Full-length transcripts facilitates Portunus trituberculatus genome structure annotation*
Fangrui LOU , Zhiqiang HAN
1 School of Ocean, Yantai University, Yantai 264005, China
2 Fishery College, Zhejiang Ocean University, Zhoushan 316022, China
Abstract Portunus trituberculatus is an ideal model for elucidating crustacean genetic networks. Here we combined single molecule real-time (SMRT) sequencing and Illumina RNA-seq to characterize the coding genes, non-coding RNAs and pseudogenes and further to improve the genome annotation information of P.trituberculatus. In this study, we assembled 9 694 non-redundancy full-length transcripts, and 658 737 307-bp repetitive sequences were identif ied in the P. trituberculatus full-length transcriptome. We also predicted the P. trituberculatus genome structure based on full-length transcripts, including 18 602 genes, 28 686 non-coding RNAs, 1 407 pseudogenes, 740 motif, and 26 434 domain. Meanwhile, 14 460, 10 211, 5 412, 7 314, and 14 448 genes had signif icant matches with sequences in the NR, KOG, GO, KEGG, and TrEMBL database, respectively. Overall, our work f irstly provided the long-read transcriptome and we believed that these data are very necessary to improve the annotation information of P. trituberculatus genome structure,and useful information for the future studies on evolution and physiological regulation of P. trituberculatus.
Keyword: Portunus trituberculatus; full-length transcripts; single molecule real-time (SMRT) sequencing
1 INTRODUCTION
The functional, physiological, and biosynthetic cellular states of crustacean are very complex, and the structural and functional genomics studies are fundamental for understanding the crustacean biology and essential to the access to high-quality genome resources. The high-throughput sequencing technologies stimulated the construction of reference genome resources for many crustaceans (Colbourne et al., 2011; Zhang et al., 2019). However, some reference genomes are often incomplete and insuffi cient to decode the annotation and structure(Choi et al., 2015; Shen-Gunther et al., 2016).Meanwhile, it seems that the encoding potential and gene expression regulation of crustaceans cannot be determined based on genomic information alone,because the post-transcriptional processing of precursor mRNAs is very diverse due to the presence of alternative splicing and polyadenylation (Kalsotra and Cooper, 2011; Elkon et al., 2013). The characteristics of transcriptome provides an opportunity to improve the accuracy and completeness of crustacean genome resources and elucidate the complexity of multiple biological mechanisms as the transcriptome information depicts the gene expression level and individual splice junction (Mortazavi et al.,2008; Wang et al., 2009).
Having obtained via high-throughput short-read sequencing (RNA-seq), transcriptome sequences of many crustaceans have been accumulated recent years (Xu et al., 2017; Lou et al., 2018). However,full-length reads benef icial for subsequent functional and transcriptional behavioral studies of crustacean important loci than short transcripts are recently considered as full-length transcripts can eff ectively predict the exon-intron structures, alternative splicing,alternative polyadenylation, and other genome structures (Ogihara et al., 2004; Soderlund et al.,2009). Since 2012, the third-generation sequencing platforms, such as Pacif ic Biosciences (PacBio) and Oxford Nanopore (ONT), have been gradually applied to long-read sequencing (Eid et al., 2009; Feng et al.,2015).
The swimming crabPortunustrituberculatus, one of the Asia’s most valuable marine crustaceans, is an ideal model for elucidating crustacean genetic networks (Qi et al., 2013). Research intoP.trituberculatusis facilitated by increasingly ref ined transcriptome knowledge of the morphological and physiological characteristics. TheP.trituberculatuschromosome-level reference genome, assembled in 2020, used a sequencing strategy combining shortreads, ONT long-reads, and high-through chromosome conformation capture (Hi-C) sequencing data (Tang et al., 2020). Although the assembly eff ect ofP.trituberculatusgenome is relatively perfect, the defects in the ONT sequencing technology are apparent (Wyman et al., 2019). Meanwhile, shortreads used forP.trituberculatusgenome annotation do not provide the full-length sequence of each RNA,limiting their utility for def ining the genome annotation ofP.trituberculatus.
