Multiple forest structural elements are needed to promote beetle biomass,diversity and abundance
Noln J.Rpp,Mihel Stb,Julin Frey,Nthlie Winiger,Alexnr-Mri Klein
a Nature Conservation and Landscape Ecology,University of Freiburg,Tennenbacherstrasse 4,79106,Freiburg im Breisgau,Germany
b Ecological Networks,Technical University Darmstadt,Schnittspahnstrasse 3,64287,Darmstadt,Germany
c Forest Growth and Dendroecology,University of Freiburg,Tennenbacherstrasse 4,79106,Freiburg im Breisgau,Germany
d Wildlife Ecology and Management,University of Freiburg,Tennenbacherstrasse 4,79106,Freiburg im Breisgau,Germany
Keywords:Allometry Biodiversity Coleoptera Forest management Terrestrial laser scanning Window trap
A B S T R A C T Background:Retention forestry is a management strategy aiming to mitigate biodiversity loss by retaining structural elements such as dead trees that would otherwise be removed.Here we analyze the biomass,diversity and abundance among forest beetles collected using window traps on 128 1-ha forest sites reflecting gradients in the amount of structural elements in southwestern Germany.Results:We found that beetle biomass increased with mean diameter at breast height(a measure of tree size),and decreased with stand structural complexity.Biomass of individual feeding guilds responded differently to forest structural elements,namely lying deadwood,understory complexity,tree basal area and stand structural complexity.Beetle family diversity increased with the effective number of layers,i.e.1-m forest strata occupied by vegetation assessed via terrestrial laser scanning.Abundance of feeding guilds responded to only elevation and share of deciduous trees.Community composition in terms of biomass was structured by forest elements similar to biomass of individual feeding guilds,with the addition of lying deadwood.This differed from community composition in terms of abundance of feeding guilds,which was structured by primarily standing deadwood volume and share of deciduous trees.
1.Introduction
Retention forestry is a relatively recent advent in the field of natural resource management(Gustafsson et al.,2012)that emphasizes actions to preserve forest structural elements which otherwise would have been removed or altered.The goal of preserving these elements is the integration of biodiversity conservation into forest management for sustainable use.Such elements include dead wood as a resource for forest specialist taxa(Storch et al.,2020;Eckerter et al.,2021),stands of living trees for habitat(Gustafsson et al.,2012)or the maintenance of continuous forest cover(Gustafsson et al.,2019).Retention,thus,contrasts previously dominating approaches to utilizing forest resources which have been based more or less on timber yield and often involved clear-cutting(L¨amås et al.,2015).
Understanding how various metrics of beetle biodiversity may increase with more forest structural elements can provide insights in how best to support forest insect communities.Beetles are the largest and most diverse animal order(Stork,2018),representing numerous feeding guilds which can be broadly categorized as carnivores,fungivores,herbivores,omnivores,palynivores,parasitoids,saprovores,and saproxylics based on feeding modes(Freude et al.,2009).Apart from their importance in forest ecosystem processes(Schoenly et al.,1991;Schigel,2011;Skelton et al.,2019),beetles comprise the majority of saproxylic insects in forests(Ulyshen andˇSobotník,2018).Saproxylic taxa are sensitive to forest management which has involved the removal of deadwood(Seibold and Thorn,2018),having additional negative impacts on other feeding guilds(Sandstr¨om et al.,2019).Furthermore,variation in forest structural elements can result from direct management actions such as selective harvesting,removal of deadwood,and quarantine cutting(Ammer et al.,2010),having cascading effects on beetle feeding guilds.
Few studies have focused on the biomass of insects as related to forest management(Seibold et al.,2019)and to the best of our knowledge none have analyzed biomass,diversity and abundance of feeding guilds together along retention forestry structural gradients.While several studies focused on anthropogenic drivers of changes in forest insect community metrics(Netherer and Schopf,2010;Sall′e et al.,2014;Seibold et al.,2014,2019;Pureswaran et al.,2018;Harris et al.,2019),knowledge on specific forest retention measures,which have the potential to support forest insect communities via influencing forest structural elements is scarce.Several studies have shown that forest structural elements such as multi-layered vegetation(Knuff et al.,2019),the amount of deadwood left in forest stands(Eckerter et al.,2021;Haeler et al.,2020)and stand size and connectivity(Correcher et al.,2019)can increase relative abundance of insects.Many studies however often exclusively use metrics such as diversity,abundance,or biomass of single functional groups or feeding guilds to study these trends(Eckelt et al.,2017;Lachat and Müller,2018;Forister et al.,2019;Grinde et al.,2020;Eckerter et al.,2021).The parallel and partly synonymous consideration of different metrics has led to mixed conclusions as to which actions may be the best for promoting insects as these metrics do not necessarily respond to the same drivers(Macgregor et al.,2019;Vereecken et al.,2021).Therefore,one metric of insect communities may not be exclusively better than the others for measuring management actions on insect biodiversity,as these may respond quite differently.Biomass for example is highly influenced by the sizes of the insects sampled(Sample et al.,1993)whereas abundance may be strongly influenced by the presence of locally numerous taxa(Gaston and Lawton,1988).Diversity can also be strongly influenced by ubiquitous species,but unlike abundance the presence of less common taxa will have an effect on the measure used.Varying responses from individual insect community metrics are especially important when considering feeding guilds or classifications based on feeding/resource-use(Lassau et al.,2005),which may respond differently to the same environmental elements(Pilskog et al.,2016;Wetherbee et al.,2020).As a consequence,little research has examined the potential of forest management strategies to promote forest insect biodiversity by examining the varying effects that components of such strategies have on diversity as well as the biomass and abundance of feeding guilds.
