Characterization of grass carp FosB, Fosl2, JunD transcription factors in response to GCRV infection
Fei Yu, Wanjuan Li, Longlong Wang, Shunzheng Que, Liqun Lu,*
aNational Pathogen Collection Center for Aquatic Animals, Shanghai Ocean University, Shanghai, 201306, China
bInstitute of Marine Biology, College of Oceanography, Hohai University, Nanjing, 210098, China
cKey Laboratory of Agriculture Ministry for Freshwater Aquatic Genetic Resources, Shanghai Ocean University, Shanghai, 201306, China
dNational Experimental Teaching Demonstration Center for Fishery Sciences, Shanghai Ocean University, Shanghai, 201306, China
Keywords:
Grass carp reovirus
FosB
Fosl2
JunD
Co-expression
In flammation
A B S T R A C T
Activating protein-1 (AP-1) composed of Jun and Fos family proteins, which are frequently activated by pathogens to induce cellular inflammation and immune responses. In this study, by using RNA sequencing (RNA-seq)technology, we found that FosB, Fosl2, and JunD expression was co-induced 12 and 20 h in grass carp (Ctenopharyngodon idella) kidney cell line (CIK cells) after grass carp reovirus (GCRV) infection. Full-length cDNA of the three genes were cloned and phylogenetic analysis indicated that the corresponding proteins shared high homology with those in higher vertebrates. The tissue distribution analysis of the three genes showed they were ubiquitously expressed in various tissues of healthy grass carp. In infection assay, FosB, Fosl2, and JunD were upregulated and co-expressed 8 h after GCRV infection in CIK cells. Protein interaction and co-expression network analysis revealed that FosB, Fosl2, and JunD showed similar functions and were co-expressed in mammals and fish. Taken together, these results indicated that the co-expression of the AP-1 family proteins in grass carp uniformly responds to GCRV infection. These findings provided insights into the further study of AP-1 transcription factors function during grass carp reovirus infection.
1.Introduction
Transcription factor AP-1 is a dimer complex composed by DNA-binding proteins of the Fos (FosB,c-Fos, Fra-1/Fosl1, and Fra-2/Fosl2), Jun (JunD,c-Jun, and JunB), and other transcription factor families (Gazon et al., 2018). These proteins are key regulators of a wide range of cellular processes including cell proliferation, cell differentiation, cell death, oncogenesis, inflammation, and immune responses(Fennewald et al., 2007). AP-1 proteins commonly contain a leucine-zipper motif that shows the ability to dimerize and contains a basic domain for interaction with the conserved DNA sequence (b-zip domain) (Li, Li, et al., 2015). Thus, Fos proteins can form heterodimeric complexes with Jun proteins, and Jun proteins themselves can form homodimers and dimerize with each other to drive the promoters of many target genes (Uluçkan et al., 2015). In addition, the Fos and Jun families have been shown to dimerize with some members of the b-zip or non-b-zip transcription factor families CREB/ATF, NF-κB, MyoD,p300/CBP, and the TATA-binding protein, providing a functional link between the binding of AP-1 proteins to their target sequences and the transcriptional regulation of target genes (Foletta et al., 1998).
In mammals, AP-1 proteins have been reported to be important regulators of cytokine expression and play prominent roles in inflammatory diseases. Variations in AP-1 expression can cause pathological changes in various organs, such as bone and the hematopoietic system(Wagner, 2010).FosBknockdown was reported to significantly decrease the mRNA and protein levels of inflammatory chemokines, interleukin 6, interleukin 8, CD38, and tumor necrosis factor in HEp-2 and THP-1 cell lines (Wang et al., 2013).JunDbelongs to the proto-oncogene Jun family, but unlikec-Jun, whose expression influences cell growth by affecting the S/G2 and M phases of the cell cycle,JunDhas been shown to negatively modulate cellular proliferation (Jia et al., 2007).JunDis also associated with macrophage activation (Behmoaras et al., 2008),hypertrophic growth, and angiogenesis in the heart (Hil fikerkleiner et al., 2005). In hepatic stellate cells, theJunDheterodimer induced interleukin 6 promoter activity at an alternative DNA-binding site of the promoter (Smart et al., 2001). Aberrant expression of Fosl2 in adult T-cell leukemia cells (ATL cells) could alter the expression of 49 genes,includingMDM2,BCL-6, andc-Myb(Nakayama et al., 2008).
