al.,2021).In this study, we used the bioinformatics database JASPAR to identify 44 putative HOXA5 binding sites in the promoter region (–3000 to 0 bp) of humanFAM3Aand four putative Hoxa5 binding sites in the promoter region(–3000 to 0 bp) of mouseFam3a.Furthermore, our ChIP and luciferase reporter assay results verified that Hoxa5 can bind to the promoter region ofFam3ato promote its transcription and its protein expression, thus regulating hippocampal neuron apoptosis.There have been several other reports that HOX family transcription factors play a positive role in transcriptional regulation.For example, Hoxa13 activatesFAM3Atranscription and inhibits CCI-induced hippocampal neuronal apoptosis (Liu et al., 2019).In addition,Wu et al.(2017) found that Hoxa13 promotesBMP7transcription in gastric cancer by targeting the gene’s promoter region.Finally, Hoxa13 targets theAldh1a2promoter and upregulates its expression to regulate limb morphogenesis (Shou et al., 2013).To further clarify the mechanism by whichcircRap1bandHoxa5coregulate HT22-AIS apoptosis, we cotransfectedcircRap1boverexpression,Hoxa5siRNA, and their negative control plasmids into HT22-AIS cells and found thatcircRap1boverexpression inhibited cell apoptosis, whileHoxa5knockdown promoted apoptosis.Hoxa5knockdown could reverse the inhibitory effect ofcircRap1boverexpression alone on cell apoptosis.In response tocircRap1boverexpression,Hoxa5andFam3amRNA and protein expression levels were up-regulated, andHoxa5knockdown reversed the effects ofcircRap1boverexpression onFam3aexpression, suggesting thatcircRap1bregulatesFam3aexpression throughHoxa5.Finally, we verifiedin vivothat the rate of hippocampal neuron apoptosis was lowest in response to combined overexpression ofcircRap1bandHoxa5.In summary, our findings suggest thatcircRap1bacts as a modular scaffold to recruit KAT7 to the promoter region ofHoxa5, which then increases the level of H3K14ac modification to activateHoxa5transcription and expression, ultimately suppressing hippocampal neuronal apoptosis.Naturally protective circRNAs can be overexpressedin vivoutilizing lentiviral or adenovirus vectors that encode microcassettes with exons, as well as endogenous splicing donor and recipient sites, and inverted repeats of flanking introns that support RNA reverse folding (Holdt et al.,2018).Microinjection ofcircDLGAP4lentivirus markedly improved ischemic stroke prognosis (Bai et al., 2018).Our study showed the mechanism by which thecircRap1b/Hoxa5/Fam3aaxis regulates neuronal apoptosis, and may provide a new approach and wider therapeutic target for developing an effective treatment for AIS.However, there was a design limitation to this study in that we only used male mice.To improve the clinical significance,future studies should consider using female mice.The translational value of our findings was not explored in this study, and future studies should also investigate this potential to improve the treatment of stroke.
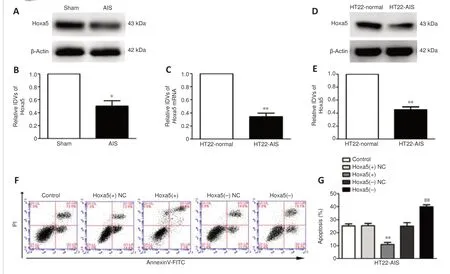
Figure 2|Endogenous Hoxa5expression and its effect on neuronal apoptosis
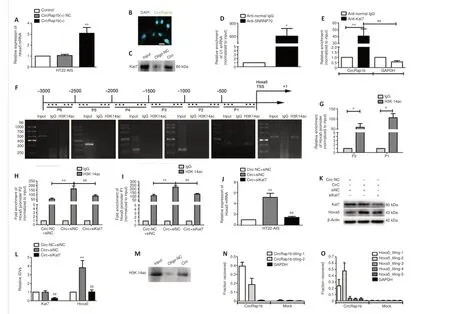
Figure 3|CircRap1binduces H3K14ac modification in the Hoxa5 promoter region by recruiting the acetyltransferase Kat7.