In the present study,using a space-for-time approach(Blüthgen et al.,2022)we test whether biomass and abundance among feeding guilds as well as family diversity are related to retention structural variables associated with forest heterogeneity,deadwood amounts and understory structure.We hypothesize that beetle diversity will increase with increasing forest heterogeneity,measured using variables such as tree species richness,deadwood diversity,stand structural complexity and canopy gap fraction.We hypothesize that beetle biomass and abundance will increase with increasing amount of forest habitat,measured by retention forestry metrics such as forest cover and forest vegetation strata.We also hypothesize that both metrics of more specialized feeding guilds will increase with the increasing abundance of resources used exclusively by those groups.For example,fungivores will be related to decaying deadwood,which is a substrate for fungal resources,or herbivores will be related to herb cover/understory complexity,while saproxylics will increase with deadwood volumes.Furthermore,we hypothesize that the biomass and abundance of more generalist feeding guilds(carnivores,omnivores,saprovores)will increase with increasing forest habitat(greater forest cover,more forest vegetation strata).We expect these biomass and abundance increases to be due to increased resources provided by greater amounts of forest habitat such as larger amounts of prey items and more decaying non-woody plant material.For beetle community composition,in terms of both biomass and abundance,we hypothesize that the forest variables related to resources will structure communities.
2.Materials and methods
2.1.Study region & plots
The study was conducted on 135 1-ha plots in the southern Black Forest in Baden-Württemberg,Germany.These plots were established in 2016 by the‘Conservation of Forest Biodiversity’(ConFoBi)project(Storch et al.,2020).The Black Forest consists of mixed forest comprised of mainly Norway spruce(Picea abiesL.),European beech(Fagus sylvaticaL.),Silver fir(Abies albaMill.),maple(Acerspp.),and oak(Quercusspp.),covering a complete gradient from completely coniferous to deciduous stands.Research plots range in elevation between 443 and 1,334 m above sea level,with an average of 819±183 m(mean±SD).They reflect variations in slope(1°‒34°,15°±9°)and aspect(3°‒360°).Tree communities vary in number of trees with diameter at breast height(DBH)greater than 7 cm(98–1,212,425±205),tree basal area(BA)(9–73 m2,34±9 m2),tree species richness(2–15,5.5±2.2),and the proportion of deciduous species(0–96%,28%±25%).Management regimes have created gradients ranging from even-aged stands of individual species(mainly planted spruce that naturally would not occur in high abundance),to uneven-aged stands of European beech and unmanaged forest reserves.In the past,economic management of the Black Forest has resulted in large swaths of Norway spruce,while more recent management regimes have focused on converting forests to Beech-dominated,representing the potential natural vegetation of the area.Additionally,conservation initiatives have focused on the retention of potential habitat trees(Storch et al.,2020),and standing deadwood structures to support biodiversity.Plots reflect such conservation/retention measures in a design that uses variation in space to(also)infer variation in time(sensu Blüthgen et al.,2022)along gradients of the number of standing dead trees,standing and lying deadwood volumes as well as a plethora of structural and compositional forest variables.For more detailed information on the ConFoBi plot selection and forest variables see Storch et al.(2020).For a map of the study area see supplementary material(Fig.S1).
2.2.Forest variables
Plot-level forest variables were measured during full forest inventories conducted in 2017 and 2018.From these inventories mean diameter at breast height(DBH),tree basal area(BA),deciduous tree share,standing/lying deadwood volumes,and plot elevation were obtained.Standing and lying deadwood diversity indices used in the present study were calculated by Knuff et al.(2020)following the procedure described in Siitonen(2001).These indices represent all combinations of deadwood species,types and decay stages,for each unit of standing or lying deadwood greater than 7 cm diameter present at each plot:the type(coniferous vs.broadleaved),decay class(1–2,3,4–5),and diameter in 10 cm classes.For example,a standing dead conifer decayed at class 1–2 with a diameter of 10–19 cm would represent one unique deadwood type in the standing deadwood diversity index.The standing and lying deadwood diversity indices were summed to create combined deadwood diversity used in analyses.These indices were combined to account for the potential additive effect of different deadwood types on beetles(Parisi et al.,2018).Herb cover and understory species richness were measured from six 5 m×5 m subplots in 2017(Helbach et al.,2020).Canopy gap fraction(percentage of open area in tree canopy)was measured from images taken during unmanned aerial vehicle flights over plots(Frey et al.,2018).Forest cover(percentage of forested area in 1 km2surrounding plot centers)values were calculated using aerial image data by Storch et al.(2020).The remotely sensed indices,stand structural complexity index(SSCI),effective number of layers(ENL)and understory complexity index(UCI)were derived from terrestrial laser scans at insect trap locations(Frey et al.,2019),which were the northwest and southeast corners within each plot.Mean values for each index were calculated from the two trap locations to generate one value per plot.The SSCI is a measure of geometric complexity of forest stand vegetation.SSCI correlates with stand structure,microclimatic variance and can be used to differentiate among forest types(Ehbrecht et al.,2016).Scanning is not limited in height,and relays the points of a vertical scanline of the whole plot.Points on the scanline are connected to a polygon,the fractal dimension of which is expressed as the ratio of area to perimeter.The ENL is an index for measuring the vertical height of vegetation layering,or forest strata is correlated with more diverse and more evenly layered stands.ENL algorithms consider 3D space in voxels,marking them filled when containing at least one point from the laser scan.The ratio between filled and unfilled voxels is then summarized in 1 m thick layers,providing a histogram of space filled by vegetation as a function of height.A value for the ENL is then an inverse Simpson index of the diversity between these layers(Ehbrecht et al.,2016).The UCI represents an index similar to the SSCI but for vegetation around trap locations at specific strata.It is computed using laser scans on a horizontal plane,from which a ratio of perimeter to area is calculated(Willim et al.,2019).For additional information about the remotely sensed indices used for this study see Knuff et al.(2020).For summary information of forest variables see Table 1.
