Improving the resistance of the rice PTGMS line Feng39S by pyramiding blast, bacterial blight, and brown planthopper resistance genes
Dabing Yang, Lizhong Xiong, Tongmin Mou, Jiaming Mi
National Key Laboratory of Crop Genetic Improvement and National Centre of Plant Gene Research (Wuhan), Huazhong Agricultural University, Wuhan 430070, Hubei, China
Keywords:Bacterial blight resistance Blast resistance Brown planthopper resistance Genomic breeding PTGMS line Two-line hybrid rice
A B S T R A C T Knowledge of rice(Oryza sativa L.)genes and various DNA markers can be used in genomic breeding programs aimed at developing improved elite rice cultivars.We used an efficient genomic breeding approach to pyramid four resistance genes(Pi2,Xa23,Bph14,and Bph15)in the popular photoperiod-and thermosensitive genic male sterile(PTGMS)rice line Feng39S.We performed foreground selection for the target genes,followed by recombinant selection and background selection.This process reduced the sizes of the genomic segments harboring the target genes (566.8 kb for Pi2, 1143.9 kb for Xa23, 774.7 kb for Bph14,and 1574.9 kb for Bph15) and accelerated the recovery of the recurrent parent genome to proportions ranging from 98.77%to 99.16%,thus resulting in four near-isogenic lines.To assemble the four resistance genes in Feng39S, we performed a double-way cross combined with foreground and background selection to generate two improved lines of Feng39S (Pi2 + Xa23 + Bph14 + Bph15) with a recurrent parent genome recovery of 98.98%. The two lines showed agronomic performance, grain quality, and fertilitysterility transition characteristics similar to those of the original Feng39S line. The newly developed PTGMS lines and corresponding hybrid combinations were resistant to various field blast isolates and seven representative isolates of bacterial blight. At the seedling stage, the lines also showed resistance against brown planthopper. This study provides an efficient and accurate genomic breeding approach for introducing desirable traits into PTGMS lines.
1. Introduction
Rice (Oryza sativaL.) is a staple food for over half the world’s population[1].To ensure food security,great effort has been made to increase rice yield during the last several decades[2].For example, an effective two-line hybrid breeding system was established based on photoperiod- and thermo-sensitive genic male-sterile(PTGMS)lines,resulting in major increases in rice yield[3,4].However, the use of high-yielding two-line hybrid cultivars has led to the overuse of pesticides, owing to their lack of resistance against major diseases and insect pests,such as blast,bacterial blight,and brown planthopper, resulting in environmental damage and increased production costs [5-9]. Breeding two-line hybrid cultivars with superior resistance to major diseases and insect pests would stabilize high yield in rice while protecting the environment.
The genetic basis of rice resistance against blast, bacterial blight, and brown planthopper has been well characterized.Twenty-five blast resistance genes, 12 bacterial blight resistance genes, and 14 brown planthopper resistance genes have been cloned from cultivated and wild rice [10-13]. Of these genes, the blast resistance genePi2, the bacterial blight resistance geneXa23, and the brown planthopper resistance genesBph14andBph15have been widely used in marker-assisted backcross breeding programs [14-21]. However, undesirable genes can be transferred to rice by linkage drag, and unknown genetic background effects can negatively influence agronomic traits, leading to low yield or poor grain quality [20-22].
To overcome these challenges,genomic breeding strategies that allow multiple genes to be simultaneously introduced with minimal introgression segments can be used to create improved breeding lines. This strategy, representing an improvement over standard pyramiding strategies, could be used to achieve durable resistance to diseases and insect pests in hybrid cultivars[23].Target genes conferring resistance to diseases or pests can be pyramided via foreground selection, which takes advantage of recombination events between target genes and flanking DNA markers to reduce the size of the donor segment containing the target gene,as well as background selection to accelerate recurrent parent genome(RPG)recovery.These selection steps remove linkage drag and unknown genetic background effects to create a pyramided line with multiple or broad-spectrum resistance to various diseases and pests [9,23]. Wang et al. [24] recently developed five lines with brown planthopper resistance in theindicarestorer rice line Wushansimiao by transferring two resistance genes,Bph14andBph15, via genomic breeding.
Feng39S is an elite PTGMS line that is widely used for two-line hybrid rice breeding in China for its high general combining ability and grain quality and its relatively low critical sterility temperature point. However, Feng39S is susceptible to blast, bacterial blight, and brown planthopper, limiting the cultivation of hybrid cultivars derived from Feng39S. The objective of this study was to increase the resistance of Feng39S against rice blast, bacterial blight, and brown planthopper by pyramiding four resistance genes from three PTGMS donors into Feng39S using genomic breeding.
2. Materials and methods
2.1. Plant materials and breeding procedure
Feng39S,an eliteindicarice PTGMS line developed by Hefei Fengle Seed Co., ltd. of China, was used as the recipient parent. Three Guangzhan 63-4S-derived PTGMS lines,Hua1201S,Hua1015S,and Hua1165S (https://www.ricedata.cn/variety/varis/617488.htm),were used as donors ofPi2,Xa23, andBph14+Bph15, respectively[16,27].Analysis of the pedigrees of these donors[28-31]indicated thatPi2,Xa23, andBph14+Bph15originated from the nearisogenic lines (NILs) C101A51, CBB23, and B5, respectively.
NILs containingPi2,Xa23,Bph14,andBph15were developed by the marker-assisted backcross breeding procedure described in Fig.1.Feng39S was crossed with each of the three resistant donors,and the F1plants were backcrossed to the recurrent parent Feng39S. In each backcross generation, all plants were genotyped for foreground selection. In BC1F1, parental polymorphic simple sequence repeats (SSR) markers covering all the chromosomes were employed for background selection. In BC2F1and the subsequent generations, plants containing the target genes were subjected to recombinant selection to minimize the size of the donor chromosome segment. The selected recombinants were further detected using the SSR markers, followed by genotyping with the high-density RICE6K SNP array for background selection,to rapidly recover RPG. Advanced backcross plants with homozygous or heterozygous genotypes were obtained as NILs.
