miR398b negatively regulates cotton immune responses to Verticillium dahliae via multiple targets
Yuhun Mio, Kun Chn, Jinwu Dng, Lin Zhng, Wirn Wng, Ji Kong, Stvn J. Klostrmn,Xinlong Zhng, Alifu Airxi,, Longfu Zhu,
a National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan 430070, Hubei, China
b Hubei University of Chinese Medicine, Wuhan 430065, Hubei, China
c Oil Crops Research Institute, Chinese Academy of Agricultural Sciences, Wuhan 430062, Hubei, China
d Institute of Economic Crops, Xinjiang Academy of Agricultural Sciences, Urumchi 842000, Xinjiang, China
e United States Department of Agriculture, Agricultural Research Service, Salinas, CA 93905, USA
Keywords:Cotton miR398b NBS-LRR ROS homeostasis Verticillium dahliae
A B S T R A C T MicroRNAs (miRNAs) play essential roles in plant defense responses, although such roles have not been identified in cotton in response to the plant pathogenic fungus Verticillium dahliae.In this study,the functions of miR398b and its target genes in cotton-V. dahliae interaction were investigated. The transcript levels of miR398b were down-regulated by V. dahliae infection and miR398b overexpression in cotton made the plants more susceptible to V. dahliae. The results suggest that miR398b negatively regulates cotton resistance to V. dahliae. This may occur by miR398b repression of some CC-NBS-LRR genes via translational inhibition,interfering with defense responses and leading to cotton susceptibility to V.dahliae.Alternatively,miR398b may guide the cleavage of the mRNAs of GhCSD1,GhCSD2 and GhCCS,each of which functions in reactive oxygen species(ROS)regulation and homeostasis, thereby causing excessive ROS accumulation in miR398b-overexpressing plants in response to V. dahliae infection. This study suggests conserved and novel roles of miR398b in the cotton-V.dahliae interaction.These discoveries may be coupled with new strategies in cotton breeding programs to improve resistance to V. dahliae.
1. Introduction
Cotton(Gossypiumspp.)cultivation is a major fiber and seed oil crop. Verticillium wilt disease caused by the fungusVerticillium dahliaeis one of the major threats limiting cotton productivity[1]. The symptoms of this disease include wilting, stunting,chlorosis, vascular discoloration, and early senescence. Strains ofV. dahliaeisolated from cotton have been characterized as causing a defoliating or non-defoliating phenotype, and the genetic basis for the defoliating phenotype was recently established [2]. There are few resistant germplasm sources or efficient management measures available to control the pathogen, especially postinfection. It has a broad host range, infecting over 200 plant species, and survives for years in the soil, precluding crop rotation as a strategy for disease control [3].
Plants have evolved two major innate immunity systems in the ongoing ‘‘arms race” with pathogens. One of these systems is known as microbe-associated molecular pattern (MAMP)-triggered immunity (MTI), which serves as the first barrier to the invasion of pathogens [4]. Another is known as effector-triggered immunity (ETI), which is carried out by dominant resistance (R)proteins that recognize pathogen effector proteins directly or indirectly,to trigger gene-for-gene resistance[5].The canonical R proteins contain a nucleotide-binding site (NBS) and leucine-rich repeat (LRR) domains. The core nucleotide-binding domain in NBS-LRR proteins is known as the NB-ARC domain because of its presence in APAF-1(apoptotic protease-activating factor-1),R proteins, and CED-4 (Caenorhabditis elegansdeath-4 protein) [6]. In plants, NBS-LRR proteins mediate pathogen-specific effectortriggered immunity, and the genes that encode them are widely used as markers in plant breeding to generate disease resistance.A drawback ofR-mediated resistance is that it occurs at the expense of fitness [7], which suggests that the expression ofRgenes must be strictly regulated and inactivated in the absence of pathogens.
Reactive oxygen species (ROS) also play multiple roles in MTI and ETI. ROS can function as secondary messengers directly or indirectly to activate the expression of defense-related genes and induce programmed cell death during the hypersensitive response(HR)[8,9].However,excessive ROS impairs many cellular functions by causing oxidative damage to DNA, RNA, proteins, and membranes [10]. In response, plants have evolved many antioxidative systems to eliminate ROS, including enzymatic and nonenzymatic mechanisms. Enzymatic ROS scavenging mechanisms in plants include superoxide dismutase (SOD), glutathione peroxidase(GPX), ascorbate peroxidase (APX), and catalase (CAT). SODs act as the first line of defense against ROS. There are three types of SODs in plants and their names correspond to the metal ligands with which they associate: copper/zinc SOD (Cu/Zn-SOD, also known as CSD), manganese SOD (Mn-SOD) and iron SOD (Fe-SOD)[11].There are three CSD isozymes,which are localized in different cellular compartments inArabidopsis: cytosolic CSD1,chloroplastic CSD2, and peroxisomeic CSD3. InArabidopsis, there is a copper chaperone for superoxide dismutase(CCS,which delivers copper to the CSD) to activate all three CSD isozyme activities[12]. Overexpression ofCSD1andCSD2increases the tolerance of transgenic plants to UV and high light treatment, salt, and heavy metal stresses [13,14]. SODs have also been reported to play roles in HR during cotton-Xanthomonas campestrisinteraction and barley-Blumeria graminisinteraction [15,16].
MicroRNAs (miRNAs) are approximately 21 or 22 nucleotide(nt) long non-coding RNAs that mediate gene silencing by targeting mRNA for cleavage or by translational repression in both plants and animals[17,18].Primary miRNAs(pri-miRNAs)are transcribed by RNA polymerase II but contain an imperfect stem-loop or hairpin structure [19,20]. miRNAs are released from their pre-miRNAs by RNase III-like Dicer-like enzymes in plants, and then associate with Argonaute (AGO) protein to inhibit gene expression at the level of transcriptional gene silencing (TGS) or posttranscriptional gene silencing (PTGS) [21]. Hundreds of miRNAs have been discovered via deep-sequencing and genetic approaches,and they function in plant development [22-25] and responses to biotic and abiotic stresses [29,27,28].
In plant defense,miRNAs act as regulators ofRgene expression.Two miRNAs, nta-miR6019 and nta-miR6020, were reported [29]to guide the cleavage of the tobacco mosaic virus(TMV)resistance gene,N, which is a toll and interleukin-1 receptor-NBS-LRR immune receptor. miR482/2118 is another well-known miRNA that targetsNBS-LRRresistance genes [30]. miR9863 members guide the cleavage ofMla1, encoding a coiled-coil (CC)-NB-LRR receptor that trigger isolate-specific immune responses against powdery mildew in barley [31]. Just recently [32], aBrassicamiRNA, miR1885, was found to target both theRgeneBraTNL1and a photosynthesis-associated geneBraCP24to balance plant growth and defense responses against viral infection.In particular,miRNA-NBS-LRRinteractions can trigger the production of phasiRNAs, which function synergistically with miRNAs in eithercisortransto suppress the expression ofRgenes [33]. Thus, miRNAmediated repression ofNBS-LRRgenes is an important mechanism for plants to balance the tradeoff between growth and defense.The miRNA known specifically as miR398b has been reported [34,35]to act in response to various abiotic stresses by modulating the expression of its target genes. miR398b was the first miRNA reported to be downregulated in response to biotic stress(byP.syringaeinfection) inArabidopsis[36,37]. In our previous work [34],we determined by miRNA and degradome sequencing that miR398b functions under temperature stress . Four targets of miR398b have been identified by computational prediction and sequence analysis:CSD1,CSD2,CCS, andCOX-5b(encoding a subunit of the mitochondrial cytochromecoxidase) inArabidopsis[38,39]. miR398 negatively regulates MTI and resistance to pathogenic bacteria [37]. However, whether miR398 can participate in ETI and resistance to fungal pathogensV. dahliaeis unknown.The objective of the present study was to investigate the role of miR398b in cotton-V. dahliaeinteraction.
