A new gain-of-function OsGS2/GRF4 allele generated by CRISPR/Cas9 genome editing increases rice grain size and yield
Wenshu Wng, Weipeng Wng, Ynlin Pn, Cho Tn, Hongjing Li, Y Chen, Xingdn Liu,Jing Wei,b, Nin Xu, Yu Hn,b, Hn Gu,b, Rongjin Ye,b, Qi Ding,b,, Chonglie M,b,
a Life Science and Technology Center, China National Seed Group Co., Ltd., Wuhan 430000, Hubei, China
b State Key Laboratory of Crop Breeding Technology Innovation and Integration, China National Seed Group Co., Ltd., Wuhan 430000, Hubei, China
Keywords:Genome editing GS2/GRF4 Grain size Yield Rice
A B S T R A C T Grain size is one of the most important factors affecting rice grain quality and yield, and attracts great attention from molecular biologists and breeders.In this study,we engineered a CRISPR/Cas9 system targeting the miR396 recognition site of the rice GS2 gene, which encodes growth-regulating factor 4(OsGRF4) and regulates multiple agronomic traits including grain size, grain quality, nitrogen use efficiency, abiotic stress response, and seed shattering. In contrast to most previous genome editing efforts in which indel mutations were chosen to obtain null mutants,a mutant named GS2E carrying an in-frame 6-bp deletion and 1-bp substitution within the miR396-targeted sequence was identified. GS2E plants showed increased expression of GS2 in consistent with impaired repression by miR396. As expected,the gain-of-function GS2E mutant exhibited multiple beneficial traits including increased grain size and yield and bigger grain length/width ratio.Thousand grain weight and grain yield per plant of GS2E plants were increased by 23.5%and 10.4%,respectively.These improved traits were passed to hybrids in a semidominant way, suggesting that the new GS2E allele has great potential in rice improvement. Taken together, we report new GS2 germplasm and describe a novel gene-editing strategy that can be widely employed to improve grain size and yield in rice. This trait-improvement strategy could be applied to other genes containing miRNA target sites,in particular the conserved miR396-GRF/GIF module that governs plant growth, development and environmental response.
1. Introduction
CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR associated) protein 9 (CRISPR/Cas9) system has been shown in recent years to be a valuable genome-editing tool. For plant genome editing, a variety of different CRISPR/Casbased systems have been devised [1]. These systems have been employed not only in gene function study but also in crop trait improvement and novel germplasm creation [2-6].
Grain shape and size are important factors for grain quality and are related to grain yield.Several genes controlling grain size have been cloned over the past decades[7-15].Among them,GRAIN SIZE ON CHROMOSOME 2(GS2) encodes growth-regulating factor 4(OsGRF4) which belongs to the protein family of GRF transcriptional regulators [10,11,16]. OsGS2/GRF4 interacts with GRFinteracting-factor 1(GIF1)to positively modulate grain size in rice,and is subjected to post-transcriptional regulation by miR396,facilitating the decay ofGS2transcripts and inhibitingGS2function[17].In several studies[10,11,16],it was found that semi-dominant alleles with a 2-bp TC/AA substitution at the miR396 binding site inOsGS2/GRF4perturbed post-transcriptional regulation by miR396c,resulting in large grain and increased grain yield. Besides regulating grain size, GS2/GRF4 also influences panicle length, seed shattering [18], cold stress response [19], and metabolism of carbon and nitrogen [20]. Considering the multiple beneficial traits conferred byGS2function enhancement in rice and its semidominant effect, it will be of great value for rice improvement.Moreover, most GRFs can interact with GIFs, and are subjected to post-transcriptional regulation by miR396. This miR396-GRF/GIF module plays pivotal roles in regulating rice growth,development,and environmental response[21-26].It is evolutionarily conserved across seed plants [27]and has been proposed as an excellent target for breeding and biotechnology [21]. Thus, harnessing this module, particularly by manipulating the miR396 target site ofGRFswith CRISPR/Cas technology, holds great potential for crop improvement.
In addition,the broad adoption of hybrid rice cultivars has contributed greatly to world food security. However, the high cost of hybrid seed production is one of the major factors hindering its application, especially as labor expense increases and labor availability decreases in some countries. Development of mechanized seed production methods to cut seed production costs has become necessary for hybrid rice production. One such methods involves the use of small-grain sterile lines combined with large-grain restorer lines. The difference in seed thickness of parents allows mechanized seed production [28]. A disadvantage of this system is that genes controlling small grain are mainly recessive,reducing the yield of hybrids carrying them. Using dominant or semidominant genes conferring large grain size will solve this problem.However, such dominant or semi-dominant genes are scarce in present rice gemplasm.