To pyramid the four resistance genes, pairwise crosses were performed using the four NILs, leading to the generation of the double-cross F1population. Finally, two lines containingPi2+Xa23+Bph14+Bph15were produced by self-pollination,foreground selection, and background selection. These two improved PTGMS lines were then separately crossed with the elite restorer line Huanghuazhan (HHZ) to obtain the corresponding hybrid combinations for disease-resistance and agronomic evaluations.The pyramided line HHZ(Bph14+Bph15),carrying the brown planthopper resistance genesBph14andBph15in the HHZ genetic background,was also crossed with these two PTGMS lines for further assessment of the genetic effect of the homozygous genotypeBph14Bph14+Bph15Bph15in a hybrid genetic background.
2.2. Foreground and recombinant selection
Four flanking markers for each of the four target genes were used for foreground and recombinant selection (Fig. 2; Table S1).Among these, two tightly linked markers located upstream and downstream of the target gene were used to track the target gene for foreground selection. The other two flanking markers were used to reduce the sizes of the undesirable segments linked with the target gene by recombinant selection. The physical distances between adjacent markers are shown in Fig. 2. Genotyping of the selection markers was performed as described by Mi et al. [32].Amplified DNA products were electrophoretically resolved on a 4% denaturing polyacrylamide gel in 0.5× TBE buffer.

Fig. 1. Genomic breeding scheme for Feng39S (Pi2 + Xa23 + Bph14 + Bph15). The recombinant selection followed by the ‘‘+” symbol indicates that recombinants were identified, whereas ‘‘-” indicates that no recombinants were identified.
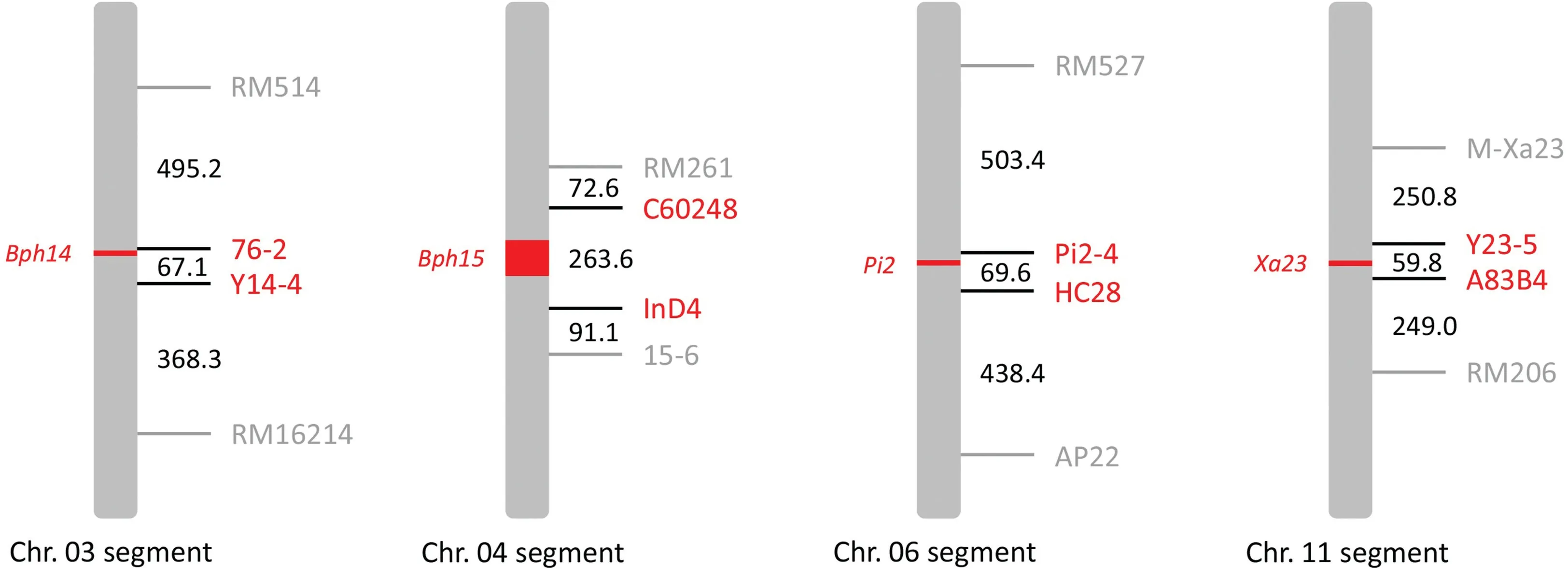
Fig. 2. Gene-specific marker system for foreground and recombinant selection. The numbers to the right of the chromosome segments indicate the physical distances (kb)between selection markers.
2.3. Background selection
Whole-genome background selection was performed based on SSR markers and SNP arrays. A group of 455 SSR markers distributed evenly throughout the genome were investigated for polymorphisms (http://www.gramene.org/). Of these markers, 19, 27,and 30 showed polymorphism between Feng39S and Hua1201S,Feng39S and Hua1015S, and Feng39S and Hua1165S, respectively.These markers were used for background selection from BC1F1to BC3F1. The high-density RICE6K SNP array [33], containing 5102 SNPs, was used to detect the genetic backgrounds of the selected plants in the advanced backcross generation and the higherdensity SNP array GSR40K, containing 36,584 SNPs, was used for background selection during the development of the pyramided lines. An additional set of selection markers were also developed based on the RICE6K and GSR40K assays to eliminate the residual background genome from donors in the BC3F1, BC4F1, and MF3(multiple-cross filial, MF) generations (Table S1). SNP array genotyping was performed as previously described [33].
2.4. Evaluation of rice blast resistance
Resistance to rice leaf and panicle blast was assessed in the natural blast nursery located in Wangjia village, Yichang city, Hubei province, China (31°05′N, 111°64′E, 654 m altitude). Blast evaluations were performed for each test line and the corresponding hybrid combination according to the technical specifications for identification and evaluation of blast resistance in rice variety regional tests[34].The improved PTGMS lines were also subjected to resistance spectrum analysis via artificial inoculation at the seedling stage in the Plant Protection Institute of Guangdong Academy of Agricultural Sciences, China. Thirty-four isolates ofM. oryzaecollected from various rice-growing areas of China were used for artificial inoculation of leaf blast at the seedling stage in the greenhouse. Inoculation and evaluation were performed as described by Bonman et al. [35]. Disease reaction was evaluated using a 0 to 9 rating system, as described in the Standard Evaluation System for Rice[36],where 0 to 3 is resistant and 4 to 9 is susceptible. The rice cultivar CO39 was used as a susceptible control.Blast resistance was represented by resistance frequency, which was defined as the number of incompatibleM.oryzaeisolates/total number ofM. oryzaeisolates.