2. Material and methods
2.1. Plant materials and V. dahliae proliferation
Seedlings ofGossypium hirsutumcv.YZ1,G.barbadensecv.7124,and transgenic lines derived fromG.hirsutumcv.J668 were grown in a controlled environment chamber under a 16 h light/8 h dark cycle at 25 °C for treatment and sampling.V. dahliaestrain V991 was cultured on potato dextrose agar(PDA)medium for 3-4 days,and hyphae were transferred into Czapek’s medium for 3 days at 25 °C to prepare suspension solutions.
Two-week-old seedlings ofG. barbadensecv.7124 andG. hirsutumcv. YZ1 were infected withV. dahliaeconidial suspensions(1 × 106conidia mL-1) by root dip inoculation, and control plants were mock-inoculated with sterile water. Roots were harvested at 0, 6, 12, 24, and 48 h post-inoculation, frozen in liquid nitrogen,and stored at -80 °C for RNA extraction.
2.2. Plasmid construction and genetic transformation
To study the expression patterns of miR398b, a 1233 bp promoter region of miR398b was cloned into the pGWB433 vector and used to drive expression of theGUSreporter gene inArabidopsis. To localize CSD-family proteins, cDNA sequences lacking stop codons corresponding toGhCSD1,GhCSD2,GhCCSandGhCSD3were inserted into the N-terminal GFP-fusion expression vector PMDC84[40] by Gateway Cloning (Invitrogen Thermofisher, Shanghai,China). To express the VdNLP1:GFP protein, the cDNA sequence ofVdNLP1lacking the stop codon was inserted into the NterminalGFP-fusion expression vector PMDC84 by Gateway Cloning. To examine whether miR398b represses the expression of theNBS-LRRgene, the 35S promoter with duplicated enhancers was cloned into the reporter vector pGreenII 0800-LUC to generate the vector pGreenII 0800-35S:LUC. A 100-bp fragment ofGh_A04G0380andGh_D05G3257containing the predicted binding sites was amplified from cotton genomic DNA and ligated into pGreenII 0800-35S:LUCat theNotI site. The 21-bp target sites ofGh_D05G3257(5NBts) and the mutated target sites ofGh_D05G3257(5NBmts) were amplified using the primers 5NBts-F/R and 5NBmts-F/R, respectively, as described previously [41].The isolated fragments were ligated into the N-terminal GFPfusion expression vectorpMDC84-35S:GFPat theKpnI site. For Virus-induced gene silencing(VIGS)vector construction,fragments ofGhCSD1,GhCSD2,GhCCS, andNBS-LRRwere amplified from root samples ofG. hirsutum. PCR products were digested with two restriction endonucleases,BamHI andKpnI, and ligated to the TRV vector as reported previously [42]. To study the function of miR398b,a 313 bp genomic sequence containing the miR398b precursor was cloned and ligated into a pK2GW7.0 vector to overexpress the miR398b precursor. The miR157 overexpression vector was generated previously [43]. A miR398b-resistant version ofGhCSD2(rGhCSD2) was generated by introducing six mutations into the target sites ofGhCSD2through recombinant PCR and cloned into the vector pK2GW7.0. The primer sequences used in vector construction are listed in Table S1. All vectors were transferred toAgrobacterium tumefaciens(strain GV3101).
For genetic transformation,A. tumefaciensGV3101-mediated transformation was used to transform hypocotyl sections ofG.hirsutumcv.J668 as previously described[44].Null plants containing no transgenes were separated from self-pollinated35S:rCSD2hemizygotes TN and self-pollinated35S:miR398bhemizygotes (ON).
2.3. Histochemical assay
Ten-day-old transgenicArabidopsisplants were inoculated withV. dahliaeconidial suspension (1 × 106conidia mL-1), 2 μmol L-1peptides of either flg22 or nlp20, for 12 h. Peptides of flg22 and nlp20 were synthesized in GenScript(Nanjing,China).Plants were incubated in the GUS staining solution at 37°C for 6 h,followed by decolorization in 75% alcohol and then photographed under a stereoscopic microscope (MZFLIII, Leica, Wetzlar, Germany) following Sun et al. [45].
2.4. Reverse transcription quantitative-PCR (RT-qPCR) analysis
Stem-loop RT-PCR was used to quantify miRNAs and target genes in a previous study [43]. Briefly, 3 μg RNA was reversetranscribed to cDNA using stemloop primers and SuperScript III RT (Invitrogen). The reaction procedure was as follows: 16 °C for 30 min; 60 cycles of 30 °C for 30 s, 42 °C for 30 s; and then 70 °C for 5 min to inactivate the reverse transcriptase.
RT-qPCR was performed using the ABI Prism 7500 system(Applied Biosystems, Foster City, CA, USA) following the manufacturer’s protocol. The thermocycling program was as follows:95°C for 1 min; 40 cycles of 95 °C for 10 s, 60 °C for 40 s.
2.5. RLM-RACE analysis
The 5′RACE procedures were performed with a GeneRacer Kit(Invitrogen, https://www.lifetechnologies.com). Five micrograms of total RNA from an equal mixture ofG. hirsutumV991-inoculated samples andG. barbadenseV991-inoculated samples were ligated to an RNA adapter and reverse-transcribed into cDNA using the GeneRacer Oligo-dT primer. The 5′adaptor primers and 3′gene-specific primers (Table S1) were used to amplify cDNA ends according to the manufacturer’s instructions.
2.6. Luciferase reporter, green fluorescence protein (GFP) reporter assay and subcellular localization analysis
For Luciferase reporter assay, 100 μL ofAgrobacteriumstrain GV3101 containing thepGreenII 0800-target site-LUCplasmid were combined with 900 μL ofAgrobacteriumstrain GV3101 containing either the35S:primiR398b or35S:primiR157plasmid. The mixture was incubated(28°C,200 r min-1,shaker)to a final concentration of OD600=1.0,and resuspended in infiltration buffer(10 mmol L-1MES,pH 5.6,10 mmol L-1MgCl2and 150 μmol L-1acetosyringone)as described previously [46]. The same volume of mixed suspensions was injected into leaves ofNicotiana benthamianaby agroinfiltration. After 48 h, LUC luminescence was examined using a cryogenically cooled CCD camera as described previously [47].LUC and RLUC activities were measured as counts per second using a multimode plate reader (Perkin Elmer) using the Promega kit(N1630).
For GFP reporter assay,A.tumefaciensstrain GV3101 containing one of the binary vectors (35S:5NBts-GFPor35S:5NBmts-GFP,35S:miR398b,35S:miR157) was cultured in LB medium overnight at 28 °C. TheA. tumefacienssamples with different vectors were collected and mixed at the indicated optical density (O.D.) using a DU800 spectrophotometer (Beckman Coulter). TheA. tumefaciensharboring different vectors were infiltrated into tobacco leaves and the transfected cells were imaged by confocal laser scanning microscopy(Olympus FV1200)60 h after inoculation.For Western blot analysis,100 mg of infiltrated leaf tissues was ground into fine powder in liquid nitrogen and homogenized in 400 μL of tobacco protein extraction buffer (25 mmol L-1Tris-HCl, pH = 7.5;150 mmol L-1NaCl;1 mmol L-1EDTA;0.5%Triton X-100;5%glycerol;1%protein inhibitor;2 mmol L-1phenylmethanesulfonyl fluoride). The same amount of extracted total protein was separated on a 15%SDS-PAGE gel and transferred to a polyvinylidene fluoride membrane (Millipore). A 10,000-fold-diluted Rabbit Anti-GFP sera(Ab290, Abcam) and Secondary-Goat Anti-Rabbit IgG H&L (HRP)(Ab205718, Abcam) were used to detect GFP accumulation and the total protein was stained with Coomassie brilliant blue(R250) for a loading control.
