The chloroplast-localized protein LTA1 regulates tiller angle and yield of rice
Xiowu Pn, Yongcho Li, Hiwen Zhng, Wenqing Liu, Zheng Dong, Licheng Liu, Snxiong Liu,Xinnin Sheng, Jun Min, Rongfeng Hung,, Xioxing Li,
a Rice Research Institute, Hunan Academy of Agricultural Sciences, Changsha 410125, Hunan, China
b Biotechnology Research Institute, Chinese Academy of Agricultural Sciences, Beijing 100081, China
Keywords:Rice Yield Tiller angle Gravitropism Chloroplast development
A B S T R A C T Plant architecture strongly influences rice grain yield. We report the cloning and characterization of the LTA1 gene, which simultaneously controls tiller angle and yield of rice. LTA1 encodes a chloroplastlocalized protein with a conserved YbaB DNA-binding domain,and is highly expressed in photosynthetic tissues including leaves and leaf sheaths. Disrupting the function of LTA1 leads to large tiller angle and yield reduction of rice. LTA1 affects the gravity response by mediating the distribution of endogenous auxin, thereby regulating the tiller angle. An lta1 mutant showed abnormal chloroplast development and decreased chlorophyll content and photosynthetic rate, in turn leading to reduction of rice yield.Our findings shed light on the genetic basis of tiller angle and provide a potential gene resource for the improvement of plant architecture and rice yield.
1. Introduction
Rice(Oryza sativaL.)is a staple food crop worldwide,especially in Asia. Owing to the increasing demand for rice production, the pursuit of high yield is the eternal theme of rice breeding. Yield is a complex quantitative trait affected by many factors, including plant architecture and photosynthetic efficiency.Breeding elite rice varieties with ideal plant architecture and high photosynthetic efficiency is the most promising strategy for increasing rice yield.
Tiller angle,one of the determinants of plant architecture,influences photosynthesis by affecting light harvesting and vulnerability to insects and diseases [1]. Tiller angle is defined as the angle between the main culm and the outside tiller, varying widely among rice species and subspecies. Wild rice (O. rufipogonGriff.)tends to have a prostrate growth habit, whereas cultivated rice has an erect growth habit. The transition from prostrate to erect growth is controlled by a domestication genePROG1[2-4]. A 110-kb deletion in theRPADlocus,which is genetically linked withPROG1, may be involved in the parallel domestication of plant architecture in Asian and African rice [5].TIG1, encoding a TCPfamily transcription factor, promotes cell elongation and enlarges the tiller angle in wild rice [6]. Among cultivated rice accessions,the tiller angle is generally wider inindicathan injaponicarice,and thisindica-japonicadifferentiation has been shown to be controlled by theTAC1gene [7]. A mutation from ‘‘AGGA” to ‘‘GGGA”in the splicing site of a 1.5-kb intron results in lower expression ofTAC1and in a compact plant architecture with erect tillers.The function ofTAChomologs is highly conserved among plants,with all involved in controlling tiller or branching angle [8-10].TAC3andqTAC8have also been shown [11,12] to control natural variation of tiller angle among rice cultivars.
Several genes that control rice tiller angle have been identified by using mutant with tiller-spreading phenotype.LAZY1(LA1)regulates shoot gravitropism and rice tiller angle by affecting polar auxin transport (PAT) [13,14]. Screening for suppressors of thelazy1phenotype [15] has shown that multiple genes involved in strigolactone biosynthesis and signaling affect the tiller angle.The geneLPAinfluences tiller angle and gravitropism by regulating the sedimentation rate of amyloplasts, a mechanism distinct from that ofLA1[16]. These known genes all participate in gravity response,suggesting its critical role in the regulation of tiller angle.In plants, gravitropism involves gravity perception and signal transduction cascades that ultimately lead to an asymmetric distribution of auxin and differential organ responses at the shoot base[17]. Via large-scale transcriptome analysis, the upstream positive regulator HSFA2D and downstream transcription factors (WOX6 and WOX11) were identified [18] as being involved in the LA1-mediated shoot gravitropism pathway.OsHOX1 and OsHOX28 can suppress the HSFA2D-LA1 pathway and thus negatively regulate tiller angle establishment [19]. In response to gravity, the asymmetric distribution of auxin has been proposed [13] to be a central component of gravitropic signaling. Overexpression of the auxin efflux carrier OsPIN2 or signal factor OsIAA4 increased the tiller angle of rice [20,21].
In plants, chloroplasts, as the photosynthetic apparatus, are indispensable for growth and development and participate in synthesis of amino acids,lipids,pigments,and a wide range of precursors of various phytohormones [22]. Chloroplast biogenesis and development are regulated by the coordinated expression of plastid-encoded and nucleus-encoded genes [23,24]. The majority of chloroplast proteins, including abundant photosynthetic proteins, are encoded by nuclear genes and imported posttranslationally through translocons complexes at the outer (TOC)and inner (TIC) envelope membranes of the chloroplast [25,26].Chloroplast proteins affect numerous biological processes. Among them, multiple nucleus-encoded chloroplast proteins identified in albino and yellow leaf rice mutants influence chloroplast ultrastructure[27-29].Chloroplast-localized PHD1 regulates grain production by affecting chloroplast biogenesis and photosynthetic activity [30].ARE1encodes a chloroplast protein and mediates grain yield by modulating nitrogen use in rice[31].Several chloroplast proteins simultaneously regulate chloroplast development and plant architecture by affecting the biosynthesis of phytohormones [32-34]. For example,HTD12encodes a chloroplastlocalized 15-cis-ζ-carotene isomerase (Z-ISO), whose mutation results in reduced carotenoid-derived strigolactones, in turn affecting photosynthesis and plant architecture [33]. Chloroplastlocalized or plastid-localized proteins are also involved in gravitropism[35-37].Changes in chloroplast morphology affected gravitropic response [38,39]. According to a recent report [40],chloroplast protein LA2 interacts with the starch biosynthetic enzyme OspPGM and regulates shoot gravitropism and rice tiller angle.