2.3.Insect sampling/identification
Window traps were used to sample beetle communities(Knuff et al.,2019).Traps were constructed using two 50 cm×24 cm acrylic glass rectangles with a cone at the top and a modified plastic container at the bottom.Traps used no bait or scents,with funnels camouflaged in green to avoid biased sampling of certain insect groups and to ensure that only random flying insects were collected.Two traps were placed on each study plot~75 m apart from each other toward the northwest and southeast corners.Sampling was conducted continuously with four-week collection intervals between March and August of 2017.After collection,insects were sorted to order level,with Coleoptera identified to family(Freude et al.,2009).Bark beetles were identified to sub-family(Scolytinae)level to allow for differentiation from true weevils(Curculionidae),and considered as one distinct group in diversity analyses.While a high taxonomic resolution is always desirable,when comprehensively sampling many plots it is often logistically not feasible to identify all collected specimens to species level.In such cases,surrogacy by a higher taxonomic level(e.g.family)can provide reliable information about species diversity(Williams and Gaston,1994;Williams and Gaston,1994;Balmford et al.,1996;Zou et al.,2020).During identification,specimens were classified according to body length(mm)into one of seven size classes:<2.5,2.5–5,5–10,10–15,15–20,20–25,>25.In addition to morphological identification,metabarcoding data of the same samples from bottom collection units were used to assign unidentified specimens(~15%)of predominantly the<2.5 mm(43.4%)and 2.5–5 mm(48.7%)size classes to families.This was done by cross-referencing the number of unidentified specimens in each size class with the proportion of OTU reads from species within the same size classes.The proportions of OTU reads from families within one size class were applied to the number of unidentified specimens in the same size class to assign family identifications.This method was used for only those families exclusively revealed by metabarcoding which had previously not been identified morphologically in a sample,and were therefore among the unidentified specimens.If no additional families were revealed by the metabarcoding dataset,unidentified specimens(~7%)were excluded from further analyses.For additional information on metabarcoding procedures see the supplementary material.
Families were assigned to guilds according to their most common modes of feeding observed during the longest portion of lifecycles(Freude et al.,2009).Family level identification allows to infer common modes of feeding(Simberloff and Dayan,1991;Grimbacher and Stork,2007;Wardhaugh et al.,2012).For example,the majority of the species within the family Cerambycidae feed on deadwood and develop within deadwood structures as larvae.By comparison to the larval stage,most species of this family have short adult lifespans(Haack et al.,2017),during which they typically do not feed on deadwood.Thus,this family was considered saproxylic(feeding on dead/decaying wood)in our data.In total,eight feeding guilds were considered:carnivores(feeding on other animals),fungivores(those feeding on fungal mycelia/spores/-fruiting bodies),herbivores(feeding on living plant material),omnivores(foraging from multiple different sources),palynivores(feeding on pollen/nectar),parasitoids(requiring the death of a host as part of reproductive cycle),saprovores(feeding on decaying organic material excluding deadwood),and saproxylics.Individuals of the family Silphidae(3%)were excluded from analyses due to their attraction to decaying material present in window traps,and the related potential samplingbias.To reduce complexity,the relative abundance and biomass of parasitoid and palynivorous taxa were added to carnivores’and herbivores’respectively.For additional information about feeding guild classification,see supplementary material(Table S1).
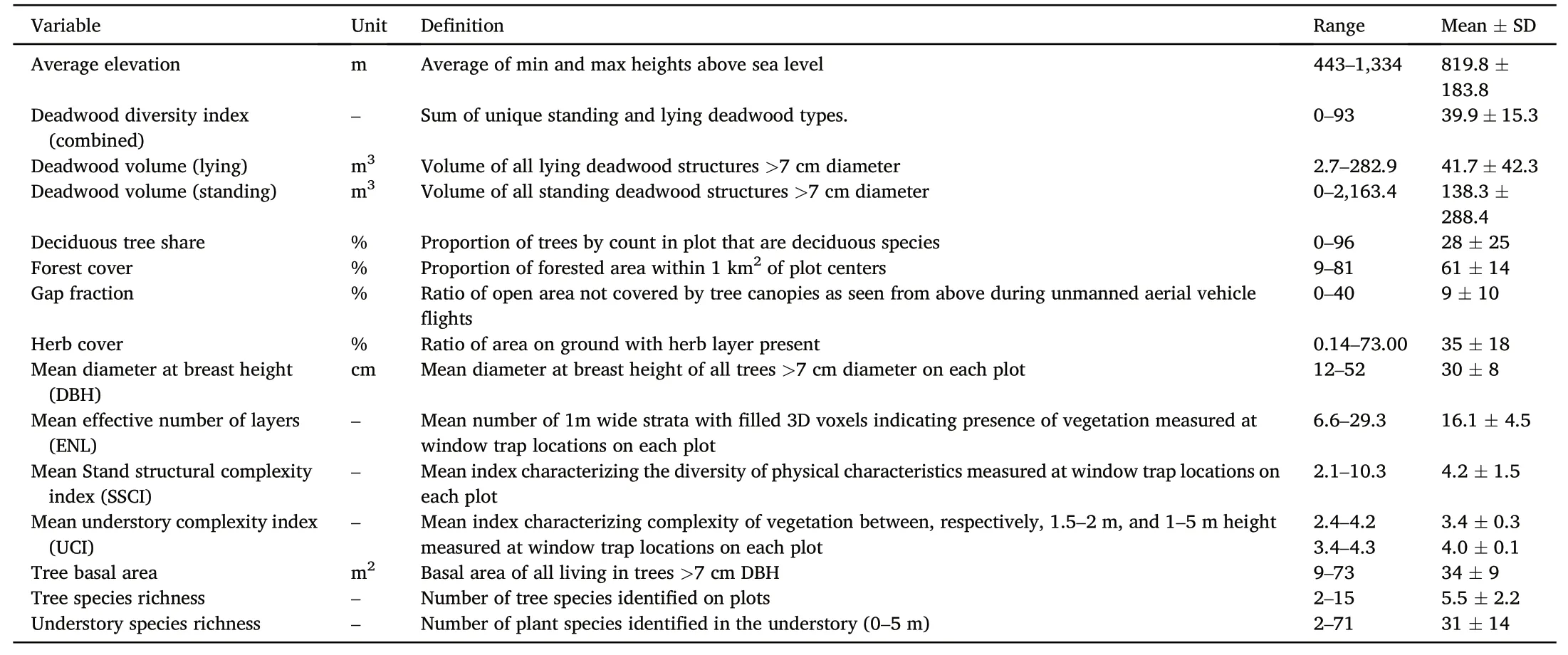
Table 1Forest explanatory variables characterizing the 128 plots(1 ha each)used for analyses,with corresponding summary statistics.Variables were excluded on the basis of assumed collinearity(ρ>0.70)following pairwise analyses,the more ecologically relevant variable being kept.A summary of Spearman's correlations among forest variables can be found in the supplement,Table S3.