We combined the Illumina and PacBio platforms to generate a more completeP.trituberculatusfulllength transcriptome with rich data of full-length cDNA sequences that extends our knowledge ofP.trituberculatustranscriptome. Our research conf irmed the ability and reliability of long-reads in the discovery of full-length cDNA transcripts and novel genes/isoforms, which will improve genome annotation effi ciency ofP.trituberculatus.
2 MATERIAL AND METHOD
2.1 Animal specimen
FemaleP.trituberculatusspecimen was collected from the coastal water of Zhoushan, China.P.trituberculatuswas kept in aquarium for 4 days with fully aerated seawater. Next,P.trituberculatuswas immediately anesthetized and tissues (gill,muscle, heart, and intestinal) were then rapidly sampled, snap-frozen in liquid nitrogen at -80 °C.
2.2 Total RNA extraction
The total RNA of each tissue was extracted using the standard Trizol Reagent Kit (Huayueyang Biotech Co. Ltd., Beijing, China) following the manufacturer’s protocol. Later, the RNAs from all tissues were pooled in equal amounts. The concentration and integrity of mix RNA were assessed using the NanoDrop 2000 system (Thermo Fisher Scientif ic,MA, USA) and the Agilent Bioanalyzer 2100 system(Agilent Technologies, CA, USA). The mRNA was obtained by depleting rRNA of mix RNA using the RNA Purif ication Beads and then was cleaned three times using Beads Binding Buff er.
2.3 Illumina sequencing library construction and sequencing
The purif ied mRNA was incubated and used in turn to synthesize the f irst- and second-strand cDNA.Afterward, A- Tailing Control and Ligation Control were applied to the tailing and adapter ligation of the double stranded cDNA, respectively. All cDNA fragments were enriched to complete the libraries construction. After the libraries were diluted to 10 pmol/L, the Agilent 2100 Bioanalyzer was used for quantitative analysis, and the qualif ied libraries were sequenced on an Illumina HiSeq 2500 across one lane with paired-end 150 bp.
2.4 Single molecule real-time (SMRT) sequencing library construction and sequencing
The purif ied mRNA was incubated and then applied to synthesize the full-length cDNA required for sequencing using the SMARTerTMPCR cDNA Synthesis Kit (TaKaRa, USA), and then high-quality large-scale library amplif ication products were purif ied to 1-6 kb using the BluePippinTM Size-Selection System (Sage Science Inc., Beverly, MA,USA). The selected library was further undergone damage renovation, blunt end ligation, and addition of SMTBell adapters, which eventually form a SMRTbell template library. Finally, polymerase was added to the SMRTbell template and the resulted polymeride was f ixed to the zero-mode waveguide(ZMW). One SMRT cell was prepared and used on the PacBio RSII platform for full-length transcriptome sequencing.
2.5 SMRT sequencing data processing
Fig.1 Remove redundancy process of consensus sequences
The SMRT analysis software v2.3 Suite (http://www.pacb.com/devnet/) was applied to f ilter out the low quality fragments in length <50 bp and accuracy<0.9 in the polymerase reads generated by the PacBio RSII platform according to the following parameters:readScore 0.90 and minLength 50. Subreads were obtained by interrupting the remaining high-quality polymerase reads at the adapter location and f iltering out the adapter sequences. Subreads sequences <50 bp were f iltered using software SMRT v2.3 with parameters: minSubReadLength at 50, and the remaining subreads are identif ied as clean reads. The Iso-seq pipeline of software SMRTLink was used to extract the circular consensus (CCS) reads from the clean reads according to the following conditions: full passes >1 and sequence accuracy >0.90. CCS reads containing the correct 5′ primers, 3′ primers, and polyA tails were identif ied as full-length sequences,or otherwise were identif ied as non-full-length sequences. The insertion sequences of CCS reads were obtained by removing cDNA primer sequences and polyA sequences, and the direction of chain synthesis was determined according to the diff erences of primers at both ends of CCS sequences. These CCS reads were divided into full-length sequences, nonfull-length sequences, chimeric sequences, and nonchimeric sequences. Next, the Iso-seq pipeline of software SMRTLink was applied to cluster those similar full-length non-chimeric sequences (multiple copies of the same transcript) to obtain consensus isoforms. The software Quiver was used to calibrate the consensus isoforms to obtain high-quality(accuracy >99%) and low-quality transcripts (Li et al., 2018). To improve low-quality transcripts accuracy, low-quality consensus sequences were corrected using clean RNA-seq data based on software Proovread (Hackl et al., 2014). Strict parameters were set in the clustering process of full-length transcripts.In order to obtain high quality consensus sequences,the possibility of multi-copy sequences of the same transcript being divided into diff erent clusters is higher than the possibility of randomly clustering two copy sequences that do not belong to the same transcript, which inevitably leads to redundant sequences. Meanwhile, the 5′ end of some reads will be degraded in the full-length transcriptome sequencing process, resulting in diff erent copies of the same transcript cannot be clustered together.Software CD-HIT (Li and Godzik, 2006) can combine sequences with high similarity, thus it was used to remove redundant sequences from high-quality transcripts (Fig.1).