2.5. Evaluation of bacterial blight resistance
Plants at the maximum tilling stage were inoculated with seven bacterial blight isolates at the rice experimental station of Huazhong Agricultural University in Wuhan, Hubei province using the leaf-clipping method [37]. Of the seven isolates, PXO61 and PXO99 are representative pathotypes in the rice-growing regions of the Philippines.ZHE173 and GD1358 are severe epidemic pathotypes in the rice-growing regions of Central and Southern China,respectively,and are used in the China National Rice Trial Program for evaluating the bacterial blight resistance levels of rice varieties[16].HeN11,FuJ,and YN24 are representatives of different virulent pathotypes[38].Plant resistance reactions to the bacterial isolates were recorded as the mean lesion length of 15 leaves three weeks after inoculation, as described by Jiang et al. [16].
2.6. Evaluation of brown planthopper resistance
Brown planthopper resistance of the improved PTGMS lines and their hybrid combinations was evaluated at the seedling stage,with B5 used as a resistant control and TN1 as a susceptible control.A modified seedling bulk test was used to evaluate the brown planthopper resistance of seedlings. Twenty seeds per test line were sown in individual 0.4 L plastic boxes (length 7 cm × width 7 cm × depth 8 cm) with three replications. The seedlings were thinned to 12-15 plants per box at the three-leaf stage, and each plant was infested with 8-10 s-to third-instar nymphs. Damage scores were recorded as 0,1,3,5,7,or 9 according to the Standard Evaluation System for Rice when the susceptible control TN1 had died [36]. The process used for seedling management was described by Wang et al. [21].
2.7. Characterization of critical sterility temperature point
Uniform, healthy PTGMS plants at the five-leaf stage were transplanted into two plastic pots, with each pot containing five plants, and labeled with plastic tags. When the main panicle was at the initiation stage(the critical stage for photoperiod or temperature sensitivity), the plants were moved to plant growth chambers with daily mean temperatures of 22, 23, 24, and 25 °C,while the light duration was set at 14 h and the relative humidity at 75% for 12 consecutive days [39,40]. The plants were removed from the chambers and grown in the natural environment until heading. Pollen grains of five florets from the upper, middle, and bottom parts of panicles were collected 1-2 days before flowering and observed under a microscope following 1% I2-KI staining. The PTGMS line was considered completely sterile when the mean pollen sterility was >99.5% under our treatment conditions [40].
2.8. Investigation of dynamic pollen fertility
One hundred seeds per line were sown at the experimental farm of Huazhong Agricultural University at 15-day intervals from April 1 to July 1 during the summer season of 2020. Fifty uniform,healthy seedlings at the five-leaf stage were transplanted to the field at a spacing of 16.7 cm between plants within a row and 26.7 cm between rows. At the heading stage, five plants per line were randomly chosen and their pollen fertility investigated. Pollen grains were collected from the top five florets of primary panicles and examined by 1% I2-KI staining [40]. The dynamic pollen fertility of all lines was investigated every-two days from July 11th to October 7th [39]. Daily minimum temperature data were provided by the Agricultural Meteorology Department of Huazhong Agricultural University.
2.9. Evaluation of agronomic traits and grain quality
The improved PTGMS lines were planted in the field with three sowing dates at intervals of 10 days, and their corresponding hybrid combinations were planted with three replications in a randomized block design during the dry season of 2020 at the rice breeding station of Huazhong Agricultural University in Lingshui county, Hainan province (18°30′N, 110°01′E, 10 m altitude). To measure the agronomic traits of the improved PTGMS lines during the sterility period, the lines were also planted in the field with three sowing dates at intervals of 15 days during the summer season of 2020 at the experimental farm of Huazhong Agricultural University (30°28′N, 114°21′E, 53 m altitude).
Fifty plants per line were transplanted to the field at a spacing of 16.7 cm between plants within a row and 26.7 cm between rows. Days to heading was recorded. At the maturity stage, five plants in the middle of the central row in each plot were used to measure agronomic traits, including plant height, panicle number per plant, panicle length, number of spikelets per panicle, spikelet fertility, 1000-kernel weight, and yield per plant. Harvested bulk seeds from each plot were used to analyze grain quality,including brown rice rate, milled rice rate, head rice rate, chalky rice rate,chalkiness degree, kernel length, kernel length/width ratio, alkali spreading value,amylose content,and gel consistency.Grain quality was classified according to the quality standards of cooking rice cultivars [41].
2.10. Statistical analysis
Two-tailedt-tests were performed to compare the agronomic traits of each improved line with those of the recurrent parent Feng39S using Microsoft Office Excel 2010(Microsoft Corporation,Redmond, WA, USA).
3. Results
3.1. Construction of the NILs
Following the breeding procedure shown in Fig. 1, the four resistance genes were transferred from Hua1201S(Pi2),Hua1015S(Xa23), and Hua1165S (Bph14+Bph15) into Feng39S using foreground, recombinant, and background selection. To develop the NIL ofPi2,Feng39S and Hua1201S were crossed to obtain F1seeds.Two true F1plants, as confirmed using the marker Pi2-4, which is tightly linked toPi2(Fig. 2), were backcrossed to Feng39S. Ten heterozygous plants were identified in the BC1F1population using the foreground selection markers Pi2-4 and HC28 forPi2(Fig. 2),followed by background selection based on 19 polymorphic SSR markers. One plant that showed an RPG recovery of 78.9% was selected for a further backcross with Feng39S.Because no recombinant in thePi2region was identified among the 72 BC2F1plants,20 plants carryingPi2were subjected to background selection. These plants showed RPG recovery values ranging from 89.5% to 100.0%based on polymorphic SSR markers.
Three plants with the highest RPG recovery values were further verified using the RICE6K SNP array.The best plant,which showed an RPG recovery of 93.3% and carried one additional introgressed segment of Hua1201S on chromosome 8, was then backcrossed with Feng39S to produce BC3F1seeds. Of the 504 BC3F1plants genotyped using HC28 and AP22 markers(Fig.2),10 recombinants with a heterozygous HC28 and Feng39S-homozygous AP22 genotype were selected. Of these plants, the best plant, which showed the elimination of the additional segment on chromosome 8 using the background selection markers YD2-1 and YD2-4, was selfpollinated to obtain the BC3F2population (Table S1). Among the 475 BC3F2plants genotyped using RM527 and Pi2-4, which is tightly linked withPi2(Fig. 2), one recombinant with a homozygous Pi2-4 and heterozygous RM527 genotype was identified,suggesting that the introgression of thePi2segment would be shortened by further genotypic selection of RM527 in the next generation. The NIL was designated as Feng39S (Pi2 Pi2), with an RPG recovery of 99.2% (Fig. S1a).