The AP-1 transcription factor family could be activated by various stimuli, such as bacteria, viruses, stress inducers, or inflammatory cytokines. Respiratory syncytial virus infection can modulate interleukin-8 production via the AP-1 signaling pathway in respiratory epithelial cells(Zhou et al., 2014), and the hepatitis E virus can activate the promoter of CXCL-8, a small multifunctional proinflammatory chemokine, via AP-1(Li, Chen, & Liu, 2015). Here, we found that grass carpFosB,Fosl2,andJunDgenes were induced by grass carp reovirus (GCRV), a typical strain of aquareovirus and the best-characterized model of aquareovirus pathogenesisin vivoandin vitro.Aquareoviruses are non-enveloped virions with a double-layer icosahedral capsid containing an 11-segment double-stranded RNA (dsRNA) genome (Attoui et al., 2002). During reovirus infection, a large number of host genes are upregulated and a few genes are downregulated (Clarke et al., 2014), which could be attributable to the activation of some specific transcription factors.Many carp genes have been found to be induced by GCRV, such as TNFα(Lu et al., 2016), interleukins (Wan & Su, 2015), interferon regulatory factors (IRFs) (Rao & Su, 2015), and interferons (Yu et al., 2017), whose transcriptional activation involves the AP-1 transcription factor family(Patil et al., 2018; Zhong et al., 2006). Thus, the evaluation of the association between the AP-1 transcription factor and GCRV infection is necessary and helpful to understand the viral pathogenesis. To understand this association, we assessed the expression of Fos-B, Fosl2, and Jun-D in a grass carp kidney cell line and attempted to determine the effect of GCRV infection on the expression of these genes.
2.Materials and methods
2.1.Fish, cell line and virus
Naive grass carp with an average weight of 200 g were obtained from a local farm and were acclimatized to laboratory conditions for two weeks in the Shanghai Ocean University fish breeding farm. TheCtenopharyngodon idellakidney (CIK) cell line (purchased from China Center for Type Culture Collection, CCTCC, China) was cultured using the M199 medium with 10% (v/v) fetal bovine serum (FBS, Thermo Fisher, USA) at 28 ℃ without CO2. The GCRV strain JX01 (GCRV-JX01)was isolated and preserved at the National Pathogen Collection Center for Aquatic Animals, Shanghai.
2.2.Tissue sampling and viral infection
Various tissues (head kidney, spleen, muscle, brain, heart, intestine,trunk kidney, gill, skin, and liver) were collected from three untreated individuals for RNA extraction. Fresh tissues were immersed in TRIzol™Reagent (Invitrogen, USA) at a ratio of 1:10 (tissue: TRIzol™ Reagent).Approximately 106CIK cells in each well of 6-well plates were infected with 1 mL GCRV JX01 at a multiplicity of infection (MOI) of 0.1 or 1. An equivalent number of cells were mock-infected as controls. The plates were incubated at 28 ℃ for 1 h, and then 2 mL of the M199 medium with 10% FBS was added to each well, and the infected cells were incubated at 28 ℃.
2.3.RNA-seq analysis
RNA sequencing and analysis were performed as previously reported(Yu et al., 2020). Brie fly, CIK cells mock-infected or infected with 1 MOI GCRV-JX01 (Mock groups, 12 h-post infection group, 20 h-post infection group) were applied to RNA sequencing.