2.6 Genome annotation analysis
2.6.1 Repetitive sequences annotation
Due to relatively low conservativeness of repetitive sequences among species, it is necessary to construct specif ic repeat database when predicting repetitive sequences forP.trituberculatus. In this study,software LTR_FINDER (Xu and Wang, 2007) and RepeatScout (Price et al., 2005) were applied to construct theP.trituberculatustranscriptome repeating sequence database based on the principle of structure prediction and de novo prediction. The database classif ied by software PASTEClassif ier(Hoede et al., 2014) was merged with Repbase database (Jurka et al., 2005) to form f inal repeating sequence database. Finally, software RepeatMasker(Tarailo-Graovac and Chen, 2009) was used to predict the repetitive sequence based on the constructed repetitive sequence database.
2.6.2 Gene structure prediction
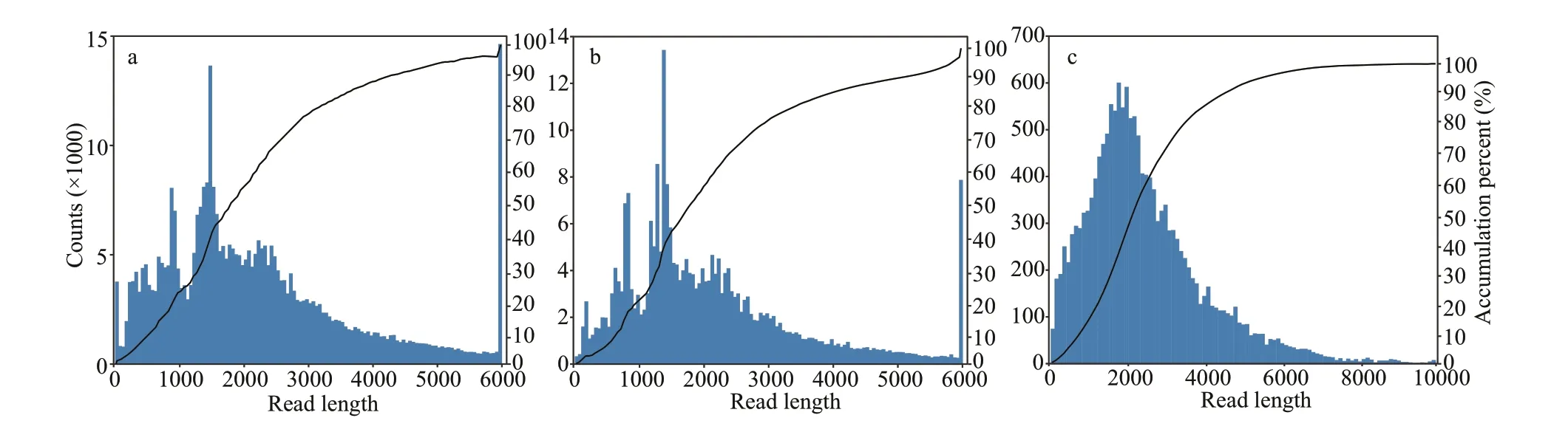
Fig.2 The length distribution of CCS (a), full-length non-chimeric reads (b), and consensus isoforms (c)
Three strategies, i.e., de novo prediction,homologous species prediction, and full-length transcripts prediction, were applied to predict the gene structure. In this study, de novo prediction was performed using software Genscan (Burge and Karlin,1997), Augustus (Stanke and Waack, 2003),GlimmerHMM (Majoros et al., 2004), GeneID(Blanco et al., 2007), and SNAP (Korf, 2004).GeMoMa software (Keilwagen et al., 2016) was used to carry out homologous species prediction based onArmadillidiumvulgare,Eurytemoraaffi nis,HyalellaAzteca,Penaeusvannamei, andTachypleustridentatus. Additionally, software TransDecoder(http://transdecoder.github.io), GeneMarkS-T (Tang et al., 2015), and PASA (Campbell et al., 2006) were used to full-length transcripts prediction. Finally,software EVM (Haas et al., 2008) was used to integrate the predicted results from three strategies.