To develop the NIL ofXa23, Feng39S and Hua1015S were crossed to obtain F1seeds. Two true F1plants, as confirmed using the marker A83B4, which is tightly linked toXa23(Fig. 2), were backcrossed to Feng39S. In the BC1F1population, 20 heterozygous plants were identified using the foreground selection markers A83B4 and Y23-5 forXa23(Fig. 2), followed by background selection based on 27 polymorphic SSR markers.One plant that showed an RPG recovery of 70.4% was selected to further backcross to Feng39S. Since no recombinant in theXa23region was found among the 72 BC2F1plants genotyped, 12 plants harboringXa23were genotyped with eight SSR markers for background selection.
Three plants showing the highest RPG recovery (88.9%) were assayed with the RICE6K SNP array. The best plant (with RPG recovery of 85.9%) was selected to backcross with Feng39S. Of the 144 BC3F1plants genotyped using the A83B4 and RM206 markers (Fig. 2), one recombinant with a heterozygous A83B4 and Feng39S-homozygous RM206 genotype was identified and backcrossed with Feng39S. RICE6K array analysis revealed that this recombinant carried four undesirable segments on chromosomes 1, 5, and 11 from Hua1015S and showed an RPG recovery of 90.9%. Among the 234 BC4F1plants, no recombinant on the other side of the target gene was identified. With the elimination of the four additional segments using background selection markers RM10695, YD23-1, RM19199, and YD23-3 (Table S1), one plant heterozygous forXa23was selected as Feng39S (Xa23 xa23), with an RPG recovery of 98.1% (Fig. S1b).
To develop the NIL ofBph14, Feng39S and Hua1165S were crossed to obtain F1seeds. Two true F1plants, as confirmed using the marker 76-2, which is tightly linked toBph14(Fig. 2), were backcrossed to Feng39S. In the BC1F1population, 27 heterozygous plants were identified using the foreground selection markers 76-2 and Y14-4(Fig.2),followed by background selection based on 30 polymorphic SSR markers.One plant that showed an RPG recovery of 60.0% was backcrossed to Feng39S. Among the 72 BC2F1plants,two recombinants between 76 and 2 and RM514 (Fig. 2) were identified and further genotyped using the RICE6K array for background selection, showing RPG recoveries of 93.9% and 94.3%,respectively.
The best plant was found to carry-three undesirable segments on chromosomes 4, 5, and 7 derived from Hua1165S, which were backcrossed with Feng39S. Among the 504 BC3F1plants, nine recombinants on the other side of the target gene were selected.After elimination of three additional segments using the background selection markers RM17673, YD14-2, YD14-7, and RM21587 (Table S1), three plants heterozygous forBph14were subjected to genetic background analysis with the RICE6K array.Of these plants, one plant lacking the additional introgression was self-pollinated to generate the BC3F2population, in which one of eleven plants homozygous forBph14was selected as Feng39S (Bph14Bph14), with an RPG recovery of 98.6% (Fig. S1c).
The BC1F1plants derived from the cross between Feng39S and Hua1165S mentioned above were also used to develop the NIL ofBph15.Twenty-seven heterozygous plants in BC1F1were identified using the foreground selection markers C60248 and InD4 (Fig. 2),followed by background selection based on 30 polymorphic SSR markers. One plant that showed an RPG recovery of 60.0% was selected to further backcross with Feng39S.No recombinants were identified by recombinant selection in the BC2F1or BC3F1generation. Of the 72 BC2F1plants, background analysis based on 12 SSR markers indicated that the RPG recovery values of 26 plants heterozygous forBph15ranged from 66.7% to 86.7%.
Two plants with the highest genome recovery value (86.7%)were selected for the RICE6K assay.The best plant with the highest RPG recovery value (82.0%), which carried four undesirable segments from Hua1165S on chromosomes 1, 4, 5, and 10, was backcrossed to Feng39S to produce BC3F1seeds. Among the 720 BC3F1plants, background selection was applied to 289 plants heterozygous forBph15using six background selection markers: RM165,RM414, RM307, RM185, YD15-2, and YD15-6 (Table S1), to eliminate the four residual segments from the donor and selected three plants for the RICE6K assay. Among these, one plant with an RPG recovery of 91.4%that carried only an introgressedBph15segment was self-pollinated to produce BC3F2seeds. Finally, one recombinant between InD4 and 15-6 (Fig. 2) was selected from the BC3F2population and designated as Feng39S (Bph15bph15), with an RPG recovery of 98.8% (Fig. S1d).
3.2. Assembly of Feng39S (Pi2 + Xa23 + Bph14 + Bph15)
To obtain Feng39S(Pi2+Xa23+Bph14+Bph15),which harborsPi2,Xa23,Bph14, andBph15in the Feng39S genetic background,two single crosses were made: Feng39S (Pi2 Pi2)/Feng39S (Xa23 xa23) and Feng39S (Bph14Bph14)/Feng39S (Bph15bph15). In the F1cross of Feng39S (Pi2 Pi2)/Feng39S (Xa23 xa23) with 27 plants,ten plants heterozygous for bothPi2andXa23were identified with a Feng39S-homozygous genotype for RM527, indicating that the linkage drag forPi2was removed in this generation.In the F1cross of Feng39S (Bph14Bph14)/Feng39S (Bph15bph15), with 84 plants,41 plants heterozygous for bothBph14andBph15were identified.