For subcellular localization,Agrobacterium tumefaciensstrain GV3101 containing binary vectors with the GFP fusion protein were infiltrated into tobacco leaf epidermal cells for transient expression analysis,and the transfected cells were imaged by confocal laser scanning microscopy at 60 h after inoculation,following Gao et al.[48].The GFP was excited at a wavelength of 488 nm,and fluorescence emission was detected at 505-530 nm. Chlorophyll autofluorescence was excited at a wavelength of 488 nm and detected at 647-745 nm.
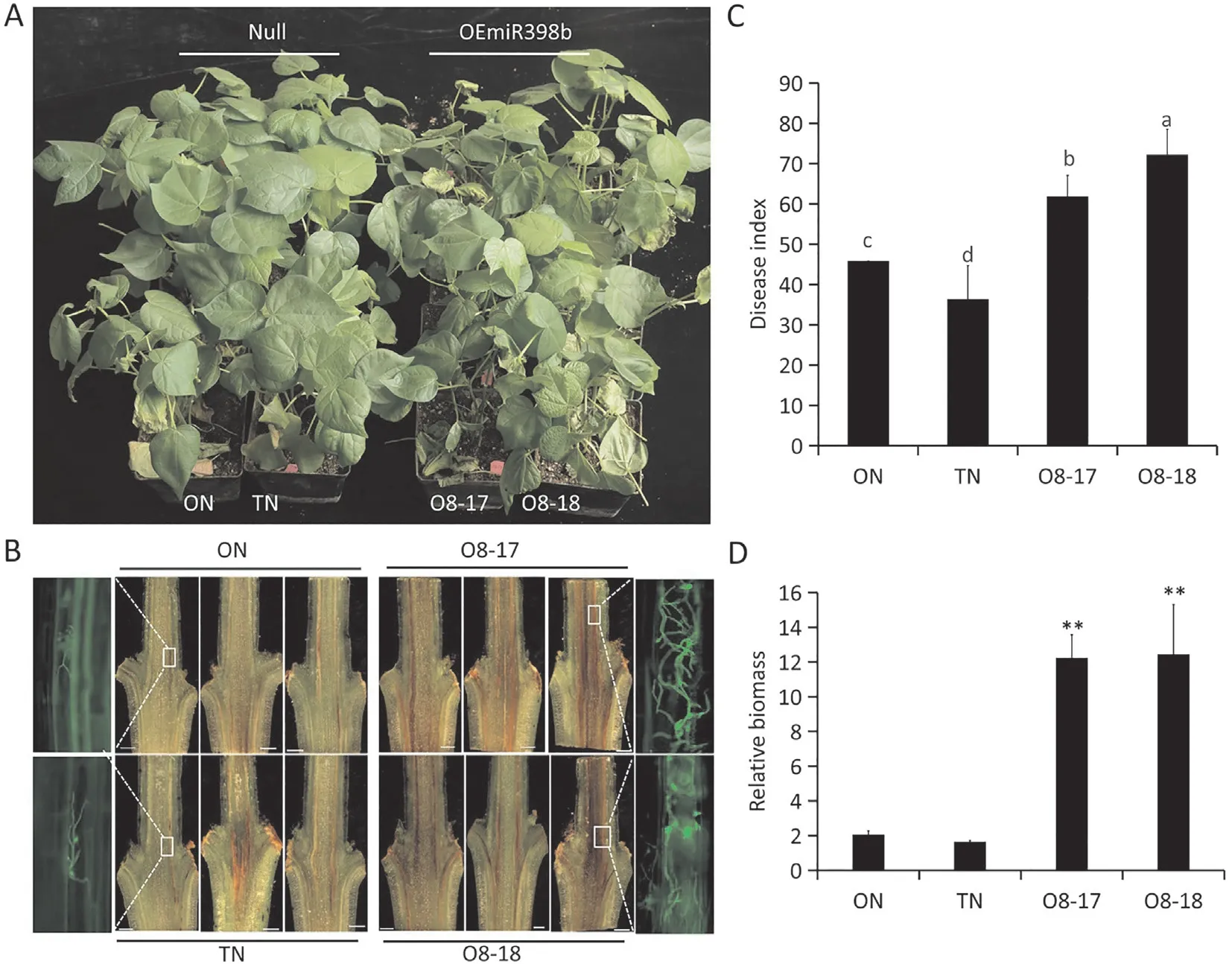
Fig.2. Overexpression of miR398b attenuates cotton resistance to Verticillium dahliae.(A)Verticillium wilt disease symptoms of null cotton plants(ON and TN)and miR398boverexpressing lines(O8-17 and O8-18)at 16 days post-inoculation with V.dahliae.(B)Dark,necrotic vascular bundles of the dissected stems of each line were photographed 16 days after V. dahliae inoculation. Fluorescence imaging shows V592-GFP hyphae in the vascular bundles of miR398b-overexpressing lines. (C) Disease index statistics of null plants(ON and TN)and miR398b-overexpressing lines(O8-17 and O8-18)at 16 d after inoculation.The bar represents the standard error from three biological repeats and different letters above columns indicate significant differences at P <0.05 (Duncan’s multiple range test). (D) PCR quantification of fungal biomass. The biomass represents the levels of amplification of the Verticillium internal transcribed spacer(ITS)region compared to the levels of cotton GhUB7 amplification.Values are means±SD(**, P <0.01, Student’s t-test).
2.7. Virus-induced gene silencing and pathogen inoculation
Agrobacterium tumefacienscells harboring TRV vectors were infiltrated into two fully expanded cotyledons of 10-day-old seedlings as described previously [42]. The primers listed in Table S1 were used to evaluate the silencing effect based on the expression levels of the target genes in leaf and root tissues.
For pathogen inoculation,3-week-old cotton seedlings of transgenic and VIGS plants (at least 30 plants for each lines) were infected withV. dahliae(V991) conidial suspension (1 × 105conidia mL-1)by root dip inoculation,and the plants were transplanted into sterilized soil for observation of disease symptoms. A fungal recovery assay and a disease index analysis were performed as described previously [49]. For fungal colonization and biomass detection, longitudinal cross sections of cotyledonary nodes were dissected at 16 days after inoculation and observed under the stereoscopic microscope. Infected stem samples were separately collected for DNA extraction and taken approximately 1 cm above the cotyledon node (CN) and first leaf node (FN). Each experiment was repeated three times.
2.8. Expression of VdNLP1 in transgenic cotton plants and ROS detection
Agrobacterium tumefaciensstrain GV3101 containing the binary vector PMDC84 encoding the VdNLP1-GFP fusion protein, or the empty vector, was infiltrated into two fully expanded cotyledons of 10-day-old transgenic and wild-type cotton plants. Lesion formation or wilting symptoms were photographed at 48 h postinfiltration.
Quantification of H2O2in leaves of NLP1-treated plants was performed with a commercial H2O2detection kit (Sangon Biotech,Shanghai, China).
3. Results
3.1. Overexpression of miR398b impairs Verticillium wilt resistance in cotton
miR398b was expressed in all organs tested, with highest expression in the root and anther (Fig. 1A). To further verify the expression pattern of miR398b, a 1233-bp upstream fragment of MiR398b was introduced into a construct upstream of the βglucuronidase (GUS) reporter gene [50] and transformed intoArabidopsisfor GUS expression analyses. The GUS staining pattern revealed that expression of miR398b was suppressed in the roots and cotyledons when pmiR398B::GUS transgenicArabidopsisplants were treated with flg22, nlp20, andV. dahliae(Fig. 1B).The expression of miR398b was downregulated uponV. dahlaieinfection in these tissues.