Although great efforts have been made to dissect the genetic basis of tiller angle and rice yield, the relationship between yield and tiller angle is still unclear,and few genes have been identified that control tiller angle and yield simultaneously.In this study,we isolated a rice mutantlta1(large tiller angle 1)with large tiller angle and reduced yield, and cloned theLTA1gene, which was allelic to the recently reportedLA2gene [40].LTA1encodes a chloroplastlocalized protein with a conserved YbaB DNA-binding domain and regulates rice tiller angle by affecting shoot gravitropism.Disrupting the function ofLTA1resulted in abnormal chloroplast ultrastructure, thereby reducing chlorophyll content and photosynthetic rate and ultimately yield.
2. Materials and methods
2.1. Plant materials and growth conditions
The ricelta1mutant is a naturally occurring mutant identified in anindicacultivar,Xiangwanxian 13.An F2population was developed from a cross betweenlta1and a compactjaponicarice cultivar,02428. The primers used for mapping are listed in Table S1.All materials were grown in either paddy fields or greenhouses in Hunan and Hainan provinces.
2.2. Plasmid construction and rice transformation
For a functional complementation assay,the full 5538-bp genomic sequence of theLTA1gene, including the 2-kb promoter sequence, the entire open reading frame (ORF), and 500 bp of downstream sequence, was amplified from the wild type (WT,Xiangwanxian 13)with specific primers(Table S2),and cloned into the pCAMBIA1300 vector using the ClonExpress II One Step Cloning Kit (Vazyme, Nanjing, China). The plasmid was inserted into thelta1mutant viaAgrobacterium-mediated genetic transformation.ForCRISPRtargeting knockout assay,two target sites were selected in the first and sixth exons of theLTA1gene. Two corresponding single-guide RNA (sgRNA)expression cassettes were constructed using target-specific primers(Table S2),and then ligated into pYLCRISPR/Cas9Pubi vector as described previously[41].The resulting plasmid was then transformed into the WT(Xiangwanxian 13).The pYLCRISPR/Cas9Pubi vector was kindly provided by Prof. Yao-Guang Liu (South China Agricultural University).
2.3. Investigation of tiller angle and yield-associated traits
A field evaluation was conducted in an experimental plot at Hunan Rice Research Institute, Hunan, China. The 25-d-old seedlings of WT andlta1were transplanted in three replications. Each replication contained 100 plants with in-row spacing and between-row distances of 20 cm each. The tiller angle was measured between the main culm and the outside tiller in 15 plants each of WT andlta1every 10 d, from 40 to 120 d after sowing.The tiller angles of transgenic plants were measured at heading stage. Five plants of each replicate were harvested after maturity and yield traits were recorded.
2.4. Histological observation
Fresh tissues of the tiller base were collected and fixed in formalin-acetic acid-alcohol solution (FAA) for a minimum period of 24 h, and then were subjected to a series of dehydration and infiltration treatments. Paraffin-embedded samples were sliced longitudinally,stained with safranin and Fast Green,and observed under bright field using a CKX41 microscope (Olympus, Tokyo,Japan).
2.5. Gravitropism assay and indoleacetic acid measurement
The gravitropic response was measured as detailed previously[13]. Rice seeds were grown at 28 °C in 1/2 MS medium (pH 5.8)after being de-husked and sterilized. Then the 5-d-old seedlings were rotated by 90° for gravistimulation. The gravitropic response was quantified by measuring the curvature angle every 12 h for two days.Upon gravistimulation for 24 h,the basal shoot was dissected into lower and upper sides and was collected for gene expression analysis and indoleacetic acid (IAA) determination.
IAA measurement was performed as previously described [15].Samples of 200 mg from the lower and upper sides of the basal shoot were collected for the assay. After extraction and purification,the samples were subjected to LC-MS/MS analysis using a system consisting of an Ultra Performance Liquid Chromatograph(Shim-pack UFLC SHIMADZU CBM30A, Shimadzu, Kyoto, Japan)and a tandem mass spectrometer(Applied Biosystems 6500 Quadrupole Trap, Applied Biosystems, Foster City, CA, USA).
2.6. RNA extraction, cDNA preparation, and quantitative RT-PCR
Seedlings of WT andlta1were grown in half-strength Yoshida nutrient solution in a growth chamber. Leaves of two-week-old seedlings were sampled and immediately frozen in liquid nitrogen.Total RNAs were isolated from frozen samples using Trizol Reagent(TransGen,Beijing,China)and were used for cDNA synthesis using RT SuperMix (Vazyme). Subsequently, the cDNA was used to perform quantitative RT-PCR on a LightCycler 96 system (Roche,Rotkreuz, Switzerland) using SYBR qPCR Master Mix (Vazyme).Gene-specific primers are listed in Table S3. TheActingene was used as an internal control. Relative gene expression was calculated by the 2-ΔΔCTmethod.