2.6.3 Non-coding RNAs prediction
Non-coding RNAs are those who do not encode proteins, including miRNA, rRNA, tRNA, and other RNAs with known functions. According to the structural characteristics of diff erent non-coding RNAs, diff erent strategies were adopted to predict diff erent non-coding RNAs. In this study, we have identif ied the microRNA and rRNA based on the Rfam database using Blastn (Griffi ths-Jones et al.,2005). Meanwhile, software tRNAscan-SE (Lowe and Eddy, 1997) was used to predict the tRNA.
2.6.4 Pseudogenes annotation
The sequence of pseudogenes is similar to those of functional genes, but pseudogene loses its original function due to insertion, deletion, and other mutations. We used the predicted protein sequences and searched for homologous gene sequences(possible genes) on theP.trituberculatusgenome by BLAT (Kent, 2002) alignment. Furthermore,pseudogenes were obtained by using software Genewise (Birney et al., 2004) to search for immature termination codon and frame shift mutation in gene sequence.
2.6.5 Gene function annotation
All the predicted gene sequences were applied to analyze the gene ontology and orthologous classif ications based on NR, KOG, GO, KEGG, and TrEMBL database using the BLAST (Altschul et al.,1990) and the parameter E-value <0.00001.Meanwhile, software InterProScan (Zdobnov and Apweiler, 2001) was applied to annotate the sequence motif based on the PROSITE, HAMAP, Pfam,PRINTS, ProDom, SMART, TIGRFAMs, PIRSF,SUPERFAMILY, CATH-Gene3D, and PANTHER databases.
3 RESULT
3.1 P. trituberculatus full-length transcriptome sequencing using PacBio platform
High-quality total RNA was extracted from the multiple tissues ofP.trituberculatusand full-length transcriptome sequencing was then performed on the PacBio RSII platform. After f iltering out the low quality sequencing data and removing the adapter sequence, 42.6-Gb subreads were obtained form 1 SMRT cells. After f iltering out subreads below 50 bp in length, 23-Gb clean subreads were obtained in this study. Furthermore, 366 770 CCS reads were screened out from the clean subreads, corresponding to 847 034 414 bp. Meanwhile, the mean read length and the mean sequencing depth of CCS reads was 2 309 bp and 39 passes, respectively, among which 261 362 (71.26%) were considered as full-length nonchimeric sequences. Similar sequences in all the fulllength non-chimeric sequences were clustered into 15 832 consistent isoforms, with the mean length of 2 493 bp. After correcting the consistent isoforms,15 710 (99.23%) high-quality and 109 low-quality transcripts were obtained. The length distribution of CCS, full-length non-chimeric reads, and consensus isoforms are shown in Fig.2. Finally, CD-HIT (Li and Godzik, 2006) was applied to remove redundant sequences from high-quality transcripts, and 9 694non-redundant full-length transcripts were yielded and used for the following analyses.