2.4.Biomass measurement
While drying and weighing are standard for biomass measurement,this was not possible with all specimens collected(41,867).Therefore,after identification and size classification,representative samples of each family were taken using as many individuals as were available,without taking more than 60 for any single representative group.Each individual in representative samples was measured for precise length(front of the head to tip of abdomen)and width(pronotum at widest point)values(nearest 0.01 mm).While interspecific variation results in a margin of error when calculating family level allometries,accounting for the size and region of each individual reduces this error,as values calculated are based on allometries of similarly sized specimens(Ganihar,1997).These specimens were then dried at room temperature under a fume hood for no less than 72 h and dry weight recorded(at 0.0001 g)using a precision balance.Individuals of the same taxon weighing less than 0.0001 g were weighed together,with mean values taken to represent each.From these data,allometric regressions were performed to create two power curves for each taxon(Wardhaugh,2013):one for length-width and one for length×width-biomass.The length-width allometry was used to calculate a width value for each size class and taxon.The calculated width value was then multiplied by the median length value of each size class to generate length×width values for each size class and taxon.The length×width values were then used in allometries to calculate biomass of each individual in each size class and taxon.The use of length×width values has been shown to more accurately predict the biomass of invertebrates than length values alone(Wardhaugh,2013).While many equations are available for calculating allometries,power functions have been found to best estimate the dry weight of Coleoptera adults(Ganihar,1997).Biomass of each family per sample was calculated by multiplying the number of specimens by the estimated biomass value for its respective size class,then summing values from size classes.When fewer than five individuals of a family were available for measurement in representative samples,the power curve of its superfamily was used.If the power curve of a superfamily was also not available,a power curve using all measured Coleoptera specimens of the relevant size class was used.For summary information and allometric formulas for each family,see supplementary material(Table S2).
2.5.Statistical analyses
Beetle samples were pooled per plot.Shannon diversity values were calculated for each plot using the number of individuals collected from each family(R package vegan,Oksanen et al.,2017).Total beetle biomass was calculated by summing the biomass of all families.All fixed effects were assessed for collinearity using Spearman's coefficient(Dormann et al.,2013).Spearman's coefficient was chosen as not all forest variables selected are normally distributed.If a pair of variables was determined to be collinear(ρ>0.70),only one of them was retained for analysis(Table 1,Table S3).This was the case for combined deadwood diversity(correlated with both lying and standing deadwood diversity,combined deadwood diversity retained).Plots with missing values for forest variables at one or both trapping sites were omitted(seven plots).Prior to analyses,biomass values and fixed effects were log-transformed(log10(x+1))to increase normality and homoscedasticity.Following log-transformation,fixed effects were scaled(mean=0,SD=1)in all models.Initial full models were calculated including all fixed effects(Table 1).Abundances of Coleoptera(one model)and individual feeding guilds(six models)were analyzed using negative binomial generalized linear models.Biomass of Coleoptera(one model),feeding guilds(six models)and family diversity(one model)were analyzed using linear models.Prior to model averaging,Moran's I tests were used to assess initial models for spatial autocorrelation,comparing residuals with a dissimilarity matrix of plot geolocations(latitude,longitude)(Table S4).Final models of each response variable were constructed using the model averaging approach outlined in Symonds and Moussalli(2010)and Grueber et al.(2011)(R package‘MuMIn’,Barton,2020).This approach first creates a set of candidate models(from all fixed effects in the full models)using all possible combinations of fixed effects.Each candidate model is assigned an Akaike weight based on its AICc value relative to other candidate models.This weight is indicative of the explanatory power of individual candidate models with a specific combination of fixed effects.For example,a candidate model with an Akaike weight of 0.10 has a 10%probability of being the best model to describe the data.Candidate models with the highest weights were selected until reaching a cumulative weight of 0.95.Models in the selection were then averaged to produce a model including all fixed effects that appeared in the averaged model(accounting for the respective weights)and thus explained the most variation in the response variables.
To analyze the relationships between Coleoptera composition and fixed effects,principal component analysis(PCA)using the‘rda’function in the vegan package(Oksanen et al.,2017)was calculated separately on biomass and abundance values of feeding guilds at plot level.Species scores were plotted for each response variable following ordination.All fixed effects included in Table 1 were applied to the rda using the‘envfit’function with 10,000 permutations.
3.Results
In total 41,867 Coleoptera specimens comprising 71 families and 8 feeding guilds were collected and identified(Table S2).Herbivores were the largest guild by abundance(38%),followed by omnivores(21%),saproxylics(14%)and carnivores(11%).The remaining feeding guilds(4)jointly comprised less than 25% of individuals sampled.Omnivores represented~57%of the total sampled biomass,followed by saproxylics(15%)and herbivores(13%).The remaining feeding guilds(4)combined represented less than 15%of the total biomass(Table S1).
Averaged linear models revealed total beetle biomass increased with mean tree DBH(z=3.199,p=0.001)(Fig.1a),while decreasing with SSCI(z=1.581,p=0.004)(Fig.1b).Shannon diversity of families increased with mean ENL at window trap locations(z=2.965,p=0.003)(Fig.1c).Among feeding guilds,the biomass of carnivores decreased with tree BA(z=2.500,p=0.012)(Fig.2a).Fungivore biomass increased with deciduous tree share(z=2.032,p=0.004)(Fig.2b)and elevation(z=3.363,p≤0.001)(Fig.2d)while decreasing with both gap fraction(z=3.102,p=0.002)(Fig.2c)and tree BA(z=2.217,p=0.027)(Fig.2e).Herbivore biomass increased with both elevation(z=2.700,p=0.007)(Fig.2f)and UCI at 1–5 m(z=2.110,p=0.035)(Fig.2g).Omnivore biomass decreased with only herb cover(z=1.999,p=0.046)(Fig.2h).Saprovore biomass increased with both DBH(z=2.333,p=0.031)(Fig.2i)and elevation(z=2.333,p=0.019)(Fig.2j).Saproxylic biomass increased with lying deadwood volume(z=2.720,p=0.007)(Fig.2k)while decreasing with SSCI(z=2.338,p=0.019)(Fig.2l)(Table S5).