2.7. Protein sequence and phylogenetic analysis
Arabidopsis thalianaprotein sequences were acquired from the Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org/index.jsp) using the LTA1 sequence as a query. The sequences of other LTA1 protein homologs were obtained by searching the database of NCBI non-redundant protein sequences (https://www.ncbi.nlm.nih.gov/).The protein sequences were aligned with ClustalX 1.83, and the aligned sequences were used to construct a phylogenetic tree with MEGA 4.1 by the neighbor-joining method.
2.8. Subcellular localization
The full-length coding sequence ofLTA1was amplified from WT(Xiangwanxian 13) using specific primers (Table S2) and cloned into the pBWA(V)HS-GFP vector to generate thep35S:LTA1-GFPconstruct. The vector pBWA(V)HS-GFP (as the control) andp35S:LTA1-GFPwere introduced into rice protoplasts as described previously [42]. After 20 h incubation at 28 °C, the GFP fluorescence of chloroplasts was measured under a confocal laser scanning microscope (FluoView FV1000, Olympus, Tokyo, Japan).
2.9. Chloroplast ultrastructure observation
The top second leaves of two-week-old seedlings were used for microscopic observation.Leaf tips were dissected and fixed in 2.5%glutaraldehyde fixing solution at 4°C for 2-4 h,rinsed,and further fixed with 1%OsO4in 0.1 mol L-1PBS(pH 7.4)for 2 h at room temperature. Leaf samples were then dehydrated in an ethanol gradient and infiltrated slowly with an epoxy resin. The embedded samples were cut into ultrathin sections (60-80 nm) with an UC7 ultramicrotome(Leica,Weztlar,Germany),followed by staining with uranyl acetate and lead citrate in turn. The chloroplast ultrastructure was examined under a HT7700 transmission electron microscope (Hitachi, Tokyo, Japan). The length and width of chloroplasts were measured with ImageJ software (https://imagej.net/software/fiji/downloads) and used to calculate the length/width ratio, as an indicator of chloroplast shape.
2.10. Measurement of chlorophyll content and photosynthetic rate
Fresh leaves (0.5 g) of two-week-old seedlings were collected and used to measure chlorophyll content.The leaves were homogenized in 80% acetone and centrifuged at 3000×gfor 20 min. The absorbance of the supernatants was determined with a Epoch microplate spectrophotometer (Biotech, Vermont, USA) at wavelengths of 645 and 663 nm.The total chlorophyll content was calculated asCT=20.21A645+8.02A663.The photosynthetic rate was determined in flag leaves at the heading stage at 9:00 AM using an Li-6400 portable photosynthesis system(LI-COR,Lincoln,NE,USA)according to the manufacturer’s instructions.
3. Results
3.1. Characterization of a large tiller-angle mutant lta1
lta1showed a significantly greater tiller angle than the WT at the tillering and ripening stages(Fig.1A,B).The tiller angle of both the WT andlta1increased rapidly at the vegetative stage,reaching their maximum 70 days after sowing,and then remained relatively stable at the late growth stage(Fig.1C).The maximum angles were 19.4°in the WT and 49.7°inlta1.However,there was no significant difference in leaf angle between the WT andlta1(Fig. S1). Close observation showed that the larger tiller angle oflta1was caused by greater outward bending at the tiller base than in WT(Fig. 1D). Longitudinal sections of the bending joint revealed no significant difference in cell size at the side nearest the ground(NG) between WT andlta1(Fig. 1E,F). However, the cell length oflta1at the side farthest from the ground (FG) was significantly longer than that of WT (Fig. 1E,F). These results suggested that the larger tiller angle oflta1might be due to the increased cell length on the FG relative to WT.
As shown in Fig. 1G, the grain yield per plant oflta1(35.21 g)was significantly lower than that of WT (48.77 g), with a decrease of 27.8%.Panicle number(PN),spikelet number(SN)and thousandgrain-weight(TGW)did not differ between WT andlta1(Table S4).However,the seed setting rate(SSR)was significantly lower inlta1than in WT(Fig.1H-I).Thus,the yield reduction oflta1was caused mainly by the lower seed setting rate.
3.2. The lta1 mutant showed reduced gravity response
The tiller-spreading phenotype of most rice mutants is closely associated with reduced shoot gravitropism [13,15-16]. To test whetherlta1was also involved in this process, we compared the gravity response of WT andlta1at the seedling stage. The shoot of WT began to bend after 12 h of gravistimulation and then rapidly bent and grew upward, and the curvature angle reached 63.5°after 48 h of gravistimulation(Fig.2A,B).The curvature angle oflta1was about half that of WT, indicating that the gravity response oflta1was lower than that of WT (Fig. 2A,B). The asymmetric distribution of auxin plays a key role in gravity response of rice plants [43,44]. To explore the involvement of auxin inLTA1-mediated gravitropism, we first measured the expression of the auxin-responsive marker geneOsIAA20in WT andlta1seedlings.Before gravistimulation,the expression ofOsIAA20was comparable in the upper and lower side of WT andlta1.However,OsIAA20was expressed asymmetrically in WT after gravistimulation, and its expression was much higher in the lower than in the upper side(Fig. 2C). Its expression in the lower side and upper side oflta1showed no significant difference (Fig. 2C). We further measured the endogenous indoleacetic acid (IAA) content 24 h after gravistimulation. In agreement with the results of gene expression, the IAA content was significantly higher in the lower than in the upper side of WT, whereas there was no significant difference between the lower and upper sides oflta1(Fig.2D).These results suggested thatlta1might regulate gravity response and tiller angle by affecting lateral auxin distribution.