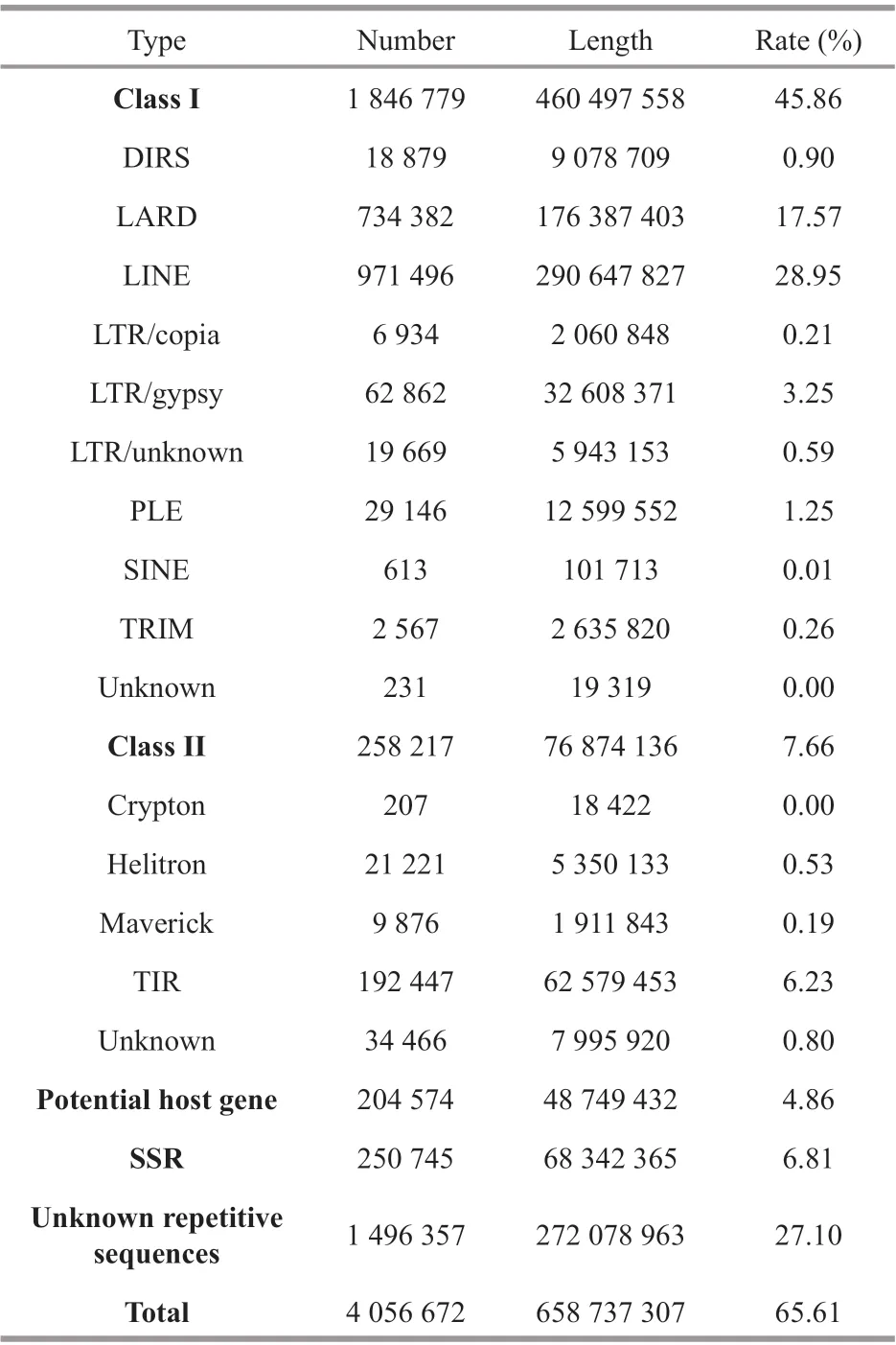
Table 1 The statistical information of repetitive sequences
3.2 The distribution of repetitive sequences
The cumulative sequences of repetitive sequences identif ied in theP.trituberculatusfull-length transcriptome occupied 65.61% of the genome,corresponding to 658 737 307 bp (Table 1). Among all the repetitive sequences, simple sequence repeats(SSRs) and potential host gene occupied 6.81% and 4.86% of theP.trituberculatusgenome in total length of 68 342 365 bp and 48 749 432 bp, respectively.Meanwhile, the transposable elements (TEs) occupied 53.52% of theP.trituberculatusgenome in total length of 537 371 694 bp. The detected TEs included mainly retrotransposons (Class I; 460 497 558 bp)and DNA transposons (Class II; 76 874 136 bp). Class I retrotransposons could be divided into eight groups,including DIRS (9 078 709 bp), LARD(176 387 403 bp), LINE (290 647 827 bp), LTR(40 612 372 bp), PLE (12 599 552 bp), SINE(101 713 bp), TRIM (2 635 820 bp), and 19 319-bp unknown retrotransposons. Class II DNA transposonscan be divided into f ive groups, including Crypton(18 422 bp), Helitron (5 350 133 bp), Maverick(1 911 843 bp), TIR (62 579 453 bp), and 7 995 920-bp unknown DNA transposons. Additionally,272 078 963-bp repetitive sequences could not be classif ied at the present study.