Fungivore,omnivore,saproxylic and total abundances showed no significant relationship with any forest variable.Averaged generalized linear models showed that the abundances of only carnivores,herbivores and saprovores were related to forest variables(Table S6).Carnivore(t=2.333,p=0.019)and saprovore abundance increased with elevation(t=2.923,p=0.003),while herbivore abundance increased with deciduous tree share(t=4.232,p≤0.001)(Fig.3 a‒c).Results were in almost all cases not related to space,with potential autocorrelation detected for only carnivore biomass(Moran'sI=0.035,p=0.001)and saproxylic abundance(Moran'sI=0.017,p=0.049)(Table S4).
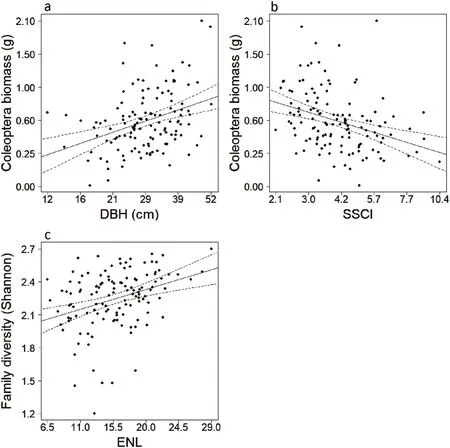
Fig.1.Log-transformed(log10(x+1))Coleoptera biomass and significant(at p<0.05)log-transformed(log10(x+1))fixed effects(a,b).Shannon diversity of families and significant fixed effects(c).a)DBH is the mean tree diameter at breast height.b)SSCI is the stand structural complexity index.c)ENL is the mean effective number of layers(ENL)or forest strata occupied by vegetation.Axes in each figure display real values for biomass(g)and fixed-effects.Solid lines give model predictions(95% confidence intervals as dashed lines).
Principal component analysis revealed for beetle community composition by biomass similar relationships to forest variables as individual biomass analyses,with significant effects from mean tree DBH(R2=0.138,p≤0.001),elevation(R2=0.110,p≤0.001),SSCI(R2=0.124,p≤0.001),and UCI at 1.5–2 m height(R2=0.091,p=0.003).Additionally,standing deadwood volume(R2=0.052,p=0.039)and tree BA(R2=0.076,p=0.010)were revealed to be significant(Table S8).Herbivores,saprovores,and carnivores comprised greater proportions of communities on plots with higher mean DBH.Saproxylics and omnivores were present in larger proportions on plots with lower SSCIs.Plots at higher elevations contained greater proportions of fungivores.The first two PCs accounted for 52.85% of the variation among sites(Fig.4a).Principal component analysis of beetle community composition by abundance revealed similar significant effects from deciduous tree share(R2=0.193,p≤0.001)and elevation(R2=0.073,p=0.009)as observed in individual analyses of several feeding guilds.Additionally,significant for community composition by abundance were lying deadwood volume(R2=0.154,p=0.005),standing deadwood volume(R2=0.264,p≤0.001),SSCI(R2=0.084,p=0.007),and tree BA(R2=0.062,p=0.028)(Table S10).The first two PCs accounted for 56.00% of the variation among sites(Fig.4b).For tables of species’scores of principal components and results from fitting of forest variables see supplementary material(Tables S7-S10).
4.Discussion
The joint analyses of beetle biomass,diversity and abundance indicates that these metrics respond differently to structural elements in forests.Retention forestry can potentially influence all structural variables in ways which promote beetles.The differences among feeding guild biomass in our models can be related to nutritional resources in several cases,as hypothesized.This is directly observed for saproxylic taxa and increasing lying deadwood(Lindhe et al.,2005;Bouget et al.,2011,2013)and herbivorous taxa and understory complexity(UCI).Increasing UCI may indicate greater availability of host plants for herbivorous taxa(Willim et al.,2019).The remaining feeding guilds and all Coleoptera together showed less direct responses than hypothesized,and their relationships to structural elements can be mediated by the resources such structural elements relate to.Fungivore biomass for example,decreased with increasing gap fraction,which has been experimentally shown to reduce the observations and species richness of fungi in temperate forests(Brazee et al.,2014).Due to increased fungal bodies observed on deadwood(Blaser et al.,2013)we expected to see higher fungivore biomass with greater deadwood volumes,which was not the case.We found,however,that the proportion of deciduous trees,which has been shown to positively scale with the amount of leaf litter and coarse woody debris(Pedlar et al.,2002)and thus the amount of fungi(Unterseher et al.,2013;Purahong et al.,2015),had a positive effect on the biomass of fungivores.The unexpected findings that both fungivore and herbivore biomass increased with elevation could possibly be due to higher growth rates of some central European tree species(Pretzsch et al.,2020)combined with denser stands(Maz′on et al.,2020)at higher elevations,providing more resources for both feeding guilds.Increased growth rates and denser forest stands may also partially explain the increase in saprovore biomass observed with elevation,via decomposition driven by denser tree canopies(Wallace et al.,2018),though our data cannot support such conclusions and these relationships remain unclear.Similarly,it is unclear how omnivore biomass decreases with increasing herb cover.The points in Fig.2h show much greater variation at higher herb cover indicating this relationship could be partly driven by stochasticity.
Saproxylic and overall biomasses decreased with stand structural complexity(SSCI)likely due to microclimatic variations correlated with higher forest strata(Willim et al.,2019)and structural complexity(Ehbrecht et al.,2016),reducing flying beetle activity and decreasing their presence in samples.