After these two single crosses were made, a double cross was performed to produce MF1seeds. Among the 55 MF1plants, one plant was identified with heterozygousPi2,Xa23,Bph14, andBph15;this plant was self-pollinated to generate MF2seeds.Among the 2160 plants of the MF2generation,four plants homozygous forPi2,Xa23,Bph14,andBph15were selected and genotyped using the high-density SNP array GSR40K. Genetic background analysis revealed diverse genetic backgrounds among the four plants,which were not detected with the lower-density RICE6K assay.Finally, two plants with an RPG recovery of 96.82% with only one residual introgressed segment on chromosome 4 were separately harvested to produce MF3seeds. Background selection with the marker YD-M04 was applied to remove the residual segment on chromosome 4 among the 21 MF3plants(Fig.1;Table S1).Finally,two plants were separately harvested to produce two pyramiding lines of Feng39S (Pi2 + Xa23 + Bph14+Bph15) with four homozygous resistance genes, which were designated as DB18129-34-268-38 and DB18129-34-303-6, respectively.
These two lines were genotyped with the high-density GSR40K SNP array to confirm the numbers,positions,and sizes of the introgressions. This analysis showed thatPi2,Xa23,Bph14, andBph15were introduced on segments of length 566.8, 1143.9, 774.7, and 1574.9 kb, respectively, and that the genetic backgrounds of both lines were identical to that of the recurrent parent Feng39S(Fig.3).
3.3. Blast resistance of Feng39S (Pi2 + Xa23 + Bph14 + Bph15) and hybrid combinations
The patterns of resistance against the 34 isolates ofM. oryzaediffered between Feng39S (Pi2 + Xa23 + Bph14+Bph15) and Feng39S. The improved lines DB18129-34-268-38 and DB18129-34-303-6 showed broad-spectrum resistance to rice blast, with high resistance frequencies of 94.12% and 97.14%, respectively,whereas Feng39S showed a low resistance frequency of 35.29%(Table S2). Markedly increased resistance of the two improved PTGMS lines and the corresponding hybrid combinations derived from HHZ in the natural blast nursery in comparison with Feng39S and the original Feng39S/HHZ were observed. Feng39S showed level 7 leaf blast incidence and level 9 neck blast incidence under heavy blast disease pressure, as this line is highly susceptible to rice blast.
The two improved PTGMS lines (DB18129-34-268-38 and DB18129-34-303-6)harboring thePi2gene showed different levels of resistance. DB18129-34-268-38 expressed resistance to leaf blast (level 1) and neck blast incidence (level 1), while DB18129-34-303-6 showed high resistance to leaf blast (level 0) and neck blast incidence (level 0). In comparison with the blast resistance of DB18129-34-303-6 and DB18129-34-268-38,the corresponding hybrid combination (DB18129-34-303-6/HHZ) showed lower resistance to leaf and panicle blast, while DB18129-34-268-38/HHZ showed the same level of leaf blast resistance and lower panicle incidence (Fig. 4). These results indicate that the blast resistance genePi2increased blast resistance in the NILs, with differing reaction patterns in the Feng39S and Feng39S/HHZ genetic backgrounds.
3.4. Bacterial blight resistance of Feng39S(Pi2 + Xa23 + Bph14 + Bph15) and hybrid combinations
The two improved lines of Feng39S (Pi2 + Xa23 + Bph14+Bph15), along with the hybrid combinations, were tested under artificial-inoculation conditions to assess the resistance ofXa23against seven bacterial blight isolates. Feng39S showed moderate resistance to isolate HeN11, with lesion length of 3.7 cm, whereas it showed a susceptible reaction to the other six bacterial blight isolates, with lesion lengths ranging from 5.6 to 22.5 cm. The restorer line HHZ showed resistance to bacterial blight isolate PXO61 and moderate resistance to ZHE173, with lesion lengths of 2.8 and 3.6 cm, respectively, whereas it showed a susceptible reaction to the other five bacterial blight isolates, with lesion lengths ranging from 6.2 to 15.0 cm.However,the hybrid combination Feng39S/HHZ was susceptible to seven bacterial blight isolates, with lesions ranging from 5.7 to 22.2 cm long (Fig. 5).These results show that the genes for resistance to HeN11 in Feng39S and the genes for resistance to POX61 and ZHE173 in HHZ are recessive.
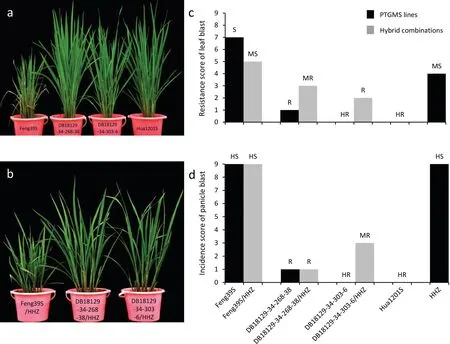
Fig. 4. Leaf and panicle blast disease reactions of the two improved PTGMS lines and their hybrid combinations in a blast nursery in Yichang in 2020. (a, b) Leaf blast resistance performance. (c) Resistance scores of leaf blast. (d) Incidence scores of panicle blast. HR, highly resistant; R, resistant; MR, moderately resistant; MS, moderately susceptible; S, susceptible; HS, highly susceptible.
With the introgression ofXa23into Feng39S,DB18129-34-268-38 and DB18129-34-303-6 showed high resistance to all seven bacterial blight isolates, with lesion lengths ranging from 0.2 to 0.4 cm. The improved hybrid combinations DB18129-34-268-38/HHZ and DB18129-34-303-6/HHZ also showed high resistance to all bacterial blight isolates tested(Fig.5).Thus,both the improved PTGMS lines and their hybrid combinations bearing theXa23gene showed effective, broad-spectrum bacterial blight resistance in contrast to both Feng39S and Feng39S/HHZ.

Fig.5. Resistance reactions of the two improved PTGMS lines and their hybrid combinations to seven bacterial blight isolates.(a)Images of leaf lesions taken on day 21 after inoculation.(b)Bacterial blight resistance levels based on lesion length.Lesion length is represented as mean±SD.HR,highly resistant;R,resistant;MR,moderately resistant;MS, moderately susceptible; S, susceptible; HS, highly susceptible.
3.5. Brown planthopper resistance of Feng39S(Pi2 + Xa23 + Bph14 + Bph15) and hybrid combinations
In the seedling bulk test, seedlings of the tested lines lacking brown planthopper resistance genes were severely wilted,whereas seedlings with homozygousBph14Bph14+Bph15Bph15were healthy, as shown in Fig. 6a. Feng39S was susceptible to brown planthopper, with a resistance score of 7.0, whereas Hua1165S(the donor ofBph14andBph15) was resistant to brown planthopper,with a score of 2.6.The two improved PTGMS lines(DB18129-34-268-38 and DB18129-34-303-6) bearing homozygousBph14Bph14+Bph15Bph15showed resistance to brown planthopper, with scores of 2.0 and 2.3, respectively (Fig. 6b).