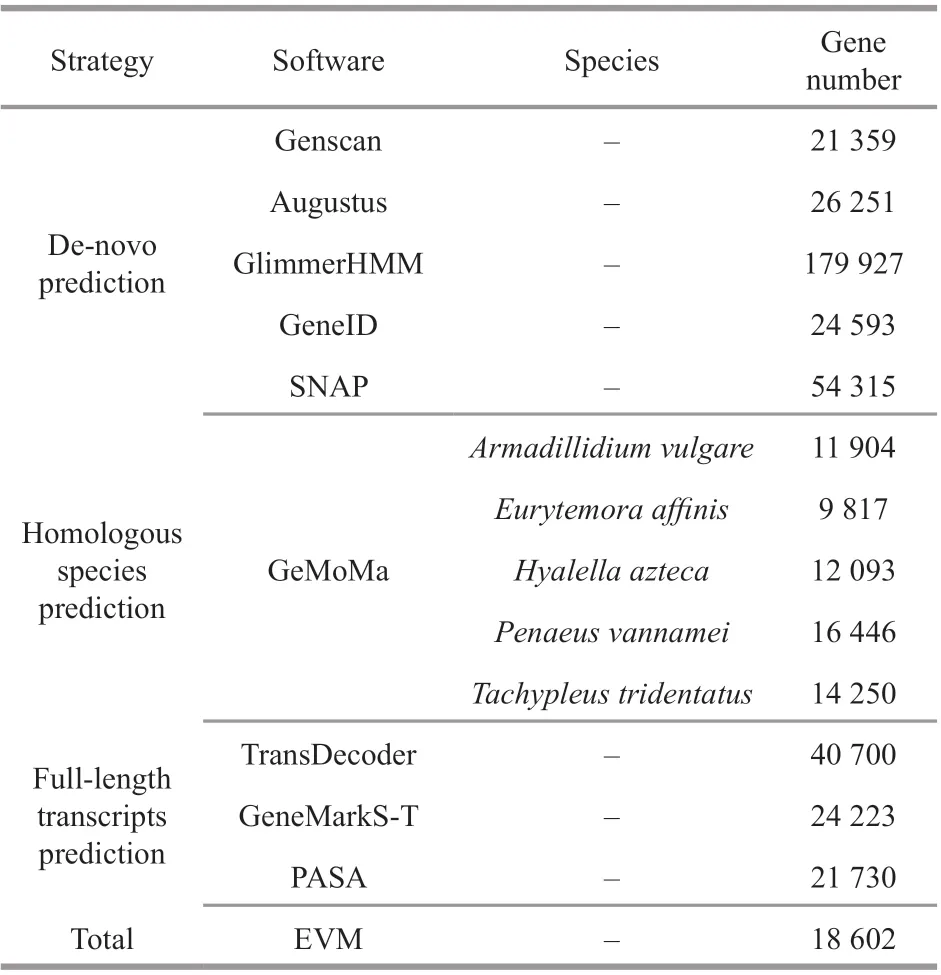
Table 2 Gene prediction results
3.3 Gene structure
3.3.1 Coding genes
In the present study, three strategies were applied to predict the coding gene. Results showed that the number of genes predicted by diff erent analysis strategies was diff erent (Table 2) and a total of 17 519,14 633, and 12 110 genes were predicted based on de novo prediction, homologous species prediction, and full-length transcripts prediction (Fig.3). In order to improve the accuracy of gene prediction, all the gene prediction results were integrated and a total of 18 602 genes were obtained in total length of 219 006 379 bp and mean length of 11 773.27 bp, respectively.Meanwhile, we also predicted 123 333 exons, 104 731 introns, and 119 274 coding genes across all genes,with a total length of 33 951 173 bp, 185 055 206 bp,and 26 236 140 bp, respectively.
3.3.2 Non-coding RNAs
In this study, 28 686 non-coding RNAs were predicted in the full-length transcriptome ofP.trituberculatus. Among all the non-coding RNAs,there are 36 miRNAs, 216 rRNAs, and 28 434 tRNAs were predicted, belonging to 23 miRNA families,4 rRNA families, and 25 tRNA families, respectively.