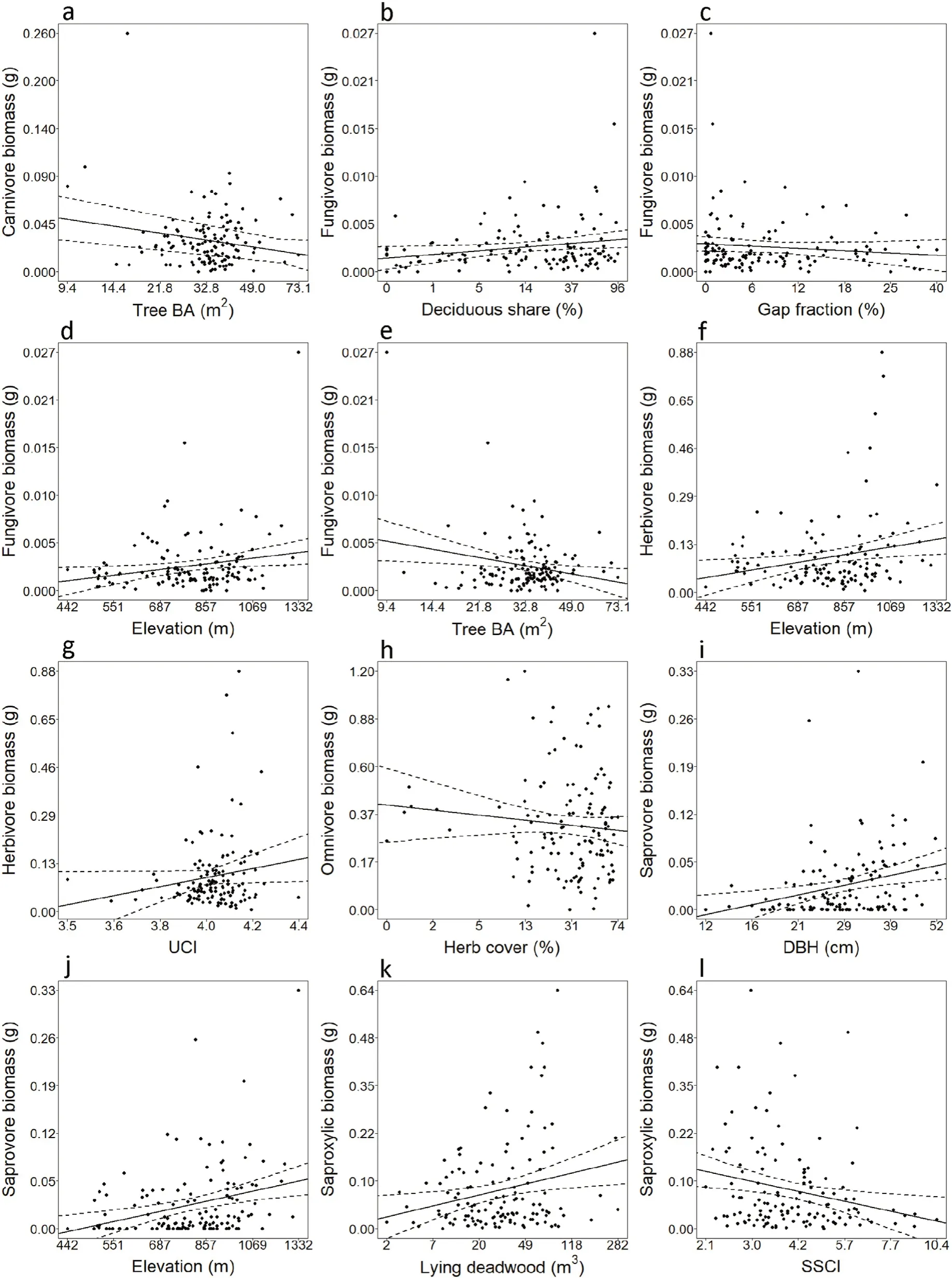
Fig.2.Log-transformed(log10(x+1))biomass of individual functional groups(a;carnivores,b‒e;fungivores,f‒g;herbivores,h;omnivores,i‒j;saprovores,k‒l;saproxylics)and significant(at p<0.05)log-transformed(log10(x+1))fixed effects.a)and e)Tree BA represents tree basal area.g)UCI is understory complexity index at 1–5 m.i)DBH is mean diameter at breast height.l)SSCI is stand structural complexity index(SSCI).Axes in each figure display real values for biomass(g)and fixedeffects.Solid lines give model predictions(95% confidence intervals as dashed lines).