Further evaluation of hybrid combinations for brown planthopper resistance revealed that the hybrid combinations DB18129-34-268-38/HHZ, DB18129-34-303-6/HHZ, and Feng39S/HHZ (Bph14+Bph15) with the heterozygousBph14bph14+Bph15bph15were moderately resistant to brown planthopper,with resistance scores of 4.7, 4.6, and 4.1, respectively. In comparison with the three hybrid combinations mentioned above,DB18129-34-268-38/HHZ (Bph14+Bph15) and DB18129-34-303-6/HHZ (Bph14+Bph15), with the homozygousBph14Bph14+Bph15Bph15,showed higher resistance to brown planthopper,with resistance scores of 1.9 and 2.4, respectively (Fig. 6b). By contrast,Feng39S/HHZ, with no brown planthopper resistance genes, was susceptible to brown planthopper damage,with a resistance score of 7.3 (Fig. 6b). Thus, a markedly increased level of resistance to brown planthopper was achieved by pyramiding homozygousBph14Bph14+Bph15Bph15into the Feng39S and Feng39S/HHZ genetic backgrounds and the hybrid combinations with homozygousBph14andBph15showed higher resistance against brown planthopper than those with heterozygousBph14andBph15, suggesting the incomplete dominance of the genetic effects ofBph14andBph15.
3.6. Fertility-sterility transition of Feng39S(Pi2 + Xa23 + Bph14 + Bph15)
The critical sterility temperature points for the fertility-sterility transition and the male sterile phase of plants were measured in the growth chamber and under natural field conditions, respectively. The critical sterility temperature points for both DB18129-34-268-38 and DB18129-34-303-6 were between 22 °C and 23 °C (Table S3). The male-sterile phase of these two improved lines lasted 80 consecutive days (from July 11 to September 29),a duration identical to that of Feng39S, as shown in Fig. S2.Feng39S(Pi2+Xa23+Bph14+Bph15)showed an equivalent sterility-fertility transition to that of Feng39S, suggesting that it could be approved based on the evaluation protocol of environmentally sensitive genic male sterile lines of rice [39].
3.7. Agronomic and grain quality traits of Feng39S(Pi2 + Xa23 + Bph14 + Bph15) and hybrid combinations
To test whether the traits of Feng39S(Pi2 + Xa23+ Bph14+Bph15) were identical to those of Feng39S, the agronomic and grain quality traits of these lines, along with their hybrid combinations,were investigated. Based ont-tests, the variation among the lines was not significant for any agronomic or grain quality trait in either Wuhan or Hainan, suggesting that Feng39S (Pi2 + Xa23 + B ph14+Bph15) and Feng39S were identical in field trials during both the fertile and sterile phases(Tables 1,S4;Fig.7).Specifically,DB18129-34-268-38 and DB18129-34-303-6 showed seed setting rates of respectively 65.6% and 67.0%, which greatly exceeded the threshold of 30% [39]. DB18129-34-268-38 and DB18129-34-303-6 showed first-class grain quality (except for brown rice percentage and amylose content) according to the quality standards of cooking rice varieties [41] (Fig. 7c). Finally, no significant differences were observed between the improved Feng39S (Pi2 + Xa23 + Bph14+Bph15)/HHZ and the original Feng39S/HHZ in agronomic traits or grain quality(Table 1).Thus,these two PTGMS lines showed increased blast, bacterial blight, and brown planthopper resistance while maintaining good agronomic performance and high grain quality in the two-line hybrid combinations.
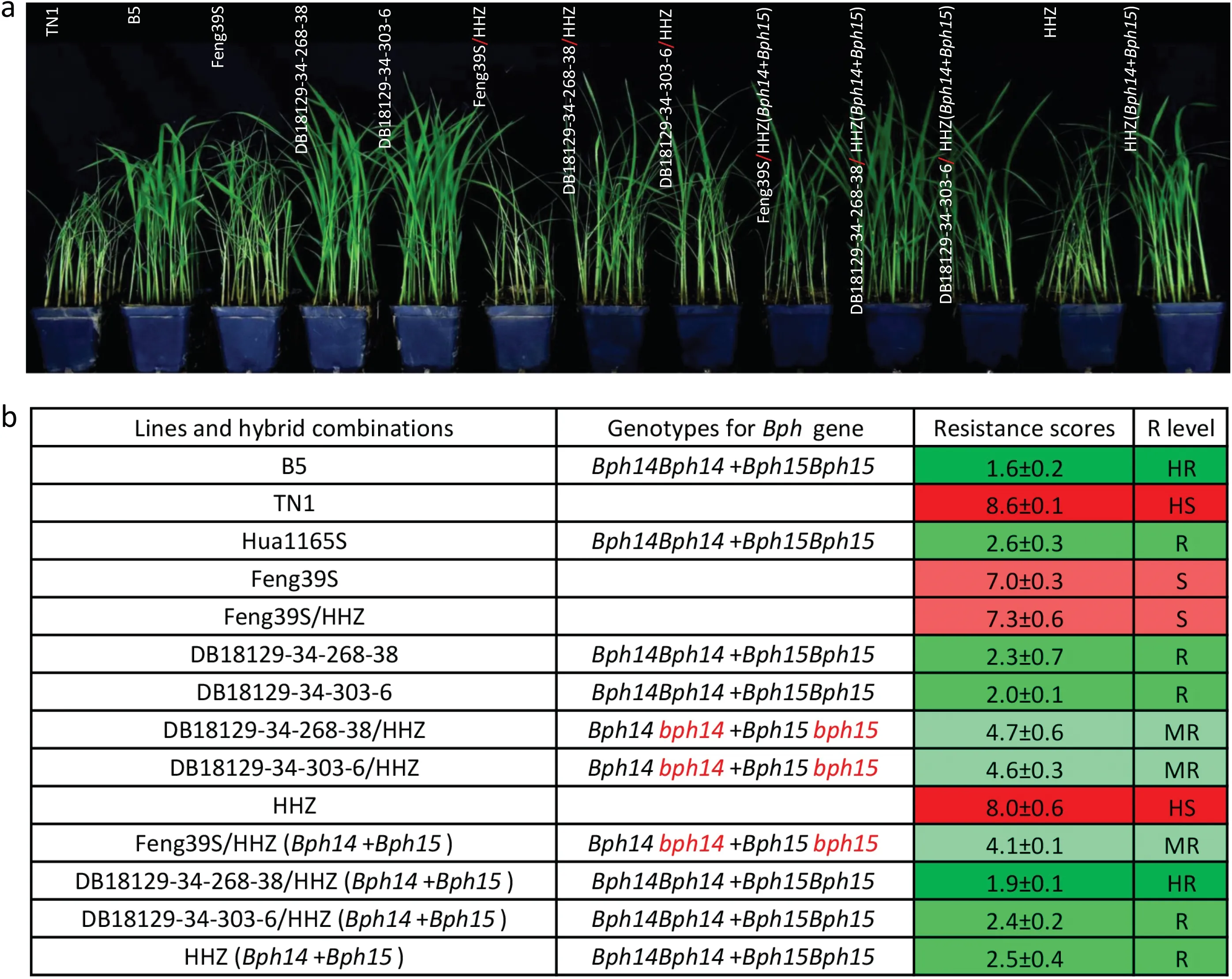
Fig. 6. Brown planthopper resistance reactions of the two improved PTGMS lines and their hybrid combinations at the seedling stage. (a) Brown planthopper resistance performance. (b) Brown planthopper resistance was scored and represented as mean ± SD. HR, highly resistant; R, resistant; MR, moderately resistant; S, susceptible; HS,highly susceptible. B5, resistant control; TN1, susceptible control.