Fig. 1. Expression patterns of cotton miR398b in different tissues and in defense responses. (A) Reverse transcription-quantitative PCR analysis for expression of miR398b in tissues.R,root;S,stem;L,leaf;P,petal;A,anther;0 d,0 d ovule;5 d,5 d fiber after flowering. The expression levels of miR398b were normalized to GhUB7.The bar represents the standard error from three biological repeats and different letters above columns indicate significant differences at P <0.05(Duncan’s multiple range test). (B) Histochemical β-glucuronidase (GUS) staining analysis of the expression patterns of pmiR398b::GUS in two independent lines of transgenic Arabidopsis leaves and roots after subjected to 12 h for flg22,nlp20,and Verticillium dahliae treatments. Scale bars, 200 μm.
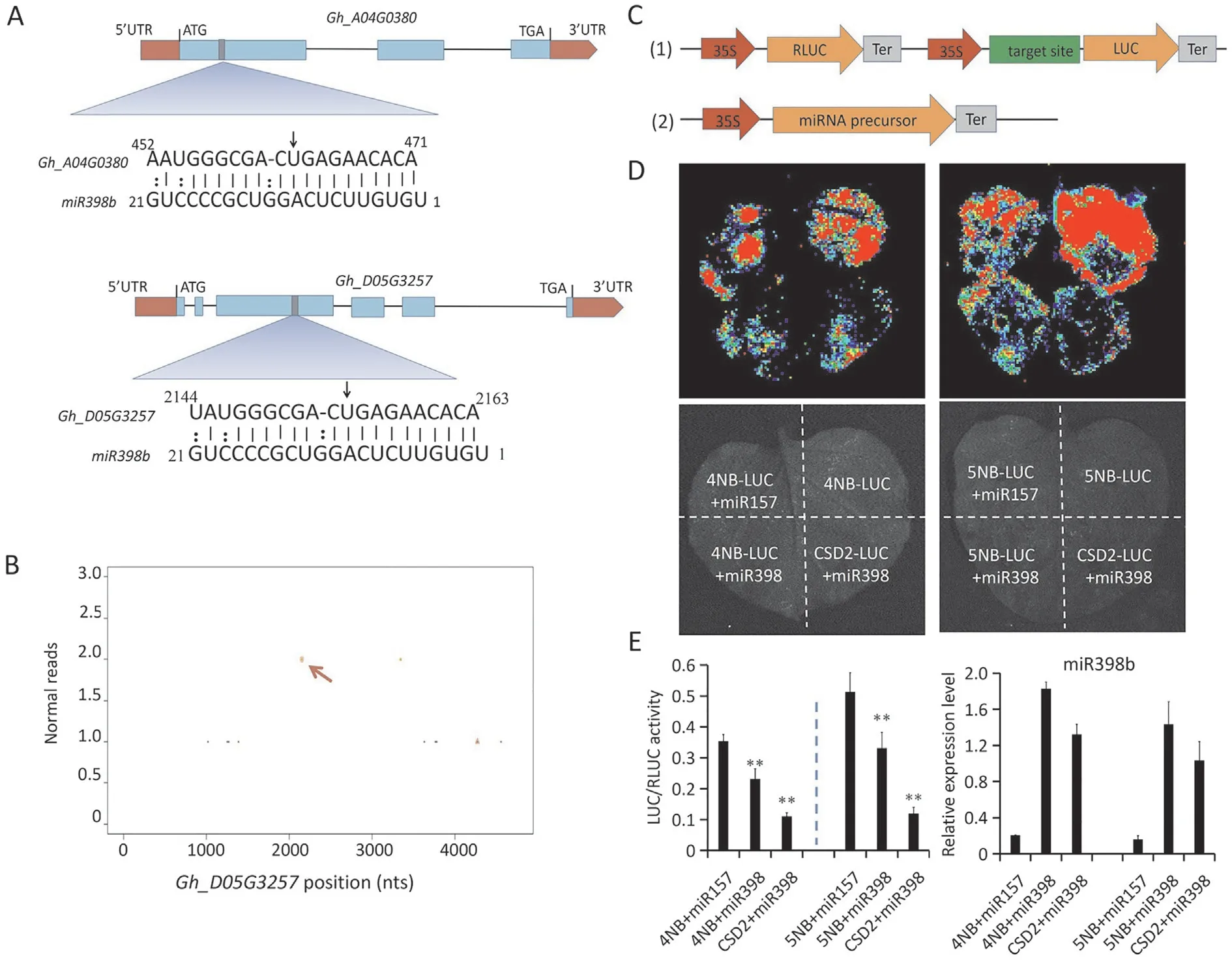
Fig. 3. The cotton miR398b inhibits the expression of NBS-LRR genes. (A and B) Schematic diagram of 4NB and 5NB target sites of miR398b validated by degradome sequencing.(C)Schematics of dual-luciferase system containing 4NB,5NB target sites in the 5′UTR region of the luciferase(LUC)gene.(D)Inhibition of the expression of NBSLRR genes by miR398b. Agrobacterium-mediated transient expression of the 4NB-LUC and 5NB-LUC fusion proteins with miR398b and miR157 in tobacco leaf cells with Agrobacterium tumefaciens cultures grown to the same optical density. After 48 h, LUC luminescence was examined using a cryogenically cooled CCD camera (Lumazome PyLoN 2048B) (E) Quantification of LUC/RLUC intensity and miR398b mRNA levels. Values are means ± SD (*, P ≤0.05; **, P ≤0.01, Student’s t-test).
Two miR398b-overexpression lines with single copy insertion(O8-17, O8-18) and two null lines (ON and TN) were obtained(Fig.S1A,B).Expression analysis showed that miR398b was highly expressed in miR398b-overexpressing plants (Fig. S1C). To test whether the expression level of miR398b affected the resistance of cotton toV. dahliae, the transgenic lines were inoculated withV.dahliaeisolate V592. Compared to the two null lines, the miR398b-overexpressing plants were more susceptible toV. dahliae,with obvious wilt symptoms and a higher disease index than the null plants (Fig. 2A-C). These observations were confirmed by quantification of the DNA levels ofV. dahliaein the tissues,where the fungal biomass was seven-fold higher in miR398boverexpressing plants than in null plants(Fig.2D).The results indicated that miR398b plays a negative role in cotton resistance toV.dahliae.
3.2. NBS-LRR genes are targets of miR398b and involved in cotton resistance to V. dahliae
In our previous degradome sequencing data [34], twoNBS-LRRgenes (Gh_A04G0380(4NB) andGh_D05G3257(5NB)) were identified as putative targets of miR398b.4NBand5NBare homologous genes with high sequence similarity.Ghir_D05G34520andGhir_A04G004640(the corresponding genes ofGh_D05G3257andGh_A04G0380in 3rd-generation genome data of cotton) contain a CC-NBS-ARC domain and two internal repeat regions.The miR398b target site is localized in the internal repeat region on the third exon ofGhir_D05G34520(Figs. 3A, S2A, B).5NBtranscripts were highly expressed in roots, hypocotyls, and stems, tissues that are heavily colonized byV.dahliae.Expression of5NBwas significantly upregulated uponV.dahliaeinfection(Fig.S3A, B).Two reads containing the cleavage site were detected in our normalized degradome data (Fig. 3B). The plant small-RNA target analysis server psRNATarget [51] was used to identify the interaction between miR398b and each of theGh_D05G3257andGh_A04G0380genes.The result predicted that miR398b represses the expression ofNBS-LRRgenes by translational inhibition(Table S2), which makes it difficult to verify cleavage by 5′random amplification of cDNA ends (RACE).