3.3. Map-based cloning of LTA1
For genetic analysis, we crossedlta1with WT and generated 397 F2plants that segregated in a 3:1 ratio(287 wt and 110 mutant plants,χ=1.55,not significant),indicating that the tiller-spreading phenotype is controlled by a single recessive nuclear gene. To isolate theLTA1gene, we developed a F2population derived from a cross betweenlta1and a compactjaponicarice cultivar,02428.We initially restrictedLTA1to the region between RM555 and RM12623 using 112 recessive plants through map-based cloning,and finally narrowed the interval to 31 kb between InDel markers ID02154 and P1 using 2449 recessive plants (Fig. S2). There were three ORFs in this region.Sequencing of these ORFs inlta1revealed a single-nucleotide polymorphism representing a transition from guanine (G) to adenine (A) in the sixth exon of ORF3(LOC_Os02g08380), resulting in an amino acid change of serine to asparagine (S159N).
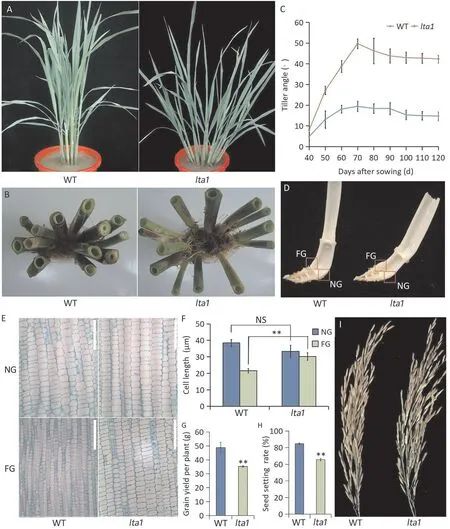
Fig.1. Phenotypic characterization of the lta1 mutant.(A,B)The plant architecture of the WT and lta1 at the tillering and the ripening stage.(C)Tiller angle changing during development in WT and lta1.Error bars represent standard deviations(SD)(n=15).(D)Magnified tiller base of the WT and lta1.NG,the side nearest the ground;FG,the side farthest from the ground.(E)Histological analysis of the longitudinal section from red boxes in(D).Scale bars,200 μm.(F)Comparison of the cell length of the bending joints between WT and lta1.Error bars represent SD(n=20).**indicates significant at the 0.01 probability level.NS indicates no significant difference at the 0.05 probability level.(G-H) Comparison of grain yield and seed setting rate between WT and lta1. Value are means ± SD (n = 3). (I) Panicles of WT and lta1 plants.
To verify the role of LOC_Os02g08380 in determining tiller angle in rice, we performed a genetic complementation test. A plasmid containing 2 kb of upstream sequence, the entire ORF,and 500 bp of downstream sequence was inserted by transformation into thelta1mutant. All lines harboring theLTA1transgene showed small tiller angle (Fig. 3A,B) and normal yield (Table S5),similar to WT. The function of LOC_Os02g08380 was further confirmed by CRISPR/Cas9-mediated gene knockout in the background of Xiangwanxian 13.We designed two sgRNA target sites,one each in the first and sixth exons of LOC_Os02g08380.Using site-specific PCR and Sanger sequencing, we identified a series of independent transgenic lines for each target site in T0plants. We selected two homozygous lines (KO#5 and KO#6) for further analysis. One(KO#5) contained a base insertion in the sixth exon close to the mutation position oflta1and the other (KO#6) harbored short deletions in both the first and sixth exons (Fig. S3). Both KO#5 and KO#6 displayed the tiller-spreading phenotype,and their tiller angles were significantly higher than that of WT(Fig.3C,D).Yieldassociated traits, except for panicle number, were sharply decreased in these lines as compared with WT (Fig. 3E-I). The yields of these knockout lines were even lower than those oflta1.The knockout lines had fewer spikelets and smaller grain weight than WT.However,there were no significant differences in spikelet number per panicle and grain weight between WT and mutantlta1.These results suggested that LOC_Os02g08380 was the geneLTA1controlling tiller angle and yield.