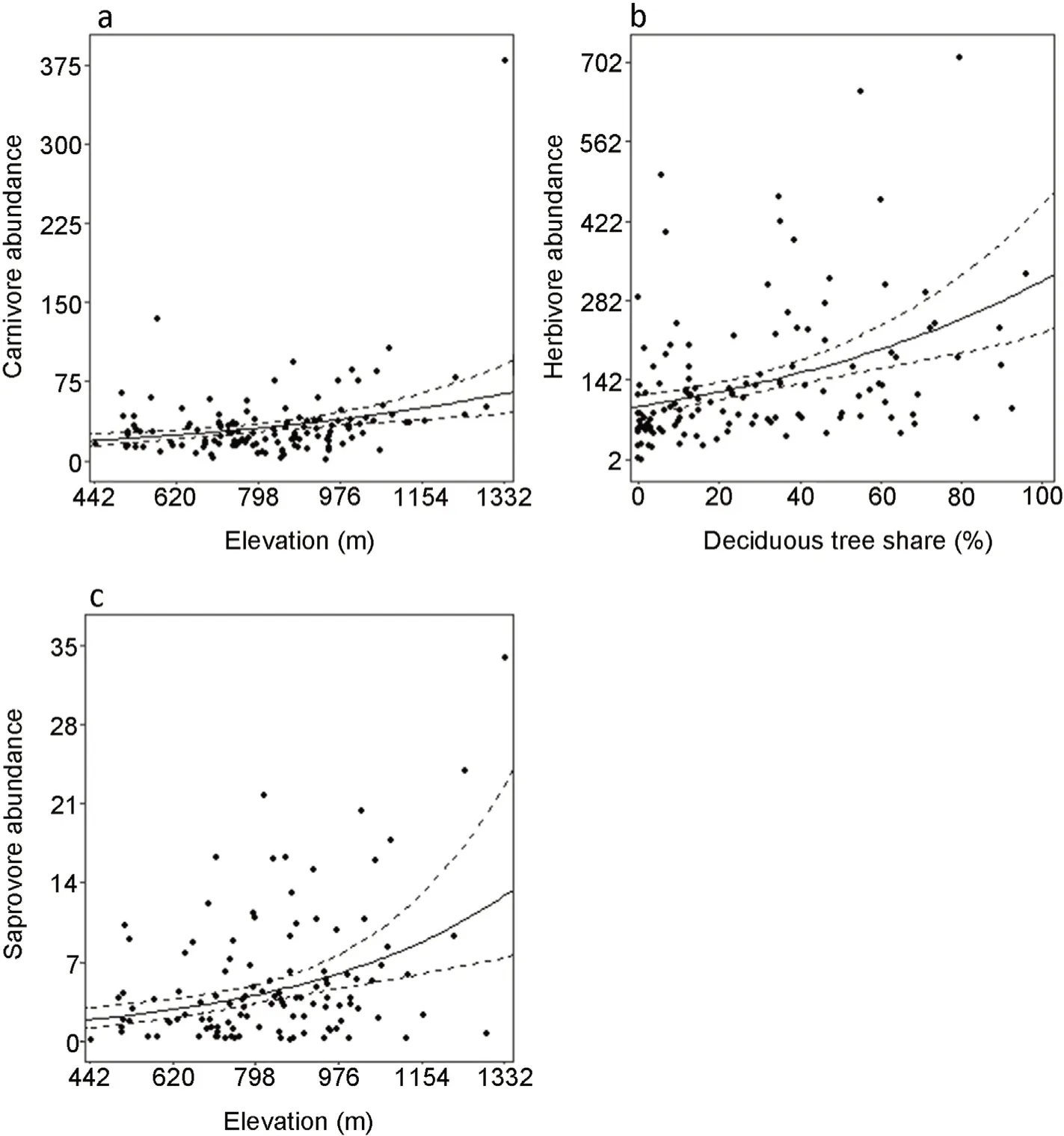
Fig.3.Abundances of Coleoptera feeding guilds(a;carnivores,b;herbivores,c;saprovores)and significant fixed effects.Solid lines give model predictions(95%confidence intervals in dashed lines).

Fig.4.Biplot of principal component analysis for a)log-transformed(log10(x+1))Coleoptera biomass,and b)Coleoptera composition by abundance.Feeding guild labels represent plotted species'scores(Table S8 biomass,Table S10 abundance).Results of environmental fitting were plotted as arrows using only significant variables(permutations=10,000,p<0.05)with arrow length corresponding to explained variance(R2)of fixed effects(Table S9 biomass,Table S11 abundance).Abbreviations are as follows:stand structural complexity index(SSCI),diameter at breast height(DBH),understory complexity index at 1.5–2.0 m(UCI),tree basal area(BA).Standing and lying deadwood represent volumes(m3).
While initially hypothesized that total beetle biomass would increase with increasing values of variables characterizing greater amounts of forest habitat such as forest cover,it increased with only mean tree diameter at breast height(DBH).Among feeding guilds,only saprovore biomass increased significantly with DBH,while the biomass of herbivores and omnivores was nearly significantly related to DBH.Tree basal area(BA)was included in models to more comprehensively examine the relationships between response variables and tree sizes/stand densities(Zeide,2005),but showed significant relationships with only carnivore and fungivore biomass,differing from mean DBH.For carnivores,this effect was significant following model averaging albeit with a signal of spatial influences according to Moran'sI.As biomass is a proxy for resource availability(Wardhaugh et al.,2012;Wardhaugh,2013),the relationship between DBH and beetle biomasses may be mediated by resources utilized by the feeding guilds measured.Feeding resources from larger trees could be increased deadwood amounts(Oettel et al.,2020;ˇSenhofa et al.,2020),increased growth of fungi(Parmasto,2001;′Odor et al.,2006),and more living tree material available.Therefore,there are positive,albeit weak,relationships between feeding guilds'biomass and DBH,most apparent when all groups are combined.Higher biomass of feeding guilds emphasizes the importance of maintaining larger trees to promote forest beetles,as larger trees are important elements for forest habitat(Larson,1995;Manning et al.,2009;Lutz et al.,2018),providing more resources and microhabitats for insects(Hor′ak,2017).The negative influence of tree BA on the biomass of carnivores and fungivores suggests that this cumulative effect requires not higher numbers of trees,but the presence of large trees,possibly in combination with uneven layering of the stand.
Only the effective number of layers(ENL)proved significant for beetle family diversity.As higher values of this index characterize more diverse and more evenly-layered stands,it indicates the presence of more heterogenous vegetation.This is possibly due to a structurally more diverse assemblage of host plants yielding a more diverse assemblage of beetles utilizing different resources.Greater host plant diversity could cascade to other similarly related feeding guilds such as carnivores,and omnivores.In addition,it has been found that diversity in the age structure of forests can benefit multiple taxa(Schall et al.,2018).Surprisingly,no other forest variables representing forest heterogeneity(including tree species richness,understory species richness,understory complexity)were found to be significant for the diversity of beetle families,contrary to our hypotheses.This could possibly be due to the coarse taxonomic unit used in analyses,as greater variation in beetle species diversity between research plots would likely result in a more nuanced relationship.In addition,our results could be influenced by the sampling method,which was conducted in only the understory,providing a snapshot of only one stratum of the forest stand.Several studies have demonstrated that the sampling of multiple forest strata(Proch′azka et al.,2018;Stone et al.,2018;Leidinger et al.,2019),multi-year sampling(Parmain et al.,2013;Chen et al.,2014)and the use of sampling methods complementary to window traps such as pitfall traps and trap nests(Staab et al.,2021)or malaise traps(Lamarre et al.,2012)can together provide more complete inventory of insects in forests.