To confirm the repression activity of miR398b onNBS-LRRgenes,a luciferase reporter system was employed with target sites about 100 bp(4NBts and 5NBts)inserted into the 5′UTR region of the luciferase (LUC) -encoding gene, where the recognized target site of CSD2(CSD2ts)was used as a positive control.A miRNA precursor sequence was cloned into another vector driven by the CaMV35S promoter (Fig. 3C). The35S:CSD2ts-LUCconstruct was co-expressed with35S:MiR398b, and the35S:4NBts-LUCand35S:5NBts-LUCconstructs were coexpressed with either the35S:MiR398bor the35S:MiR157vector byAgrobacterium-mediated transient transformation in tobacco leaves. The luminescence intensities of 4NBts-LUC, 5NBts-LUC and CSD2ts-LUC were lower when coexpressed with miR398b than when expressed alone or coexpressed with miR157 (Fig. 3D). The LUC activity and control renilla luciferase (RLUC) activity were also detected (Fig. 3E) and their ratios were consistent with the luminescence intensities shown in Fig. 3D.
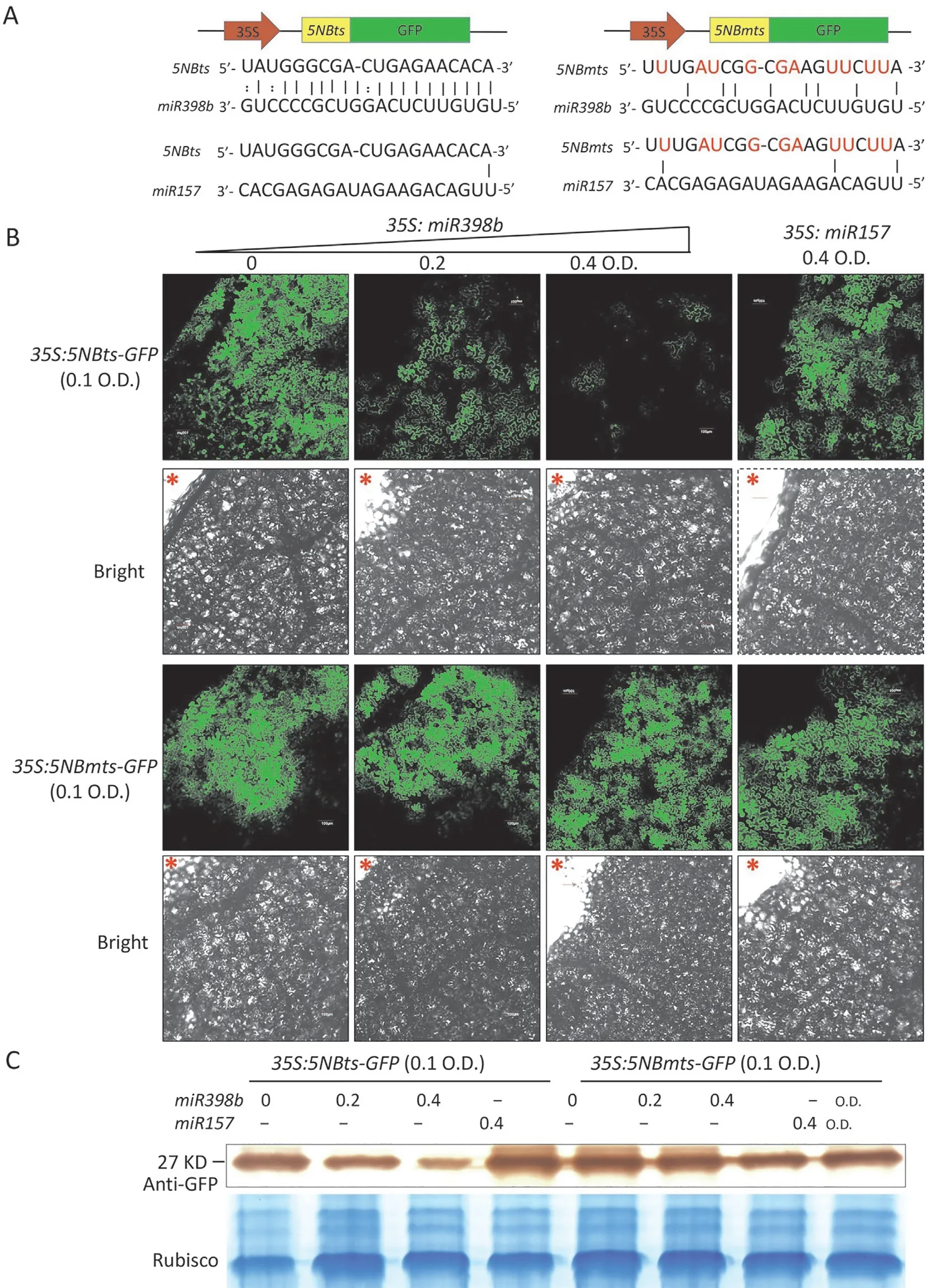
Fig.4. miR398b suppresses the expression of NBS-LRR gene in cotton.(A)Schematics of 5NBts(5NB target sites)-green fluorescent protein(GFP)and 5NBmts(5NB-mutated target sites)-GFP fusion proteins and alignment of the sequences of 5NBts and 5NBmts with miR398b and miR157,respectively.Bases in red indicate the mutated nucleotides.(B) GFP fluorescence shows that miR398b represses the expression of 5NB. Agrobacterium tumefaciens-mediated transient expression of 5NBts-GFP and 5NBmts-GFP fusion proteins with miR398b and miR157 in tobacco leaf cells at the indicated optical densities(O.D.).Laser confocal microscopy was used to observe the green fluorescence 60 h after infiltration by A.tumefaciens.*indicates the injection site,Scale bars,100 μm.(C)Western blot analysis with an anti-GFP antibody reveals the expression levels of 5NBts-GFP and 5NBmts-GFP proteins following each treatment. Total protein was stained with Coomassie brilliant blue as a loading control.
The 21 bp target sites of5NB(5NBts) and the mutated target sites (mts) of5NB(5NBmts) were fused separately at the 5′-terminus ofGFP(Fig. 4A). The35S:5NBts-GFPand35S:5NBmts-GFPconstructs were coexpressed with either the35S:MiR398bor the35S:MiR157vector byAgrobacterium-mediated transient transformation in tobacco leaves. The fluorescence intensity and protein content of 5NBts-GFP were clearly lower when coexpressed with miR398b than when expressed alone or coexpressed with miR157. However, 5NBmts-GFP intensity and protein levels were not influenced significantly when35S:5NBmts-GFPvector was coexpressed with miR398b or miR157 (Fig. 4B, C). Thus, these twoNBS-LRRgenes could be inhibited by miR398b.