Here we describe a novel strategy for increasing grain size and yield in rice via CRISPR/Cas9-mediated editing of theGS2gene. A CRISPR/Cas9 construct targeting the miR396 binding site ofGS2was designed and introduced into the cultivated rice X12. Edited lines with deletion of multiples of three base pairs were identified and selected. These would retain the in-frame translation of GRF4 protein but harbor resistance to miR396-mediated inhibition at the mRNA level. The selected edited lines showed increased grain size and yield, and like the naturalGS2alleles, the phenotypes of the edited lines showed a semi-dominant effect, inspiring us to make a cross with a TMS (thermosensitive male sterile) line, X48. The resulting F1progeny also showed increased grain size and grain yield, indicating application potential in hybrid rice.
2. Materials and methods
2.1. Plant materials and growth conditions
Rice X12 is anindicainbred and X48 is anindicathermosensitive male-sterile line. Transgenic plants and T-DNA-free mutant plants derived from transgenic plants were grown in greenhouses with temperature control located in Wuhan, Hubei, China.
2.2. Vector construction and plant transformation
The CRISPR/Cas9 vector was constructed following Liu et al.[29].The vector construct was based on a pCambia1300 backbone,and contained a Cas9 expression cassette (pUbi4::OMIGA::NLS::C as9::NLS::t35S::tNos), a CP4-EPSPS cassette (p35S::OMIGA:: CP4-EPSPS::t35S), and a sgRNA expression cassette (pU3::sgRNA::polyT::tOcs). In the Cas9 cassette, the Ubi4 promoter that droveCas9expression was cloned from sugarcane genomic DNA, and the DNA segment containing an OMEGA translation enhancer followed by a rice codon-optimizedCas9with twoOsHATNLS on each end was synthesized by Genscript Biotech (Nanjing, Jiangsu,China).The CP4-EPSPS cassette containing theAgrobacterium tumefaciensstrain CP4′s 5-enolpyruvylshikimate-3-phosphate synthase(EPSPS)gene and conferring glyphosate resistance was synthesized by Genscript Biotech Corp. (Wuhan, Hubei, China). In the sgRNA expression cassette, the OsU3 promoter, sgRNA and poly T were synthesized by Genscript Biotech and ligated into the vector throughAscI andPmeI sites at each side. The resulting plasmid was sequenced to verify correctness and then transformed into theA.tumefaciensstrain EHA105,by which the plasmid was further introduced into X12 callus. The sequences for the three cassette and primers used in vector construction are shown in the Supplementary materials.
2.3. Identification of GS2 gene-edited plants
Genomic DNA of all transgenic glyphosate-resistant T0plants was examined by PCR using the specific primers CP4-EPSPS-F/CP4-EPSPS-R (Table S1). All PCR-positive plants were then subjected to fragment-length assay.In each assay, a DNA segment including the editing target site and having length <500 bp was amplified using a pair of gene-specific primers: FLD-F (5′-FAM modified) and GS2-R (Table S1). The lengths of the PCR products were estimated by capillary electrophoresis using an Applied Biosystems 3730XL Capillary Sequencer (Thermo Fisher Scientific,Waltham, MA, USA). Finally, homozygous T1mutant lines with desired PCR product length were subjected to DNA sequencing using the PCR amplicons amplified with the gene-specific primer pair GS2-F/GS2-R (Table S1) across the target site.
2.4. Phenotype analysis
Grain width, grain length, chalky grain rate, and chalkiness degree were measured with SC-E type Seed Rice Appearance Quality Inspection Analyzer (Wanshen Co., Ltd., Hangzhou, Zhejiang,China).Grain weight was calculated based on 300 fully filled grains and converted to 1000-grain weight. Fifteen randomly selected grains from each plant were used for measuring grain width and length.
2.5.RNA extraction,cDNA preparation,and quantitative real-time PCR
Total RNA was extracted from leaves,culms,and panicles using the TRIzol reagent (Thermo Fisher Scientific). First-strand cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche, Basle, Switzerland). Quantitative RT-PCR analyses were conducted to amplifyGS2on an Applied Biosystems 7900HT instrument with 2×SYBR Green PCR Master Mix(Thermo Fisher Scientific).OsActin7was used as a control. The expression level ofGS2was calculated as 2-(CtGS2-CtActin7)and shown as the ratio ofGS2vs.OsActin7for relative quantification. The primers are listed in Table S1.