It was expected(similar to feeding guild biomass)that abundance of more specialized feeding guilds such as fungivores and saproxylics would display stronger relationships than more generalized guilds such as saprovores.It is surprising then that carnivores,herbivores and saprovores are the only feeding guilds showing significant relationships for abundance.With herbivores,it is expected that increasing deciduous tree share promotes larger abundances of leaf-feeding herbivores(Vehvil¨ainen et al.,2007),via the presence of more feeding resources of different tree species.However,higher elevation,which truncates insect flight periods(Welti et al.,2021)was not expected to increase the abundance of any feeding guilds.While abundance decreasing with elevation is a general trend with many terrestrial insects,it is not uniformly the case for all species(Hodkinson,2005;Binkenstein et al.,2017).Our observed results could then simply be driven by several species within the carnivore and saprovore feeding guilds,which are more abundant at higher elevations.Abundance and biomass differed in their relationships with forest elements as biomass can be more strongly affected by few large-bodied individuals(Sample et al.,1993),while abundance can be strongly influenced by locally numerous species(Gaston and Lawton,1988),possibly exploiting a small amount of locally available feeding/nesting resources.In this way,biomass may be more indicative of resource availability and stability as larger insects require greater amounts of resources(Ribeiro and Freitas,2011).
Principal component analysis(PCA)of composition by biomass supported the conclusions from averaged models using biomass of individual feeding guilds,with the addition of standing deadwood volume and tree BA as important variables in structuring composition.Notably,each feeding guild was present in greater proportions at plots with the highest DBH values,concurrent with hypotheses and results regarding resource availability(Welti et al.,2020).The PCA of composition by abundance was similar to,yet different from,the PCA by biomass.The absence of DBH from the PCA using abundance indicates that for the abundance of same feeding guilds,forest variables indicating the amount of resources becomes less important.Instead,composition by abundance is seemingly influenced by structural variables such as standing deadwood(Jonsell et al.,1998;Bouget and Duelli,2004;Gullan and Cranston,2014)which relate to the presence of resources.Standing deadwood and lying deadwood were similarly related to composition by abundance,which can possibly be due to similar ecological use by both structures.As seen with individual biomass models,and PCA using biomass,SSCI was significant for structuring communities by abundance.However,what is observed is greater compositional similarity at higher values of SSCI,while plots with more distinct communities plot with other variables such as standing deadwood.The clustering of points with SSCI while feeding guild species’scores plot elsewhere in the PCA,indicates high abundances of smaller individuals within multiple feeding guilds with greater SSCI.Tree BA was also important for composition by abundance,and as observed with biomass increases the proportions of saproxylics and omnivores.While significant for composition by biomass,UCI was only nearly significant for composition by abundance.Comparing PCAs using biomass and abundance separately shows that the two metrics,though similar,are not identical.The differing responses to structural elements these composition metrics show emphasizes the importance of understanding the metric used,and considering both(if possible)when developing strategies to promote forest beetles.
5.Conclusions
Management of forests at stand level determines the availability of structural elements.The effects of forest structural elements on beetle biomass,diversity and abundance are complex and interrelated,making management decisions with the intent of promoting them difficult.Structural elements can be augmented through retention forestry.Maintaining unevenly layered stands can promote beetle diversity via the creation of more forest strata occupied by vegetation.The retention of forest elements including large trees,lying and standing deadwood,and more complex understories can have positive influences on the biomass and abundance of beetle feeding guilds via direct and indirect resource availability,with strong yet varying influences on community structure,depending on the biodiversity metric used.The varying relationships with forest structural elements,and the differences in responses between biomass,diversity and abundance suggest that multiple specific elements are needed to promote forest beetles.Management strategies should therefore prioritize multiple forest elements simultaneously to account for the varying responses of different metrics to assess these efforts,and achieve greater specificity by factoring resource use of different feeding guilds.
Funding
Funding for the RTG ConFoBi has been provided by the German Research Foundation(DFG)(Grant number GRK 2123/2),which was not involved in conducting research or preparation of the manuscript.
Availability of data and material
Data and material for this work are available upon request to the corresponding author.
Code availability
Codes are available upon request to the corresponding author.
Authors contributions
NJR conceived the research question and hypotheses,conducted data collection and analyses and drafted the manuscript.MS provided conceptual and analytical advisement and reviewed drafts of this work.NW conceived of,and conducted fieldwork for,the study which collected the specimens used to produce this manuscript.JF contributed remote sensing data.AMK and MS designed the research.All authors commented the manuscript.
Ethics approval
NJR reports this work was carried out in compliance with the Guidelines for safeguarding good research practice,laid out by the German Research Foundation(DFG).
Consent to participate
Not applicable.
Consent for publication
NJR grants full consent for the publication of this manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank the conservation authorities of the state of Baden-Württemberg for sampling permits.For collecting and processing insect specimens and providing contributions to previous versions of the manuscript we thank Dr.Anna Knuff.The assistance of Laura-Sophia Ruppert during sorting and identification of beetle specimens was crucial to the completion of this work.The metabarcoding data used could not have been gathered without the assistance of Kay Lucek,who additionally provided comments on a draft of this manuscript.Matthias J¨ager provided support for this project.We thank the many scientific assistants were involved in the sorting and processing of insect samples.We are grateful for the helpful comments and insights provided by two anonymous reviewers.We thank Ilse Storch,Johannes Penner and the ConFoBi coordination team for establishing and supporting this research training group.Funding for this project was provided by the German Research Foundation.
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.i.org/10.1016/j.fecs.2022.100056.
- Forest Ecosystems的其它文章
- Diversity of click beetles in managed nonnative coniferous and native beech stands:Consequences of changes in the structural and species composition of tree stands in Central Europe
- Environmental and canopy conditions regulate the forest floor evapotranspiration of larch plantations
- Allometry-based estimation of forest aboveground biomass combining LiDAR canopy height attributes and optical spectral indexes
- Examining approaches for modeling individual tree growth response to thinning in Norway spruce
- Influence of soil and elevation on roadside cryptogam diversity in the tropical Andes
- Patterns of species diversity and its determinants in a temperate deciduous broad-leaved forest