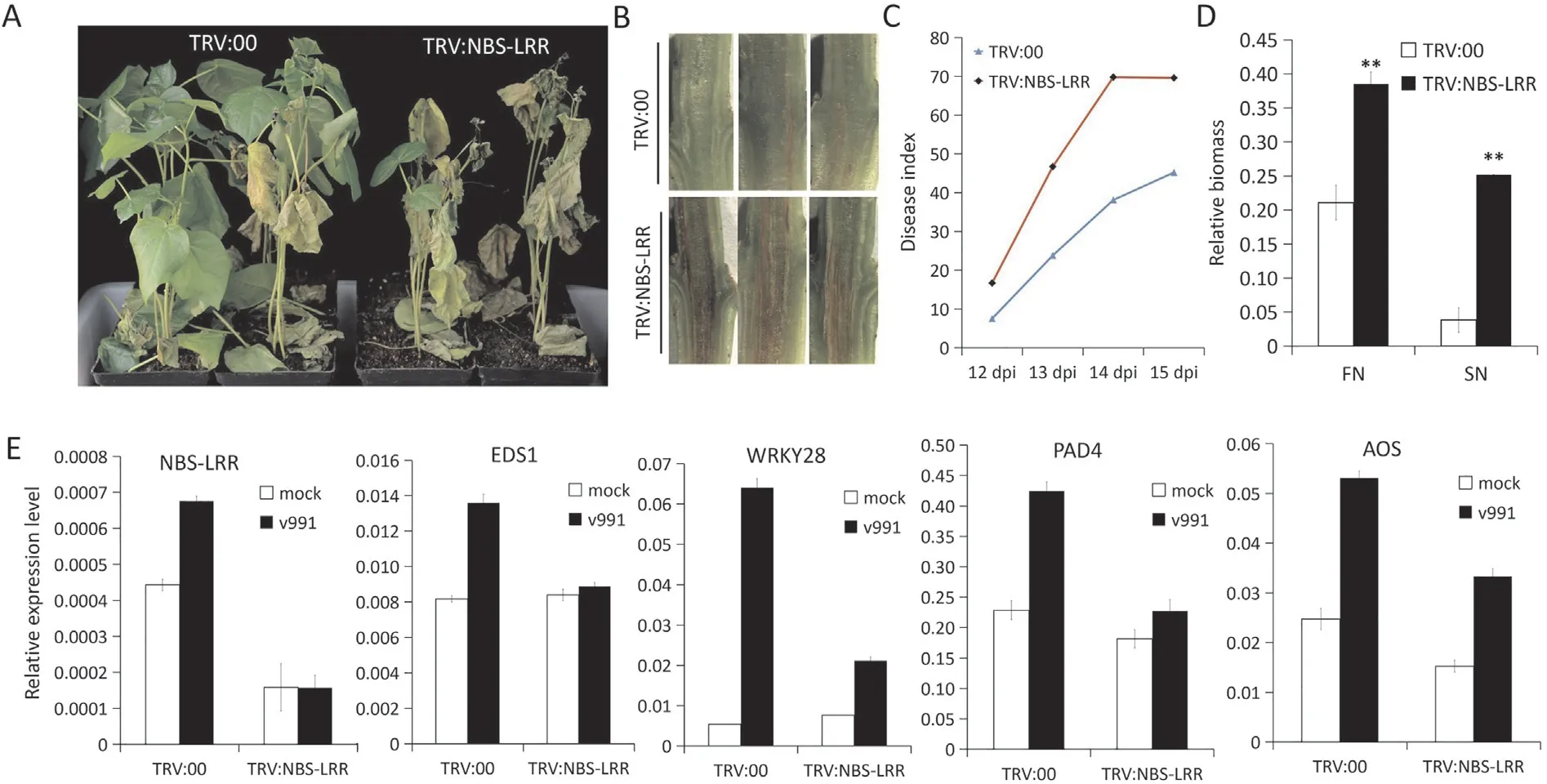
Fig.5. Suppression of cotton NBS-LRR genes increased susceptibility to Verticillim dahliae.(A)Disease symptoms of the TRV:00 and TRV:NBS-LRR plants following inoculation with isolate V991 of V. dahliae. The images were obtained at 16 days post-inoculation (dpi). (B) Dark and necrotic vascular bundles of the dissected stems from TRV:00 and TRV:NBS-LRR plants at 16 dpi with V.dahliae.(C)Disease index statistics of TRV:00,TRV:NBS-LRR plants from 12 d to 15 dpi with V.dahliae.(D)Fungal biomass determined by quantitative PCR in TRV:00 and TRV:NBS-LRR cotton plants.The relative biomass is represented by the DNA quantification levels of Verticillium internal transcribed spacer(ITS)compared to those of cotton UB7. CN, samples from the cotyledonary nodes of cotton stems; FN, samples from the first internode. Values are means ± SD (**, P ≤0.01,Student’s t-test). (E) Suppression of cotton NBS-LRR genes attenuated the expression of defense-related genes upon V. dahliae infection.
To further investigate the function of the twoNBS-LRRgenes(4NBand5NB)in cotton response toV.dahliae,VIGS was employed to knock down the expression levels of both genes. Knockdown ofNBS-LRRgenes made the plants more susceptible toV. dahliae(Fig. 5A).TRV:NBS-LRRplants showed severely necrotic vascular bundles with higher disease indices than those ofTRV:00plants(Fig. 5B, C). Also, more fungal biomass could be detected inTRV:NBS-LRRplants inoculated withV. dahliae(Fig. 5D). We examined the expression patterns of some defense-associated genes inTRV:NBS-LRRplants upon infection byV. dahliae. Expression of genesGhEDS1,GhPAD4,GhWRKY28(genes involved in the salicylic acid synthesis and signaling pathway) [52] andGhAOS(jasmonate acid synthesis-associated gene)[53]was reduced inTRV:NBS-LRRplants in comparison withTRV:00plants duringV. dahliaeinfection(Fig. 5E). Thus, knockdown ofNBS-LRRgenes attenuated the defense response and impaired cotton resistance toV. dahliae.
3.3. miR398b targeted GhCSD genes and regulated ROS homeostasis
Previous degradome sequencing data [34] and other findings[14,38] suggested that Cu/Zn SOD family membersCSD1,CSD2andCCSwere also target genes of miR398b.To determine whether these family members were similarly regulated in cotton, 5′-RNA ligase-mediated rapid RACE(5′-RLM-RACE)experiments were performed,revealing thatGhCSD1,GhCSD2,andGhCCSare also targets of miR398b and that their respective target sites are at the 5′UTR region ofGhCSD1, the fourth exon ofGhCSD2, and the last exon ofGhCCS(Fig. 6A-C).
To further investigate the function of miR398b and theGhCSDsin cotton uponV. dahliaeinfection, the constructs of a miR398bresistantGhCSD2(GhrCSD2)-overexpression vector, with six synonymous codon mutations at the miR398b-target sites were prepared (35S:GhrCSD2) and transformed into cotton (Fig. 6D).TwoGhrCSD2overexpression lines (T8-14 and T8-15) (Fig. S1A,B) were obtained. Transcript levels ofGhCSD1,GhCSD2andGhCCSwere decreased with no difference inGhCSD3andGhCOX-5bexpression in miR398b-overexpressing lines (Fig. 6E, F). The transcripts ofGhCSD2were significantly increased inGhrCSD2-overexpressing lines (Fig. 6E). Expression levels ofGhCSD1,GhCSD2andGhCCS,the target genes of miR398b,were significantly upregulated during pathogen infection compared to the mock treatments(Fig. S4A). However, transcript levels ofGhCSD3, the non-target gene of miR398b, were decreased afterV. dahliaeinoculation(Fig.S4A).Downregulation ofGhCSD1,GhCSD2,andGhCCSby VIGS rendered the plants more susceptible toV. dahliae(Fig. S4B-D).These results indicate that the members of the CSD family,GhCSD1,GhCSD2andGhCCSare required to mount appropriate defense responses in cotton following challenge withV. dahliae.
To investigate the functions of miR398b andGhCSDfamily genes in cotton resistance, we first examined the subcellular location of the CSD family proteins.GhCSD2,GhCSD3,and GhCCS were localized in the chloroplast, while GhCSD1 showed a localization pattern similar to that of free GFP protein in the cell(Fig.S5).These results demonstrate that the CSD family members localize in different cellular compartments to regulate ROS homeostasis.