3. Results
3.1. CRISPR/Cas9-mediated editing of GS2 gene
To increaseGS2expression and increase grain size by destroying the miR396-targeted site ofGS2in cultivated rice, a 20-bp nucleotide sequence near the miRNA396 recognition site was chosen as a target for CRISPR/Cas9-mediated cleavage (Fig. 1A). To reduce off-target risk, the editing target sequence was designed to include only a small part of the miR396-binding site, and thus to show low sequence similarity to the miR396-binding sites of other genes. CRISPR-P 2.0 (https://crispr.hzau.edu.cn/CRISPR2/)[29]online analysis found 24 potential off-target sites of this target sequence, among which 11 were located in exons of genes. The directed CRISPR/Cas9 vector was then constructed and introduced into the rice inbred X12.
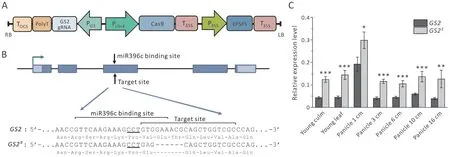
Fig.1. Creation of the GS2 gene editing line.(A)Schematic diagram of the CRISPR/Cas9 vector used in this study.RB,T-DNA right border;LB,T-DNA left border; P35S,CaMV 35S promoter; T35S, CaMV 35S terminator; EPSPS, Agrobacterium tumefaciens strain CP4′s EPSPS (5-enolpyruvylshikimate-3-phosphate synthase) gene as selection marker;PUbi4,sugarcane Ubi4 promoter;PU3,rice U3 snRNA promoter;gRNA,guide RNA;TOCS,octopine synthase gene terminator.(B)Editing strategy and sequence of the edited site.Exons and UTRs are represented by blue and light blue boxes, respectively, and green arrow marks the transcriptional start site. Protospacer adjacent motif (PAM) is underlined,while modified nucleotides are shown in red text and hyphens.(C)GS2 expression in organs of wild type GS2 plants and GS2E mutant plants.Relative expression represents the ratio of GS2 to OsActin1 expression level. Values are shown as mean±SD (n=3). *, P <0.05; **, P <0.01; ***, P <0.001, generated by Student’s t-tests.
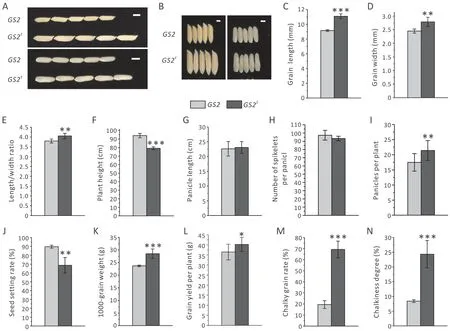
Fig.2. Comparison of agronomic traits between plants with wild type GS2 allele and those with mutant GS2E allele.(A,B)Grain morphology.Scale bar,5 mm(A),and 2 mm(B).(C)Grain length(n=15).(D)Grain width(n=15).(E)Grain length/width ratio(n=15).(F)Plant height(n=15).(G)Panicle length(n=15).(H)Number of spikelets per panicle (n=15). (I) Effective panicles per plant (n=15). (J) Seed setting rate (n=6). (K) 1000-grain weight (n=6). (L) Grain yield per plant (n=15). (M) Chalky grain rate(n=3).(N)Chalkiness degree(n=3).Data in(G,H)were collected randomly from five individual plants of GS2 or GS2E.Values are shown as mean±SD.*,P <0.05;**,P <0.01;***, P <0.001. Student’s t-tests were used to generate P values.
A total of 37 transgene-positive transformants (T0) were obtained and characterized for the editing results. There were 5 biallelic mutations (-6/+1, -11/+1, -14/+1, or -22/+1) and 30 homozygous mutations (+1/+1 or -22/-22) among the positive transgenic plants. The only desirable mutation type obtained was the -6 bp mutation in line 425D-018 (-6/+1). Twelve T-DNA-free homozygotes of 425D-018 (-6/+1) T1plants were characterized by Sanger DNA sequencing, confirming that these plants harbored a 6-bp deletion of the sequence GAAACG and a T-to-A substitution at the directed-editing site (Fig. 1B). This genotype resulted in a deletion of two amino-acid residues(Glu and Thr)and a Val-to-Glu substitution in the GS2/GRF4 protein. The 12 homozygous plants showed similar grain phenotypes, with no visible difference in plant morphology. The new allele is referred to asGS2Ehereafter.The 11 potential off-target sites in gene exons predicted by CRISPR-P 2.0 were checked inGS2Eplants and no mutation was found (data not shown). To test whether the mutation inGS2Ecaused elevated expression ofGS2, quantitative RT-PCR was performed for X12 andGS2Eplants.The results showed that the transcriptional level ofGS2inGS2Eplants increased 1- to 2-fold in all tested organs relative to X12 (Fig. 1C), confirming that the newGS2Eallele caused the higher accumulation ofGS2transcripts.