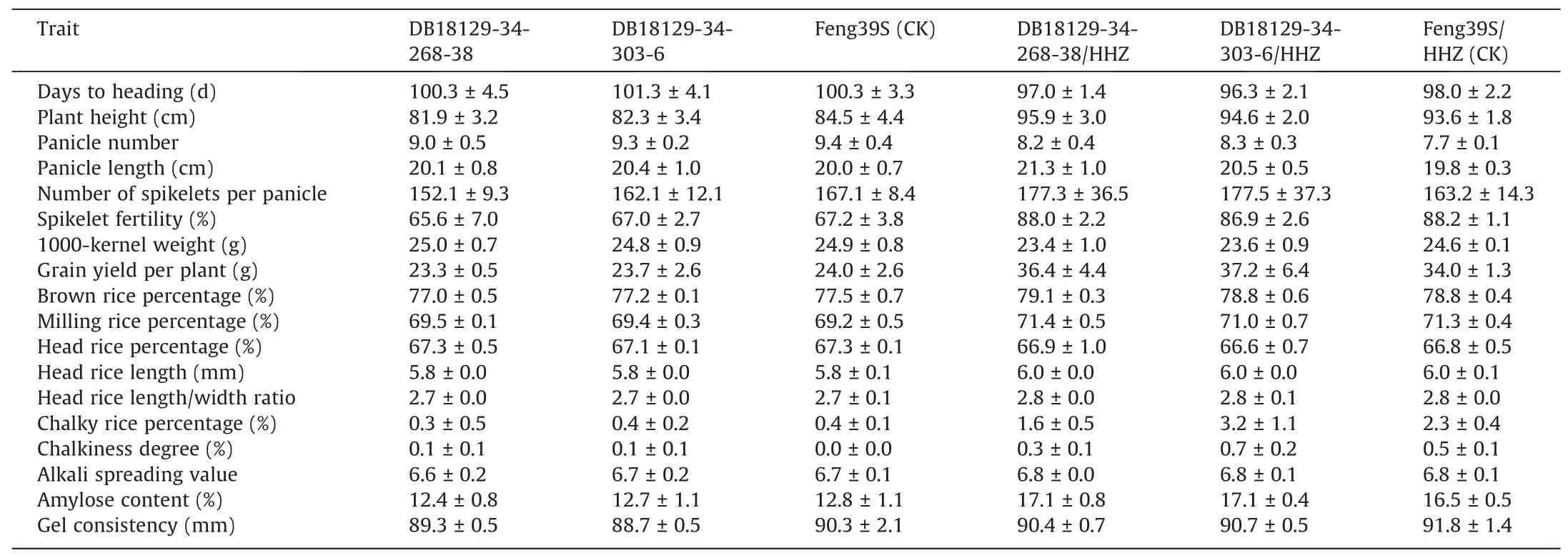
Table 1Agronomic traits of the improved lines and their hybrid combinations (Hainan, 2020).

Fig.7. Plant type and head rice appearance of the two improved lines and Feng39S.Plant type at the sterile stage in Wuhan(a)and the fertile stage in Hainan(b).Head rice appearance at the fertile stage in Hainan (c).
4. Discussion
Phenotypic selection during conventional breeding has been successfully used to develop several elite inbred or hybrid crop cultivars. However, this process is complex, and developing multiple resistant cultivars using this approach is time-consuming. Genomic breeding has been successfully used to improve single traits in restorer lines of hybrid rice [24,32]. In the present study, we demonstrated an efficient genomic breeding strategy for integrating multiple resistance traits into the elite PTGMS line Feng39S,using a combination of foreground, recombinant, and background selection.
Four major resistance genes (Pi2,Xa23,Bph14, andBph15)derived from three rice PTGMS lines were integrated into the genetic background of the susceptible PTGMS line Feng39S using the genomic breeding strategy.The two improved lines of Feng39S(Pi2 + Xa23 + Bph14+Bph15) showed increased resistance against blast,showing a mean resistance frequency of 95.59%under artificial inoculation,implying that thePi2gene is effective for blast disease control in rice. The efficacy of thePi2gene in conferring broad-spectrum, strong resistance against blast in natural blast nurseries in China and India has already been reported [14,44-46]. We detected a significant difference in the reactions of the improved line harboringPi2between the Yichang and Enshi experimental sites. Thus, each rice-growing region will require its own combination of blast resistance genes in future genomic breeding programs [47].
The strong resistance to seven bacterial blight isolates shown by the improved Feng39S (Pi2 + Xa23 + Bph14+Bph15) suggests thatXa23might confer broad-spectrum resistance against prevalent bacterial blight isolates worldwide, whereas the widely usedXa7showed high susceptibility to PXO99 in a previous breeding program [17]. Given thatXa23is rarely found in modern breeding lines [19], this finding will potentially accelerate its use in hybrid breeding programs.