To examine whether redox homeostasis was controlled by miR398b and itsGhCSDtargets in response toV. dahliae, we measured the hydrogen peroxide (H2O2) content in cotton roots after Necrosis and ethylene-inducing peptide 1 (Nep1)-like protein 1(NLP1) treatment.NLP1 is considered a conserved virulence factor inV.dahliaethat functions as a cytolytic toxin.This protein induces plasma membrane leakage, leading to cytotoxicity and necrosis[54,55]. Furthermore, the production of ROS triggered by NLPs induced the onset of lesion formation [56]. To validate the role of ROS in cotton response toV. dahliae,VdNLP1was transiently expressed in the cotyledons of miR398b- andGhrCSD2-transgenic plants. Necrosis could be observed at the infiltration position 48 h post infiltration (Fig. 7A). Thus, overexpression ofGhrCSD2attenuated necrosis in the presence of NLP1,while overexpression of miR398b resulted in accelerated and increased necrosis.Overexpression of miR398b produced more ROS after treatment with NLP1 (Fig. 7B). Together, these results demonstrate that miR398b plays an essential role in regulating ROS homeostasis by targeting CSD gene family members, family genes, and overexpression of miR398b impairs the ability of ROS scavenging to the benefit ofV. dahliaein the interaction.
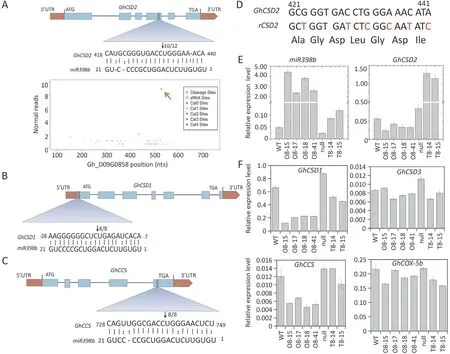
Fig. 6. miR398b can guide the cleavage of GhCSD1, GhCSD2 and GhCCS in cotton. (A) Schematic diagram of GhCSD2 target site for miR398b, validated by degradome sequencing and RNA ligase-mediated rapid random amplification of cDNA ends (RLM-RACE). Watson-Crick pairing (vertical dashes) and non-Watson-Crick pairing (colon)are indicated. The number of miR398b cleavage products is indicated by bold font and arrow. (B and C) Schematic diagram of GhCSD1 (B) and GhCCS (C) target sites of miR398b determined by RLM-RACE experiments. (D) Schematic representation of miR398b-resistant GhCSD2 (rCSD2) vector constructed by synonymous mutation. The top strand depicts the original miR398b target site of GhCSD2 from 421 to 441 nt calculated from the start codon,and the bottom strand shows the sequence post-mutation;bases in red indicate nucleotides that were replaced. (E and F) Reverse transcription-quantitative PCR analysis of the expression of miR398b, the target genes GhCSD2, GhCSD1,GhCCS, and the non-target genes GhCSD3 and GhCOX-5b in roots of wild type, null, miR398b-overexpressing (O8-15, O8-17, O8-18, O8-41) and miR398b-resistant GhCSD2 overexpressing (T8-14, T8-15) lines. Bars represents standard error of the mean of three biological repeats.
4. Discussion
Many researchers have employed miRNA sequencing to investigate the transcriptional profiles in the early (12-48 h) and late(7-10 d) stages of cotton responsive toV. dahliae[57-59]. Some miRNAs have been identified and shown to play important roles during the interaction of cotton withV.dahliae.miR482 expression was down-regulated when cotton was challenged withV. dahliae,while the expression of its target genes was up-regulated to increase cotton resistance toV. dahliae[60]. In contrast, ghrmiR477 was up-regulated when cotton was challenged withV.dahliae, leading to the low expression of its target geneGhCBP60A,and increased cotton resistance toV. dahliae[58]. miR166 and miR159 were increased and transported into the hyphae of the fungus fortrans-kingdom RNA silencing to reduce fungal virulence when cotton was infected byV.dahliae[61].In this study,we found that the expression of miR398b was down-regulated(Fig.1B),and the expression of the targets includingNBS-LRR(Gh_A04G0380andGh_D05G3257) andGhCSDs(GhCSD1,GhCSD2,GhCCS) were all significantly up-regulated during cotton uponV. dahliaeinfection(Figs. S3A, B and S4A).All these results indicate that miRNAs and their target modules play essential roles in the interaction between cotton andV. dahliae.
In the present study, we demonstrated that overexpressing miR398b made plants more susceptible toV. dahliaein cotton(Fig. 2). In support of these findings, miR398 has been reported to negatively regulate MAMP-triggered immunity [37] and the expression levels of miR398 were downregulated upon inoculation of plants with incompatible strains DC3000 (avrRpm1) and DC3000 (avrRpt2), but not the compatible strain DC3000 [36].These findings implicate the involvement of miR398 in the regulation of both MTI and ETI.In our study,miR398b targeted twoNBSLRRgenes (Gh_D05G3257andGh_A04G0380) and the target site of the genes is in the internal region, a region not completely conserved inNBS-LRRgenes.No identical target sequences were found in homologous genes inOryza sativa,Arabidopsis thaliana,Zea maize,Populus trichocarpa,orTheobroma cacao.Thus,Gh_D05G3257
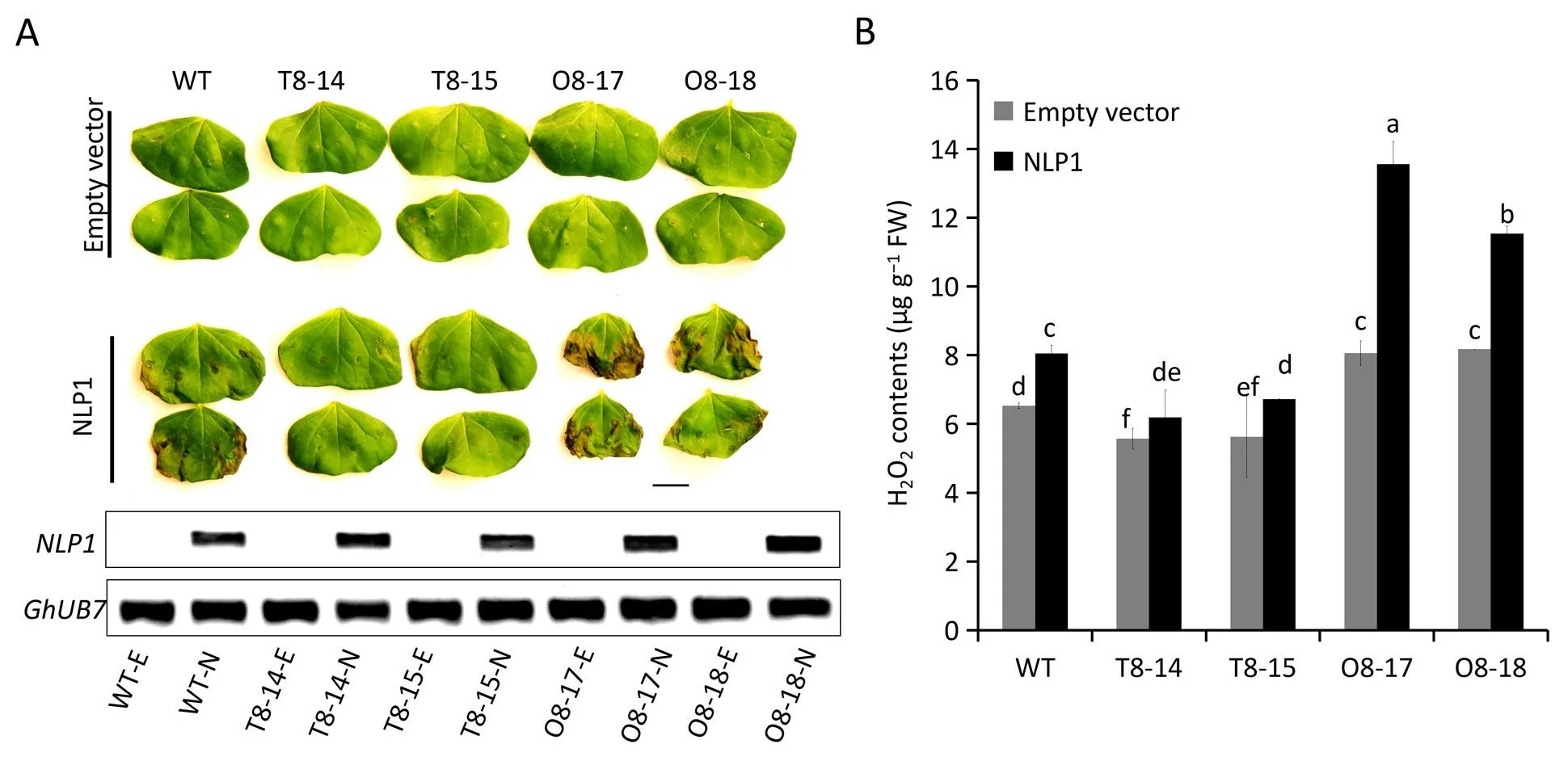
Fig. 7. Overexpression of miR398b in cotton results in constitutive reactive oxygen species (ROS) accumulation and defective ROS elimination in response to Verticillium dahliae.(A)Necrotic lesions on treated cotyledons of wild type and transgenic plants.Images were obtained 48 h post leaf infiltration of the necrosis and ethylene-inducing peptide 1-like protein 1 (NLP1) expressed via Agrobacterium tumefaciens-mediated plant transformation. The gel picture shows the expression analysis of NLP1 in cotton cotyledons by RT-PCR(E,empty vector;N,NLP1).(B)ROS quantification in NLP1-treated cotyledons of wild type and transgenic cotton.Bars represent the standard error of the mean of three biological repeats and different letters above columns indicate significant differences at P <0.05 (Duncan’s multiple range test).