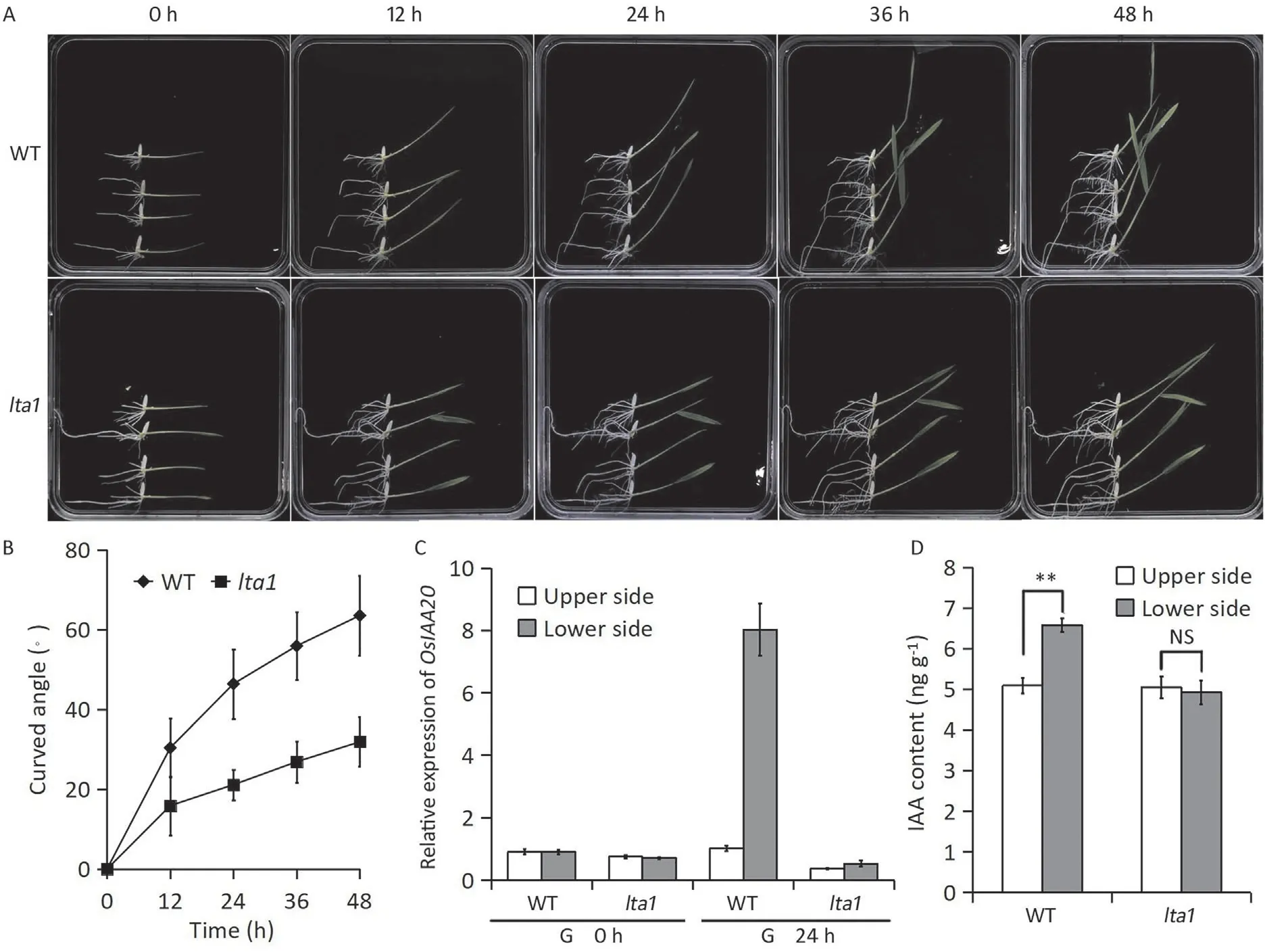
Fig. 2. Gravitropic analysis of lta1 mutant. (A) Gravity response of WT and lta1 at five time points after gravistimulation. (B) Kinetic comparison of the curvature angle of seedlings between WT and lta1 after gravistimulation. Error bars represent SD (n = 8) (C) Expression levels of OsIAA20 in the lower side and upper side of seedlings upon gravistimulation. Values are means ± SD (n = 3). (D) Endogenous IAA content in lower and upper sides of seedlings upon gravistimulation. Values are means ± SD (n = 3).** indicates significant at the 0.01 probability level. NS indicates no significant difference at the 0.05 probability level.
3.4. LTA encodes a chloroplast-targeted protein
TheLTA1gene harbors seven exons and six introns,and the fulllength cDNA consists of 889 bp, with a 97-bp 5′-UTR, 558-bp CDS,and 234-bp 3′-UTR. It is a single-copy gene in rice and encodes a putative nucleoid-associated protein of 185 amino acid residues with a predicted YbaB DNA-binding domain (amino acids 75-177). LTA1 also contains a predicted chloroplast transit peptide at the N-terminal end (http://www.cbs.dtu.dk/services/TargetP-2.0/),suggesting that it may be localized in the chloroplast.To determine the subcellular localization of LTA1,we transiently expressed GFP alone and the LTA1-GFP fusion protein under the control of the CaMV 35S promoter in rice protoplasts. The green fluorescence of GFP alone was detected in the cytoplasm, nucleus, and plasma membrane, whereas that of LTA1-GFP overlapped strongly with chloroplast autofluorescence signal (Fig. S4), supporting a chloroplast location of LTA1.This result was consistent with the location of theArabidopsishomology (STIC2), which has been previously established as a chloroplast protein [45].
To identify homologs of LTA1, a BLAST search was performed against the databases of TAIR (http://www.arabidopsis.org/index.jsp) and NCBI (https://www.ncbi.nlm.nih.gov). Homologous proteins were widely found in monocot and dicot plants but not in animals. A phylogenetic tree indicated that these proteins could be classified into two clades, and homologs from monocots such asOryza brachyantha,Triticum turgidum,T. aestivum, andAegilops tauschiishare high sequence similarity with LTA1 (Fig. 4A). Multiple sequence alignments showed that these proteins were highly conserved in the YbaB DNA-binding domain, especially in the mutant position oflta1(Fig.4B).To test whether thelta1mutation affected its expression,we measured the transcription level ofLTA1in WT andlta1by quantitative RT-PCR.LTA1was ubiquitously expressed in various rice tissues, with the most abundant expression in photosynthetic tissues including leaves and leaf sheaths(Fig. S5). However, there were no significant differences in gene expression between WT andlta1, suggesting that the mutation oflta1did not change the transcription level.