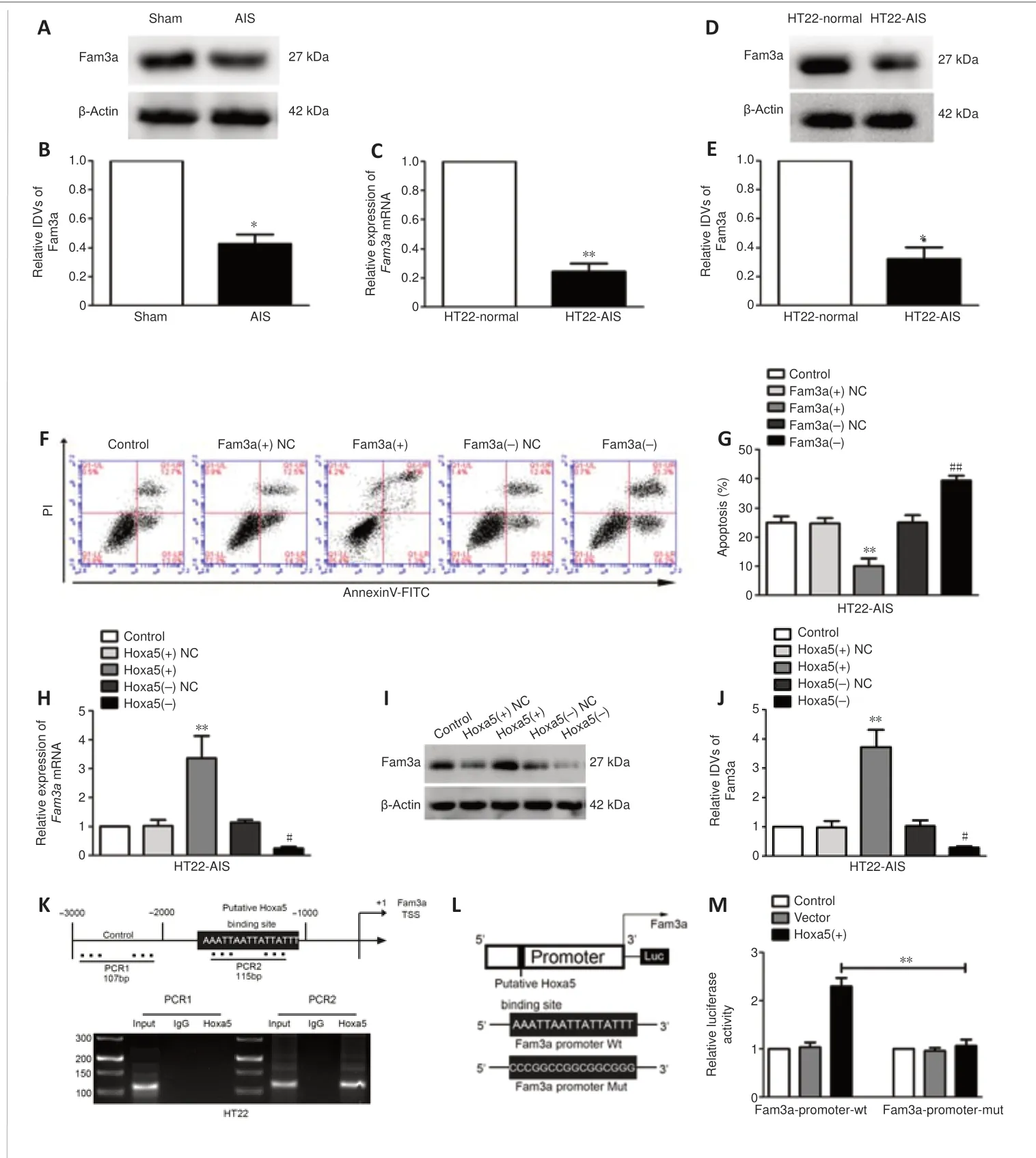
Figure 4| EndogenousFam3aexpression and its effect on neuronal apoptosis, and the mechanism by which Hoxa5regulatesFam3a.
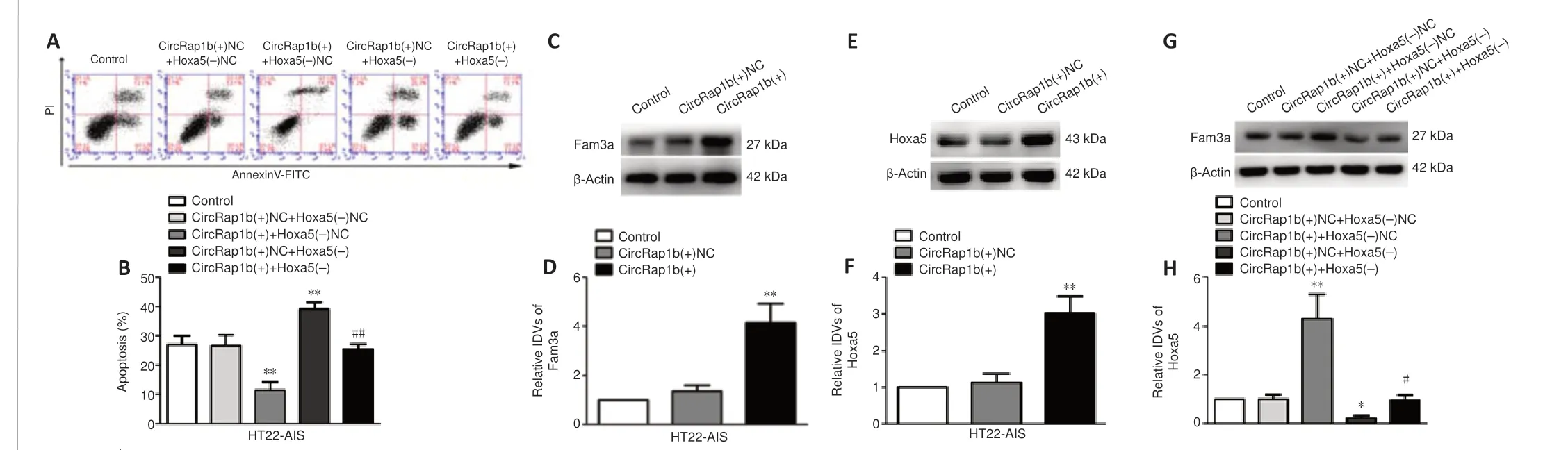
Figure 5|CircRap1bandHoxa5coregulateFam3aexpression and neuronal apoptosis.