andGh_A04G0380may be unique target genes of miR398b in cotton and contribute to cotton resistance againstV.dahliae(Figs.3,4 and 5;Table S2).It has been said that there are two modes of negative regulation of miRNA on target exist: mRNA cleavage and translational repression. Brodersen et al. [62] showed that some miRNA-target pairs (e.g., miR398-CSD1/CSD2, miR171-SCL6,miR156-SPL3) engage in both slicing and translational inhibition regulatory mechanisms. Other studies [63,64] also showed that the degree of miRNA-mRNA complementarity is a key determinant of the repression mechanism used, such that perfect complementarity enables cleavage, whereas central mismatches promote translational repression. We detected both cleavage incidence of miR398b onNBS-LRRgenes from degradome sequencing and translation inhibition from psRNRTarget analysis and the luciferase(Fig. 3) and GFP (Fig. 4) reporter systems. Considering the central mismatch at theNBS-LRRtarget site, we hypothesized that miR398b repressesNBS-LRRgenes predominantly at the translation level, as only two cleavage site reads were detected in degradome data and cleavage failed to be verified by RACE. In any case,to our knowledge, this is the first report that miR398b can target plantNBS-LRRgenes to regulate defense responses to a pathogen.
ROS is a general term for oxygen-derived metabolic intermediates or radicals including superoxide (O2·-), hydrogen peroxide(H2O2), hydroxyl radical (OH·) and singlet oxygen (1O2) [10].ROS-generation and-scavenging pathways function in both pathogen virulence and host resistance. The ROS burst is reported to be essential for penetration peg formation byV. dahliaeand is thus indispensable for virulence and colonization by this fungus [65].Excess ROS produced during plant-pathogen interactions facilitates infection and colonization by necrotrophic pathogens[66,67]. On the host side, rapid ROS accumulation occurs during the cotton-V. dahliaeinteraction but it is quickly scavenged to maintain homeostasis. Thioredoxin GbNRX1, an important apoplastic ROS scavenger, plays a positive role in the cotton response toV.dahliae[68]and silencing of anthocyanidin synthase(GbANS)in cotton increased H2O2production and cell death around the invasion sites, in turn leading to increasedV. dahliaeinfection[69]. These results suggest that ROS elimination after the initial burst is functionally important in cotton resistance toV. dahliae.
Among other players involved in ROS metabolism, superoxide dismutases (SODs) are found in all kingdoms of life and act as the first line of defense against ROS, dismutating superoxide to H2O2[70]. In the present study, overexpression of miR398b boosted H2O2production when the transgenic plants were treated with NLP1 andV. dahliae, and thereby accelerated NLP1-triggered cell necrosis(Fig.7).By contrast,overexpression ofGhrCSD2,which is resistant to miR398b, reduced H2O2content (Fig. 7B). These results were strikingly similar to those of recent reports from analyses of rice response toMagnaporthe oryzaeinfection[41].Expression levels ofGhMSD1,GhFSD2andGhRbohB(the gene putatively encoding NADPH oxidase to catalyze the production of O2·-) were decreased inGhrCSD2-overexpression lines upon NLP treatment,but increased in miR398b-overexpression lines in comparison with WT plants. Transcripts ofGhCSD1,GhCCSandGh_D05G3257(the other target genes of miR398b)were increased inGhrCSD2-overexpression lines and decreased in miR398b-overexpression lines(Fig. S6). These results suggest that a compensatory regulatory mechanism may also function for ROS homeostasis in cotton.However, the phenotypes of miR398- andGhrCSD2-overexpression plants in cotton upon pathogen infection differed from those in rice. Overexpression of miR398b in cotton reduced cotton resistance toV. dahliae, while overexpression of miR398b in rice increased resistance toM. oryzae[41]. We propose that the excessive ROS accumulation in miR398b-overexpressing plants damages the cell and facilitatesV. dahliaeinfection (Fig. 7), consistent with results of previous studies [66,68].
Taken together, these results suggest that miR398b may function in one of two different ways to regulate plant immunity.First,miR398b may function in ROS homeostasis by cleavingGhCSD1,GhCSD2andGhCCS1, and second,miR398b suppresses the translation ofNBS-LRRgenes to control defense response (Fig. S7). How miR398b expression is regulated and the relationship of the two kinds of target of miR398b(GhCSDsandNBS-LRRgenes)during cotton-V. dahliaeinteractions await further study.
CRediT authorship contribution statement
Yuhuan Miao:Writing-original draft,Methodology,Investigation, Data curation.Kun Chen:Writing - review & editing,Methodology, Resources, Conceptualization.Jinwu Deng:Writing- review & editing, Methodology, Resources, Conceptualization.Lin Zhang:Writing - review & editing, Methodology, Resources,Conceptualization.Weiran Wang:Resources, Supervision.Jie Kong:Software.Steven J.Klosterman:Writing-review&editing,Supervision.Xianlong Zhang:Writing-review&editing,Supervision.Alifu Aierxi:Conceptualization, Writing - review & editing,Supervision, Resources, Funding acquisition.Longfu Zhu:Conceptualization, Writing - review & editing, Supervision, Resources,Funding acquisition.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Key Research and Development Program of China(2018YFD0100403)and the project from the Ministry of Science and Technology of China(KY201702009). We thank De Zhu (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University,Wuhan, Hubei) for assistance with laser scanning confocal microscopy. We thank Prof. Keith Lindsey (Department of Biosciences,Durham University, Durham, UK) for language revision.
Appendix A. Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2020.xx.xxx.
- The Crop Journal的其它文章
- Research progress on the divergence and genetic basis of agronomic traits in xian and geng rice
- From model to alfalfa: Gene editing to obtain semidwarf and prostrate growth habits
- The chloroplast-localized protein LTA1 regulates tiller angle and yield of rice
- Genome-wide association study and transcriptome analysis reveal new QTL and candidate genes for nitrogen-deficiency tolerance in rice
- Advances in the functional study of glutamine synthetase in plant abiotic stress tolerance response
- Identification of microRNAs regulating grain filling of rice inferior spikelets in response to moderate soil drying post-anthesis