3.5. LTA1 affects chloroplast development and photosynthesis
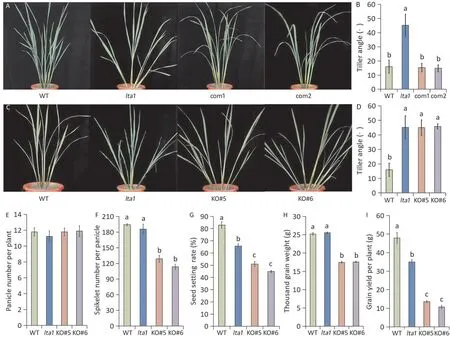
Fig.3. Functional verification of LTA1.(A)The plant architecture of WT,lta1 and the complemented transgenic lines.(B)Statistical analysis of tiller angle of WT,lta1 and the complemented transgenic lines. Error bars represent SD (n = 10). (C) The plant architecture of WT, lta1, and the CRISPR-Cas9 knockout lines (KO); KO#5 contains a base insertion in the sixth exon, whereas KO#6 has short deletions in both the first and sixth exon. (D) Statistical analysis of tiller angle of WT, lta1, and the KO lines. Error bars represent SD (n = 10). (E-I) Yield associated traits of WT, lta1, and the KO lines. Value are means ± SD (n = 3). Different letters indicate significant difference at the 0.01 probability level (Duncan’s multiple range test).
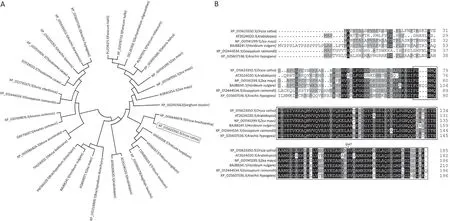
Fig.4. Phylogenetic analysis of LTA1.(A)Phylogenetic tree of the 32 protein homologs of LTA1.The black box indicates the position of LTA1.(B)Multiple sequence alignments of LTA1 and its protein homologs in Arabidopsis,Zea mays,Hordeum vulgare,Gossypium raimondii,and Arachis hypogaea;Black box indicates the conserved YbaB DNA-binding domain. * represents the mutation position of lta1.
The homologous gene (STIC2) inArabidopsishas been reported to participate in thylakoid protein targeting, with its mutation altering the morphology and structure of chloroplasts [45]. To further characterize the function ofLTA1in chloroplasts, we compared the chloroplast ultrastructures of WT andlta1and found that the chloroplast oflta1displayed an abnormal morphology. As shown in Fig. 5A and B, the chloroplasts oflta1appeared rounder,with less attachment to the cell periphery than WT chloroplasts.The shape difference between WT andlta1was quantified by measuring the length and width of chloroplast cross sections and comparing the length/width ratios (Fig. 5C). The length/width ratio oflta1was significantly lower than that of WT chloroplasts,confirming a rounder shape oflta1chloroplasts. Upon observation at higher magnification, the lamellae in the chloroplast oflta1were found to be less parallel and more loosely distributed (Fig. 5D,E),resembling the phenotype of theArabidopsis stic2mutant. Quantitative RT-PCR showed that the transcript levels of multiple genes associated with chloroplast development were significantly decreased in thelta1mutant(Fig.5F).Correspondingly,the chlorophyll content and photosynthetic rate oflta1were significantly decreased in comparison with WT (Fig. 5G,H). Thus, mutation ofLTA1resulted in abnormal chloroplast ultrastructure and reduced chlorophyll content, thereby affecting photosynthetic rate and yield.
4. Discussion
4.1. LTA1 plays a unique role in regulating tiller angle and yieldassociated traits
Tiller angle, one of the most important factors determining plant architecture, affects the photosynthetic efficiency and yield of rice[6,7].Great progress has been made in dissecting the genetic basis of tiller angle over recent years. However, how tiller angle affects yield is still largely unknown. We identified a rice mutantlta1with large tiller angle and reduced grain yield.The substitution of an amino acid in LTA1 impaired gravity response by affecting the distribution of endogenous auxin, resulting in a spread-tiller phenotype. These findings were consistent with previous report ofLA2[40].lta1is a new allelic mutant ofla2, and the mutant sites of bothlta1andla2are located in the conserved YbaB DNAbinding domain. Thelta1mutation caused abnormal chloroplast development, leading in turn to reductions in photosynthetic rate and rice yield.The effect of theLTA1gene on yield was further verified by genetic complementation andCRISPRtargeting knockout assay. However, Huang et al. [40] have shown that there were no significant differences in yield-associated traits between the wild type andla2.This paradox may be due to differences in genetic background and mutation site. Thela2mutant was in the 93-11 background, whilelta1was generated from an aromatic cultivar,Xiangwanxian 13. Different genetic backgrounds may have different effects on the fine function ofLTAI. In addition, the mutation sites ofla2andlta1were different, located at the fifth and sixth exons, respectively. Considering that the yield oflta1was greater than those of the knockout lines, we speculated that different mutations of LTA have different effects on rice yield.