3.2. Grain characteristics and agronomic traits of GS2E plants
To investigate whether theGS2Eplants gained the advantageous grain characteristics associated with elevatedGS2expression, rice grains were compared betweenGS2Eplants and wild-type (WT)X12 plants grown under greenhouse conditions. A substantial(21.3%) increase in grain length and a moderate (13.8%) increase in grain width ofGS2Eplants in comparison with X12 were found(Fig. 2A-D). These changes also brought about an increased length-width ratio (Fig. 2E), a favorable trait in the market. Thus,the advantageous grain characteristics of theGS2allele were successfully introduced into the cultivated rice X12.In contrast,otherGS2mutant lines obtained by our gene editing but containing homozygous frame-shift mutations all showed reduced grain size(Fig. S1).
To determine whetherGS2Eplants showed higher yield,GS2Eplants and WT plants were scored for agronomic traits.GS2Eplants did not differ from WT plants with respect to heading date (data not shown).Although the height ofGS2Eplants decreased by about 15 cm(15.8%)(Fig.2F),the panicle length and spikelet number per plant were not substantially different betweenGS2Eand WT plants(Fig. 2G, H). Increases in effective panicle number and thousand grain weight (TGW) were observed forGS2Eplants in comparison with WT plants, with respectively 22.5% and 23.5% increases(Fig. 2I, K). However,GS2Eplants suffered a severe reduction of seed setting rate,from 90%(WTGS2)to 69%(GS2E)(Fig.2J).Despite the seed setting rate decrease, increases in effective panicle number and TGW offset the negative effect of low seed setting rate,resulting in a 10.4% increase in grain yield per plant forGS2Erelative to WT plants (Fig. 2L). The chalky grain rate and chalkiness degree ofGS2Egrain rose markedly(Fig.2M,N),presumably owing to the enlarged grain size. Generally, the gain-of-functionGS2Emutant is a newGS2germplasm exhibiting multiple beneficial grain traits, and our novel gene-editing strategy was effective for rice grain improvement.
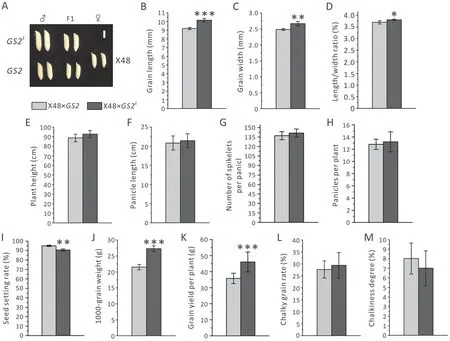
Fig.3. Comparison of agronomic traits between the F1 progenies of two hybrid combinations,X48×GS2E(dark gray columns)and X48×GS2(light gray columns).(A)Grain morphology. Scale bar, 5 mm. (B) Grain length. (C) Grain width. (D) Grain length-width ratio. (E) Plant height. (F) Panicle length. (G) Number of spikelets per panicle. (H)Effective panicle number per plant.(I)Seed setting rate.(J)1000-grain weight.(K)Grain yield per plant.(L)Chalky grain rate.(M)Chalkiness degree.n=15 in(B-H,K);n=6 in (I, J); n=3 in (L, M); Values for (F, (G) were collected from five randomly selected individual plants of GS2 hybrid or GS2E hybrid, and values are shown as mean±SD. *,P <0.05; **, P <0.01; ***, P <0.001. Student’s t-tests were used to generate P values.