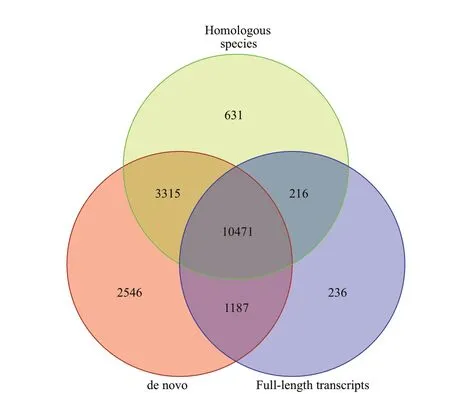
Fig.3 The integrated gene prediction results based on three strategies
3.3.3 Pseudogenes
After looking for homologous gene sequences on theP.trituberculatusgenome and then identifying the immature termination codon and frameshift mutations, 1 407 pseudogenes were predicted in theP.trituberculatusfull-length transcriptome in total length of 3 368 546 bp on average of 2 394.13 bp.
3.3.4 Gene function annotation
To obtain the gene functional information, all the predicted gene sequences were applied to compare with the sequences of f ive databases. Results show that 78.24% (14 554/18 602) genes were annotated in f ive databases. Of all homology searches, 14 460(77.73%), 10 211 (54.89%), 5 412 (29.09%), 7 314(39.32%), and 14 448 (77.67%) genes had signif icant matches with sequences in the NR, KOG, GO, KEGG,and TrEMBL database, respectively. After annotation,740 motif and 26 434 domain were also be found in the gene sequences.
4 DISCUSSION
4.1 Improvement of annotated information of P. trituberculatus genome
Tang et al. (2020) constructed aP.trituberculatuschromosomal-level genome and the genome assembly quality has been rigorously validated in a variety of methods. However, short transcripts used in genome annotation are diffi cult to provide fulllength transcript isoforms for each RNA, limiting their utility for def ining alternative splicing and alternative polyadenylation and perfecting genomic information (Au et al., 2013). Meanwhile, the quantif ication of expression levels are complicated by multiple amplif ication steps in the library preparation of RNA-seq (Sharon et al., 2013). On the other hand, short-reads transcribed by highly repetitive regions or very similar members of multigene families can also be misassembled(Schliesky et al., 2012; Li et al., 2014). Considering the shortcomings of short transcripts in the application ofP.trituberculatusgenome annotation,the well-characterized full-length transcript tags can compensate for the above def iciencies, and ultimately are benef icial for behavioral studies on subsequent function and transcription of important loci ofP.trituberculatus. Because of the longer reads, both PacBio (the read length is up to 60 kb) and ONT (the read length is up to 1 Mb) platforms can capture complete transcripts from end to end (Wyman et al.,2019). In fact, PacBio platform yields has increased dramatically in recent years, with generating up to 8 million reads per SMRT cell on the Sequel 2, far more than original RSII machines (0.15 million reads). Similar yield increases have been reported for ONT platform (Wyman et al., 2019). Meanwhile,both two platforms have the advantage of representing single molecules rather than amplif ied clusters, thus facilitating isoforms sequencing (Eid et al., 2009). However, it is worth noting that longreads sequencing from the third-generation sequencing platforms also has some disadvantages,such as high indel and mismatch error rates,especially the ONT platform (Eid et al., 2009).Compared with the ONT platform, the random errors in the sequencing process of the PacBio platform can be alleviated by self-correction of long reads,correction of short reads and some computational methods (Rhoads and Au, 2015; Lou et al., 2020).Unfortunately, most of the available tools for analyzing long-reads were not specif ically designed for direct quantif ication of long-reads. Considering that the ONT platform has long reads, and the PacBio platform has high reads accuracy, we speculate that using PacBio platforms can achieve more effi cientP.trituberculatusfull-length transcripts and aim to improve the annotated information ofP.trituberculatusgenome.