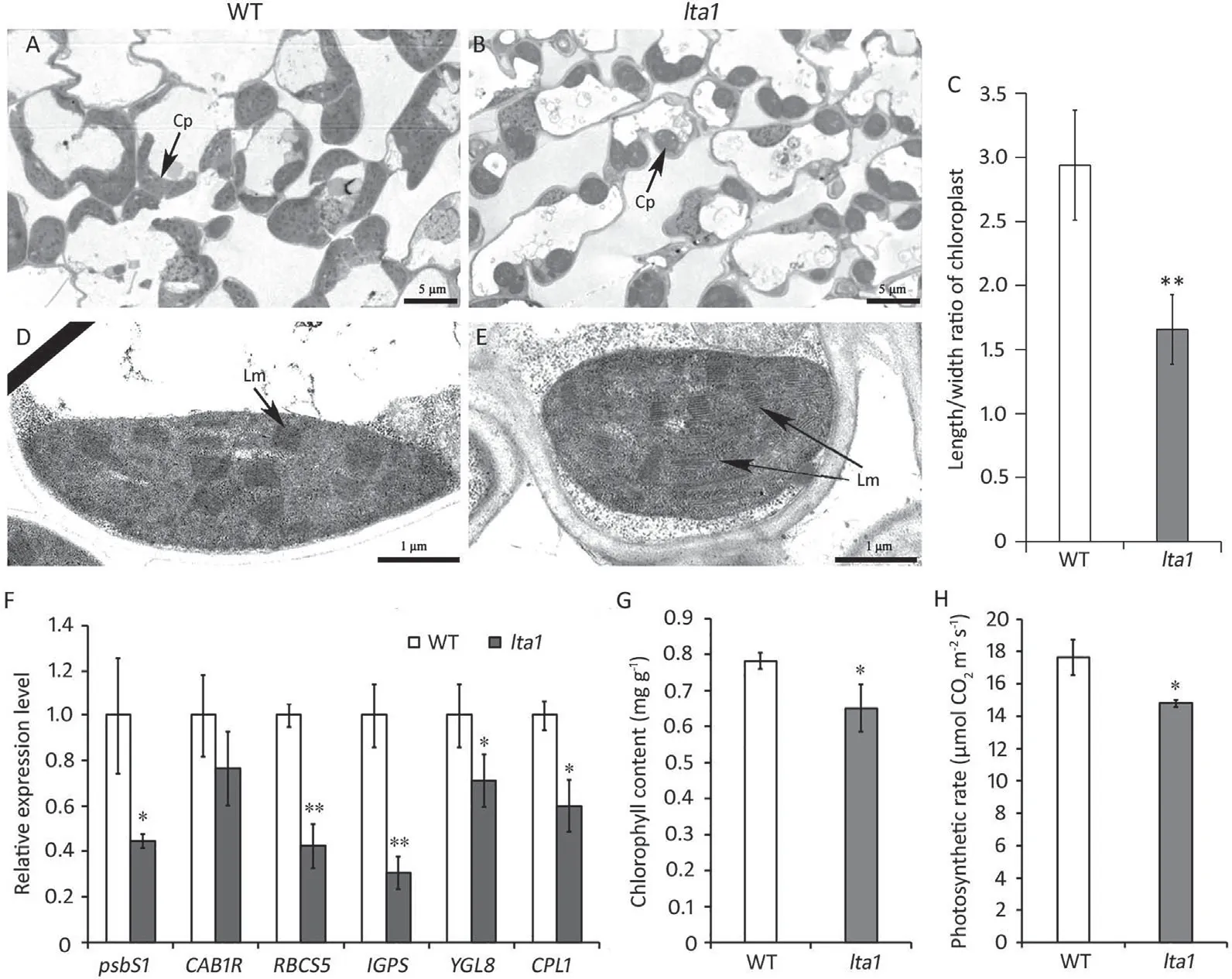
Fig.5. Chloroplast development and photosynthesis of lta1 mutant.(A,B)Chloroplast ultrastructure in mesophyll cells of WT and lta1. Samples were collected from the top second leaves of two-week-old seedlings. Cp indicates the chloroplast. Scale bars, 5 μm. (C) The length/width ratio of chloroplast in WT and lta1. Value are means ± SD(n = 30). (D,E) Chloroplast ultrastructure of WT and lta1 at higher magnification. Lm indicates the lamellae. Scale bars, 1 μm. (F) Comparison of the expression levels of chloroplast development-associated genes between WT and lta1.Values are means±SD(n=3).(G)Comparison of the chlorophyll contents of WT and lta1 at seedling stage.Value are means±SD(n=3).(H)Comparison of photosynthetic rates of WT and lta1 at heading stage.Value are means±SD(n=3).*and**indicate significant difference at the 0.05 and 0.01 probability levels, respectively.
LTA1is a homolog of bacterialybaB, which encodes a protein implicated in membrane protein biogenesis [46], DNA binding,and gene expression regulation [47,48]. The TargetP program predicted a chloroplast transit peptide at the N-terminal end of LTA1.Consistently, LTA1 protein was localized exclusively in the chloroplast and not in the nucleus, in contrast to other previously reported nucleus-localized proteins [6,7,13,16] controlling tiller angle. The nuclear location is essential for the function of LA1,and its interaction protein OsBRXL4 negatively regulates rice tiller angle by affecting the nuclear location of LA1[49].Most previously reported genes controlling rice tiller angle are expressed mainly in the tiller base,in agreement with the expectation for their activity sites [6,7,13]. The finding that LTA1 showed high expression in photosynthetic tissues suggests a unique role in regulating the tiller angle and grain yield.