The samples were used to generate sequencing libraries using the Next Ultra RNA library prep kit for Illumina (New England Biolabs, NEB)following the manufacturer instruction. Libraries were sequenced, at MajorBio Biotech Company Limited, on an Illumina HiSeq 2000 platform to generate 150 bp pair-end reads (raw data reads), which have been submitted into the NCBI Short Read Archive Database (SRA,Accession: SRP150656). Clean data was mapped to the grass carp reference genome (C_idella_female_scaffolds) by TopHat2 software. The differential expression analysis of each group was carried out by DESeq Version 1.18.0 based on reads per kilo bases per million mapped reads(RPKM), and the adjustedP-value<0.05 and fold change>2 or<0.5 was considered as a precondition for differentially expressed genes.
2.4.RNA extraction and molecular cloning
Total RNA extraction from the tissues of healthy grass carp or CIK cells was performed using 100 mg tissue or cell samples (about 1 ×106cells) with 1 mL of TRIzol™ Reagent according to the manufacturer’s instructions. The RNA yield was determined on the basis of absorbance without prior dilution by using the NanoDrop™ Spectrophotometer(Invitrogen, USA), and an A260/A280 ratio between 1.8 and 2.0 was considered indicative of purity. The isolated RNA (200 ng) was reverse transcribed into cDNA using the PrimeScript™ 1st Strand cDNA Synthesis Kit for gene cloning or the PrimeScript™ RT reagent Kit (TaKaRa,Japan) for real-time RT-qPCR. The primers used for cloning are listed in Table 1, and FosB, Fosl2, and JunD cDNA was amplified from grass carp kidney cells by PCR. The 3-Step PCR conditions were as follows: 98 ℃ for 3 min, then 30 cycles of 98 ℃ for 10 s, 55–60 ℃ for 15 s, and 72 ℃ for 1–3 min. The PCR product was cloned into the pMD20-T vector using a Mighty TA-cloning kit (TaKaRa, Japan) and further sequenced (Sangon Biotech, China). The full-length cDNA of the three genes from grass carp was obtained by fragment assembly based on cloning sequences and transcriptome data.
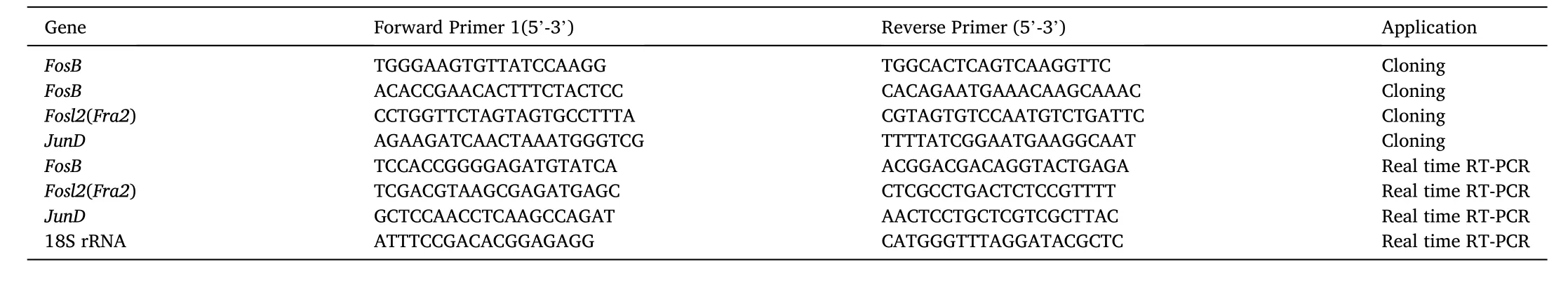
Table 1Primers used in this study
2.5.Bioinformatics analysis
The ORFs and encoding proteins were predicted using EditSeq software. The homologous sequences ofFosB,Fosl2, andJunDfrom different species were obtained from the GenBank database (NCBI, https://www.ncbi.nlm.nih.gov/). The molecular weight and isoelectric point of the proteins were determined using the online software (Compute pI/Mw tool, http://web.expasy.org/compute_pi/). Multiple alignments of amino acid sequences were produced using DNAman 6.0 software, and MEGA version 5.1 was used to construct phylogenetic trees using the neighbor-joining method. A functional protein association network analysis was conducted using an integrative database (STRING Database, https://string-db.org/).