4.2 High proportion of repetitive sequences
A total of 658.74 Mb of repetitive sequences were identif ied and accounting for 65.61% of theP.trituberculatusgenome, which is greater than 54.52% reported by Tang et al. (2020). Therefoore,we hypothesized that diff erent transcripts of the same gene may increase the number of repetitive sequences.Previous association analysis by Gao et al. (2018)between the number of whole-genome repetitive sequences and genome size in 44 plants and 68 vertebrates suggested that the proportion of repetitive sequences was positively correlated with genome size. Therefore, the high proportion (either 65.61% or 54.52%) of repetitive sequences also conf irmed the large genome size (the genome assembly size was 1.00 Gb by Tang et al., 2020) and high complexity ofP.trituberculatus. Meanwhile, we also found the TEs occupied 53.52% of theP.trituberculatusgenome.Previous studies have speculated that an increase in the number and type of TEs would contribute to the genome size expansion and evolution (Cordaux and Batzer, 2009; Sun et al., 2012). Additionally, TEs have been found to play a critical role in some biological events, such as organism development(Kano et al., 2009; Garcia-Perez et al., 2016),regulation (Elbarbary et al., 2016), diff erentiation(Morales-Hernández et al., 2016), and as promoters to activate the transcription process (Faulkner et al.,2009; Mita and Boeke, 2016). Therefore, we hypothesize that such abundant TE may help regulate some biological behavior ofP.trituberculatus.
4.3 Optimization of P. trituberculatus genetic structure information
It is worth noting that the genome structure prediction is often inaccurate due to the limitations of the short transcripts, so we used full-length transcripts to predict genome structure more accurately. In this study, three prediction strategies were used to annotate protein-coding genes and ensure the accuracy of the prediction results. We merged the prediction results and obtained 18 602 protein-coding genes, which is greater than previously predicted number (16 791;Tang et al., 2020) based on genomic data at the chromosomal level and indicate more undiscovered genes. In fact, full-length transcripts have been shown helpful in the prediction of novel genes and novel isoforms ofTachypleustridentatus(Lou et al., 2020).We believe that full-length transcripts will be necessary for future studies of novel genes and isoforms, although the genome annotated f ile in generic feature format (GFF) format has not been released to limit the prediction of novel genes and novel isoforms ofP.trituberculatus.
Meanwhile, we predicted 28 686 non-coding RNAs and 1 407 pseudogenes in theP.trituberculatusfull-length transcripts. Many studies have concluded that non-coding RNAs and pseudogenes also play important roles in the regulation of biological growth and development, abiotic stress, and other physiological functions (Schiff et al., 1985; Liu et al.,2013; Chen and Ge, 2017). Additionally, non-coding RNAs and pseudogenes appear to be more speciesspecif ic and may ultimately provide more appropriate evidence for the study of biological evolution.Therefore, we have every reason to believe that the non-coding RNAs and pseudogenes predicted in this study will contribute to the further biological research ofP.trituberculatus.
5 CONCLUSION
In this study, we have assembled theP.trituberculatusfull-length transcriptome based on SMRT sequencing technology. A total of 9 694 fulllength transcripts were generated successfully after f iltering out low-quality sequencing reads, selfcorrecting, and de-redundancy. We also successfully predicted the repetitive sequences, coding genes, noncoding RNAs, and pseudogenes. These results not only help to ref ine theP.trituberculatusgenomic annotation information, but also provide basic resources for the future evolution and physiological regulation researches ofP.trituberculatus.
6 DATA AVAILABILITY STATEMENT
All subreads in bam format were released in the NCBI Sequence Read Archive under BioProject number PRJNA749655, with accession number of SRR15248879.
 Journal of Oceanology and Limnology2022年5期
Journal of Oceanology and Limnology2022年5期
- Journal of Oceanology and Limnology的其它文章
- Comparison of three f locculants for heavy cyanobacterial bloom mitigation and subsequent environmental impact*
- Proteomic analysis provides insights into the function of Polian vesicles in the sea cucumber Apostichopus japonicus post-evisceration*
- Key physiological traits and chemical properties of extracellular polymeric substances determining colony formation in a cyanobacterium*
- Involvement of the ammonium assimilation mediated by glutamate dehydrogenase in response to heat stress in the scleractinian coral Pocillopora damicornis*
- UV-B irradiation and allelopathy by Sargassum thunbergii aff ects the activities of antioxidant enzymes and their isoenzymes in Corallina pilulifera*
- Cyanobacterial extracellular alkaline phosphatase: detection and ecological function*