4.2.LTA1 regulates rice yield by affecting chloroplast development and photosynthetic efficiency
Multiple sequence alignments showed thatLTA1shared high similarity with the homologous geneSTIC2inArabidopsis,implying similar function.STIC2was identified in a genetic screen for suppressors of chlorotictic40knockout mutant[45].TIC40 is anchored in the inner membrane of the chloroplast and is a component of the TOC-TIC machinery, by which most preproteins are imported and further targeted to their correct subcompartment in the chloroplast [50-53]. Similar to theArabidopsis stic2mutant,lta1showed an abnormal chloroplast ultrastructure including swollen appearance and loosely distributed lamellae. The finding that the expression of genes associated with chloroplast development were downregulated inlta1compared to WT further confirms the effect oflta1on chloroplast development. Compared with WT, the chlorophyll content oflta1was significantly decreased, whereas that ofArabidopsis stic2mutant was identical to those of WT plants.The mutation ofSTIC2might be compensated bySTCL[45], a close homolog ofSTIC2inArabidopsis, thus explaining the different changes of chlorophyll content betweenlta1andstic2. Abnormal chloroplast development resulted in lower photosynthetic rate inlta1, a finding consistent with observations [34,54,55] of many other mutants of chloroplast proteins.
A large tiller angle could increase shading, resulting in decreased photosynthetic efficiency of leaves. To eliminate the influence of shading on photosynthetic efficiency, we chose to measure the photosynthetic rate of the uppermost flag leaves at the heading stage. The finding that the photosynthetic rate of flag leaves was significantly decreased inlta1may account for the reductions in seed setting rate and yield. According to the latest report [40], LA2 interacts with rice OspPGM and participates in starch metabolism. The homolog of OspPGM inArabidopsis(AtPGMp) has been reported to mediate starch synthesis by controlling photosynthetic carbon flow [56]. We accordingly propose thatLTA1regulates photosynthesis and rice yield by interacting with OspPGM. That the yield reduction oflta1is much lower than those in CRISPR-Cas9 knockout lines suggests that the S159N mutation inlta1destroyed only part of the LTA1 function instead of complete inactivation, and has little effect on chloroplasts and photosynthesis compared with those of knockout lines. In fact,we observed (Fig. S6) greater declines of chlorophyll content and photosynthetic rate in knockout lines than inlta1,perhaps leading to a greater reduction in photoassimilate accumulation and in turn to corresponding decreases in spikelet number per panicle and grain weight.
4.3. LTA1 controls shoot gravitropism and rice tiller angle by influencing endogenous auxin distribution
Accumulating evidence [13,15,16,57] suggests that the tiller angle of rice is strongly associated with the shoot’s gravitropic response.Consistently,the gravitropic response was also impaired in thelta1mutant in this study. Gravitropism is a complex physiological process involved in gravitational stimulus perception,signal transduction, and the resulting differential growth that causes bending [58,59]. Among the hypotheses about gravity perception,the starch-statolith model is widely accepted [60]. In this model,amyloplasts are thought to act as the sensors of the gravitational force, so are called statoliths.A loss-of-function mutation of phosphoglucomutase(pgm1)inArabidopsisleads to a failure to accumulate starch in amyloplasts and inability to mediate normal gravitropic response [61-63]. Similarly, mutation of a starch biosynthesis gene,OsAGPL,results in impaired gravitropic response and large tiller angle in rice [64]. Initial gravity signals are translated into differential growth response, which is mediated by asymmetric auxin distribution via lateral auxin transport. In the present study,WT showed normal gravity response and asymmetric growth at the tiller base.lta1showed reduced lateral auxin transport upon gravistimulation, and the IAA content in the lower side was almost identical to that in the upper side. Correspondingly, relatively symmetric growth between NG and FG was observed at the bending joint oflta1,leading directly to a large tiller angle.
CRediT authorship contribution statement
Xiaowu Pan:Conceptualization, Investigation, Data curation,Funding acquisition,Writing- original draft.Yongchao Li:Project administration, Data curation.Haiwen Zhang:Formal analysis,Writing-review&editing.Wenqiang Liu:Investigation,Methodology.Zheng Dong:Investigation, Data curation.Licheng Liu:Investigation.Sanxiong Liu:Investigation.Xinnian Sheng:Investigation.Jun Min:Project administration.Rongfeng Huang:Resources, Writing - review & editing, Supervision.Xiaoxiang Li:Conceptualization, Project administration, Supervision.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research study was supported by the National Natural Science Foundation of China (31801335), Training Program for Excellent Young Innovators of Changsha(kq1802034),and Department of Science and Technology in Hunan Province(2019RS2047).
Appendix A. Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2021.10.005.
- The Crop Journal的其它文章
- Brief Guide for Authors
- Revisiting the role of delta-1-pyrroline-5-carboxylate synthetase in drought-tolerant crop breeding
- A new gain-of-function OsGS2/GRF4 allele generated by CRISPR/Cas9 genome editing increases rice grain size and yield
- Influence of seven levels of chemical/biostimulator protection on amino acid profile and yield traits in wheat
- Corrigendum to ‘‘De novo design of future rapeseed crops: Challenges and opportunities” [Crop J. 10 (2022) 587-596]
- Improving the resistance of the rice PTGMS line Feng39S by pyramiding blast, bacterial blight, and brown planthopper resistance genes