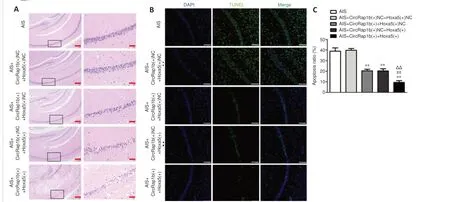
Figure 6|Combined overexpression of circRap1b and Hoxa5inhibits acute ischemic stroke (AIS)-induced neuronal apoptosis in vivo.
Additional Table 2:The primers for each polymerase chain reaction set in Chromatin immunoprecipitation assay to confirm the binding sits for Hoxa5 in the promoter region of Fam3a.
Additional Table 3:The primers of-1000 bp-TSS of Hoxa5 amplified with the DNA precipitated by H3K14ac to detect the percentage of relative to its corresponding input in quantitative real-time polymerase chain reaction.
Additional Table 4:The probes of circRap1b and Hoxa5 in chromatin isolation by RNA purification.
Additional Table 5:The primers for each PCR set in Chromatin immunoprecipitation assay to confirm the existence of H3K14ac modification in Hoxa5 promoter region.
Acknowledgments:We thank the Department of Laboratory Animals of China Medical University for the animal experiments and Sangon Biotech for offering primer design.
Author contributions:Study design: FFZ, LinZ and LiangZ; experiment implementation and data acquisition: FFZ, LinZ and YLu; data analysis: FFZ,LinZ, YLu, XD, YQL, YLi, SG, SYZ, YX, YZJ; paper discussion: FFZ, LinZ and LiangZ;manuscript drafting: FFZ; administrative and material support: LZ.All authors reviewed the manuscript and agreed the final version of this manuscript.
Conflicts of interest:The authors declare no competing interests.
Author statement:This paper has been posted as a preprint on Research Square with doi: https://doi.org/10.21203/rs.3.rs-1622225/v1, which is available from: https://assets.researchsquare.com/files/rs-1622225/v1/8b3b3263-06cd-4498-a1b8-b21c40e399ba.pdf?c=1652193425.
Data availability statement:All data relevant to the study are included in the article and uploaded as Additional files.
Open access statement:This is an open access journal, and articles are distributed under the terms of the Creative Commons AttributionNonCommercial-ShareAlike 4.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as appropriate credit is given and the new creations are licensed under the identical terms.
Additional files:
Additional Figure 1:Map of circRap1 (A) and Hoxa5 (B) overexpression plasmids in vivo experiments.
Additional Figure 2:The bands of Hoxa5 and Fam3a protein expression levels were detected by western blotting
Additional Figure 3:mRNA gene expression profile as obtained from samples of three groups in circRap1b overexpression.
Additional Figure 4:Chromatin immunoprecipitation assays and quantitative polymerase chain reaction analysis for the enrichment of H4K5ac and H4K12ac in the Hoxa5 promoter region in HT22 cells.
Additional Figure 5:Analysis for the apoptosis of HT22-AIS cells overexpression of circRap1b and Hoxa5 by flow cytometry.
Additional Table 1:The primers of lin-Rap1b, CircRap1b, Hoxa5 mRNA, Fam3a mRNA in quantitative polymerase chain reaction.
- 中国神经再生研究(英文版)的其它文章
- From static to dynamic: live observation of the support system after ischemic stroke by two photon-excited fluorescence laser-scanning microscopy
- MicroRNAs in mouse and rat models of experimental epilepsy and potential therapeutic targets
- The generation and properties of human cortical organoids as a disease model for malformations of cortical development
- Nanotechnology-based gene therapy as a credible tool in the treatment of Alzheimer’s disease
- Detection of Alzheimer’s disease onset using MRI and PET neuroimaging: longitudinal data analysis and machine learning
- A pancreatic player in dementia: pathological role for islet amyloid polypeptide accumulation in the brain