Various restorer lines carryingBph14+Bph15(conferring brown planthopper resistance) have been developed by marker-assisted selection and the corresponding hybrids released to farmers[20,21,24,42,43,48]. However, there are few reports of these resistance genes being used to improve the brown planthopper resistance of PTGMS lines[49].In the present study,we introduced two brown planthopper resistance genes,Bph14andBph15, into the PTGMS line Feng39S, producing two improved lines with increased resistance against brown planthopper at the seedling stage. Thus, Feng39S (Pi2 + Xa23 + Bph14+Bph15) showed broad-spectrum resistance to blast and bacterial blight and strong resistance to brown planthopper, indicating that we have successfully combined multiple resistance genes (Pi2+Xa23+Bph14+Bph15) in the PTGMS rice line.
In recent years, marker-assisted foreground selection coupled with backcrossing has been widely used to develop pyramided lines. The blast resistance genePi2and the bacterial blight resistance genesXa7andXa23were introduced into three elite PTGMS lines:C815S,Guangzhan 63-4S,and Feng39S,via a combination of phenotypic selection and marker-assisted selection of target genes[16,42,43]. The transfer of undesirable genes by linkage drag and the genetic backgrounds exerted negative effects on agronomic traits, such as by increasing plant height and reducing rice milling quality, as well as changing critical sterility temperature points,which are regulated by major genes and unknown minor polygenes in the PTGMS lines [15-18]. Thus, pyramiding multiple resistance genes in a single elite line using conventional markerassisted foreground selection methods is cumbersome owing to linkage drag of genes and genetic background effects.
In the present study, to address these issues, we employed a genomic breeding strategy. Feng39S (Pi2 + Xa23 + Bph14+Bph15)was developed using a two-stage genomic breeding strategy involving the construction of NILs and the assembly of pyramided lines with favorable traits[23].This strategy involves an introgression breeding scheme in which one cross, three or four backcrosses, and one or two generations of self-pollination are performed to generate NILs with desirable genes.During the introgression process, the use of gene-specific markers developed for each gene, a set of low-density SSR markers for background selection in early generations (BC1F1and BC2F1), and the RICE6K SNP array for background selection in advanced backcross generations(BC2F1, BC3F1, BC4F1) greatly facilitated selection for target genes,recombination events, and genomic background. Using this strategy, four high-quality NILs harboringPi2,Xa23,Bph14, orBph15were obtained, with the sizes of the four minimal introgression segments ranging from 566.8 to 1574.9 kb.The four NILs were further assembled to develop Feng39S (Pi2 + Xa23 + Bph14+Bph15),with greatly increased resistance against rice blast,blight bacterial,and brown planthopper. These studies provide practical examples of genomic breeding approaches for the targeted improvement of elite lines, demonstrating the superiority of this approach over conventional marker-assisted foreground selection backcrossing.
Breeding of PTGMS lines with a low critical sterility temperature point and high combining ability is a key requirement for developing two-line commercial rice hybrids [50]. Developing an elite PTGMS line using conventional breeding approaches is extremely slow, owing to the absence of efficient phenotypic selection criteria for complex traits, including low critical sterility temperature point and high combining ability [25,26]. The genomic breeding strategy used in this study allowed the best plants with the highest RPG recovery to be identified without the need for phenotypic selection. We generated Feng39S(Pi2+Xa23+Bph14+Bph15) with an RPG recovery of 98.98% by incorporating the four target genes into the genome of Feng39S.In a field test, Feng39S (Pi2+Xa23+Bph14+Bph15) showed a low critical sterility temperature point while maintaining the fertility-sterility transition pattern of the original Feng39S line.The finding that the combining ability of the hybrid combination Feng39S (Pi2+Xa23+Bph14+Bph15)/HHZ was not affected by the four introgression segments suggests that Feng39S(Pi2+Xa23+Bph14+Bph15)could be used as an upgraded PTGMS line in commercial hybrid rice breeding programs.Thus,more twoline hybrid cultivars based on Feng39S (Pi2+Xa23+Bph14+Bph15) could be developed to further test the combining ability of the improved PTGMS line on a large scale.
In this study,the combination ofPi2+Xa23+Bph14+Bph15was successfully introduced into a PTGMS line with short breeding cycles and high RPG recovery. However, various combinations of the appropriate resistance genes are essential for rice breeding in different rice-growing regions. With the development of restorer lines with alternative resistance genes using a similar genomic breeding strategy, an increasing number of resistance genes could be pyramided in two-line hybrid cultivars, further increasing the utility of genomic breeding for hybrid rice improvement.
5. Conclusions
We obtained the line Feng39S (Pi2+Xa23+Bph14+Bph15)using a genomic breeding strategy that involves foreground,recombinant, and background selection on a genome-wide scale.The improved PTGMS lines DB18129-34-268-38 and DB18129-34-303-6, with increased resistance against blast, bacterial blight,and brown planthopper, could be used as female parents of twoline hybrid rice cultivars instead of the original Feng39S, raising the multiple-resistance levels of two-line hybrid rice. This proposed strategy could pave the way for integrating desirable genes into elite cultivars for the genetic improvement of crops.
CRediT authorship contribution statement
Dabing Yang:Conceptualization, Investigation, Data curation,Formal analysis, Writing - original draft.Lizhong Xiong:Writing-review&editing,Funding acquisition.Tongmin Mou:Resources,Methodology,Writing-review&editing,Supervision.Jiaming Mi:Conceptualization, Supervision, Project administration, Writing -review & editing, Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the National Natural Science Foundation of China(31821005),Hubei Provincial Natural Science Foundation of China(2020CFB192),and the Fundamental Research Funds for the Central Universities of China (2662019QD051).
Appendix A. Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2021.11.005.
- The Crop Journal的其它文章
- Research progress on the divergence and genetic basis of agronomic traits in xian and geng rice
- From model to alfalfa: Gene editing to obtain semidwarf and prostrate growth habits
- The chloroplast-localized protein LTA1 regulates tiller angle and yield of rice
- Genome-wide association study and transcriptome analysis reveal new QTL and candidate genes for nitrogen-deficiency tolerance in rice
- Advances in the functional study of glutamine synthetase in plant abiotic stress tolerance response
- Identification of microRNAs regulating grain filling of rice inferior spikelets in response to moderate soil drying post-anthesis