3.3. The GS2E allele increased grain size and yield in hybrids
The natural-variation alleleGS2AA, derived from a dinucleotide substitution (TC-AA) in the coding sequence targeted by miR396c,is semi-dominant [16]. To test whether the newGS2Eallele behaved in the same way as the semi-dominantGS2AAallele, we separately outcrossed theGS2Eand X12 (wild-typeGS2) plants as male parent to a TMS (thermosensitive male-sterile) line, X48. As expected, the F1hybrid of X48/GS2Ecombination produced much larger grains with significantly increased length (10.30%) and width (7.30%) relative to the X48/GS2combination (Fig. 3A-C). In agreement with above results,the TGW showed a marked increase(26.80%), and the length-width ratio also increased slightly(Fig.3D,J).Unlike theGS2Eplant in the X12 background,which suffered a severe seed setting rate reduction, its X48/GS2Ehybrid returned to a normal seed setting rate of around 90%, although the X48/GS2hybrid plant still had a slightly higher seed setting rate of about 95%(Fig.3I).TheGS2Eallele greatly increased hybrid grain yield. The grain yield per plant of theGS2Eand X48 hybrid was 28.6% higher than that of wild-typeGS2and the X48 control hybrid (Fig. 3K). These findings indicated that theGS2Eallele was a semi-dominant allele.TheGS2Eand X48 hybrid had plant height,panicle length, effective panicle number per plant, and spikelet number per panicle similar to that of the control hybrid(X48×GS2) (Fig. 3E-H). The chalky grain rate and chalkiness degree also showed no marked difference (Fig. 3L-M). Together,these results suggested that the beneficial traits ofGS2Egermplasm may have great potential in the breeding of hybrid rice.
4. Discussion
Inspired by naturalGS2alleles, which contain a compromised miR396 recognition site and confer increased grain size, in this study we conducted CRISPR/Cas9-mediated genome editing that targeted the miR396 recognition site ofGS2 in anindicainbred X12. A novel semi-dominantGS2allele namedGS2Ethat carried a 6-bp deletion and a 1-bp substitution within its miR396 binding site was identified. The newGS2Eallele similarly increasedGS2expression and grain size and yield, suggesting that the mutation abolished the negative regulation of miR396 without compromising the normal function of the GS2 protein. These indicate that our novel gene-editing strategy is an effective approach for rice grain improvement. Exploiting the semi-dominant effect, thisGS2Eallele could be easily introduced into other cultivated variety through marker-assisted introgression.Given that elevation ofGS2expression is beneficial for multiple agronomic traits including grain size, yield, stress tolerance, and nitrogen usage efficiency,ourGS2gene-editing strategy may have broad application prospects. Using our CRISPR/Cas9 strategy, a similar allele can be directly created in elite restorer lines and elite inbred cultivars for fast improvement. Elite restorer lines carrying a novel semidominantGS2allele can become valuable resources for breeding of hybrid rice suitable for mechanized seed production,which will help alleviate the high-cost problem of current rice hybrid seed production.
The methodology presented in our study may have broader implications. The conserved miR396-GRF/GIF module participates in regulation of plant growth, development, and environment response [21-26], and its manipulation may hold great potential for crop improvement. Applying our strategy to edit GRF genes not only in rice but also in other crops would allow researchers to create new germplasm with higher yield and better resistance to disease and insects. Our gene-editing strategy will facilitate the genome editing of other genes with miRNA target sites but without natural gain-of-function alleles for trait improvement.
CRediT authorship contribution statement
Wenshu Wang:Conceptualization,Data curation,Investigation,Writing - original draft.Weipeng Wang:Formal analysis, Investigation.Yanlin Pan:Formal analysis, Validation.Chao Tan:Methodology, Validation.Hongjing Li:Data curation, Validation.Ya Chen:Investigation.Xingdan Liu:Investigation.Jing Wei:Investigation.Nian Xu:Investigation.Yu Han:Investigation.Han Gu:Investigation.Rongjian Ye:Methodology.Qi Ding:Conceptualization,Project administration,Writing-review&editing.Chonglie Ma:Conceptualization, Funding acquisition, Supervision,Writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
We are grateful to Ms. Li Pan, Ms. Xiaolu Teng, Ms. Zhenjin Ke,and other colleagues who contributed to this work but could not be listed as co-authors, for their invaluable help. We also thank Professor James C Nelson from Kansas State University for his great helps in quality improvement of this manuscript. This work was supported by the National Key Research and Development Program of China (2016YFD0102000) and ‘‘Breeding of Major New Varieties of Main Grain Crops” Program (2020ABA016) from Department of Science and Technology of Hubei Province.
Appendix A. Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2022.01.004.
- The Crop Journal的其它文章
- Brief Guide for Authors
- Revisiting the role of delta-1-pyrroline-5-carboxylate synthetase in drought-tolerant crop breeding
- Influence of seven levels of chemical/biostimulator protection on amino acid profile and yield traits in wheat
- Corrigendum to ‘‘De novo design of future rapeseed crops: Challenges and opportunities” [Crop J. 10 (2022) 587-596]
- Improving the resistance of the rice PTGMS line Feng39S by pyramiding blast, bacterial blight, and brown planthopper resistance genes
- Low N apparent surplus with higher rice yield under long-term fertilizer postponing in the rice-wheat cropping system