2.6.Real-time RT-qPCR
The transcription levels ofFosB,Fosl2, andJunDin grass carp tissues or CIK cells were determined in triplicate by using real-time RT-qPCR and were normalized to the 18 S rRNA level (Su et al., 2011). The real-time RT-qPCR primers were designed using a primer designing tool(https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and are listed in Table 1. Real-time RT-qPCR was performed in accordance with the instructions for SYBR® Premix ExTaqII kit (TaKaRa, Japan). The real-time RT-qPCR 2-Step conditions were as follows: 95 ℃ for 30 s, followed by 40 cycles of 95◦C for 5 s, 60 ℃ for 30 s. The relative fold changes in gene expression were determined in comparison with the corresponding controls. SPSS software was used to perform data analysis and differences with aPvalue of<0.05 and<0.01 were considered significant and highly significant, respectively.
2.7.Western blot
Western blot analysis of CIK cells was performed as described previously (Yu et al., 2017). The protein samples were separated by 10%SDS-PAGE and this followed by transferring onto 0.45 μm PVDF membranes (Merck Millipore, Germany). The membranes were blocked for 2 h at room temperature in 5% non-fat milk dissolved in PBST. After blocking, the membrane was incubated in PBST containing primary antibody and 2% non-fat milk at 4 ℃ overnight. The bound primary antibody was visualized using a secondary antibody conjugated to horseradish peroxidase (HRP) using a Chemiluminescence Kit (ThermoFisher, USA). Images were visualized using Tanon 5200 ECL Western Blotting Substrate (Tanon, China). Expression of grass carp GAPDH was used as internal control. The following antibodies were used: viral protein VP7 (He et al., 2011) (1:5000); GAPDH (1:5000, Beyotime Biotechnology, China); and HRP-anti mouse IgG (1:5000, Abmart,China).
3.Results
3.1.Characterization of FosB, Fosl2, and JunD genes
Through analyzing the RNA-seq data of infected and uninfected CIK cells,we interestingly found that, among various AP-1 genes, only theFosB,Fosl2, andJunDexpressions were induced by GCRV infection in CIK cells. The three genes in the genome of female grass carp were blasted, and putative positions and transcriptional direction were determined (Table 2). To better understand the association between the AP-1 transcription factor family and GCRV infection, we characterized the nucleic acid and amino acid sequences of grass carpFosB,Fosl2, andJunD. The full-length cDNA sequences of the three genes were submitted to the GenBank database (Accession numbers:FosB, MH107063,Fosl2,MH107064, andJunD, MH107065). The FosB cDNA obtained contained a 182-bp 5′-untranslated region (UTR), a 1149-bp 3′-UTR, and a putative 1269-bp ORF. The FosB ORF encodes a putative 422-amino acid protein with a theoretical molecular weight of 44.84 kDa and an isoelectric point of 5.19. The Fosl2 cDNA consists of a 149-bp 5′-UTR, a 571-bp 3′-UTR, and a putative 1026-bp ORF. The Fosl2 ORF encodes a putative 341-amino acid protein with a theoretical molecular weight of 37.76 kDa and an isoelectric point of 5.71. The cloned JunD cDNA is composed of a 264-bp 5′-UTR, an 824-bp 3′-UTR, and a putative 855-bp ORF. The JunD ORF encodes a putative 284-amino acid protein with a theoretical molecular weight of 31.66 kDa and an isoelectric point of 6.58.
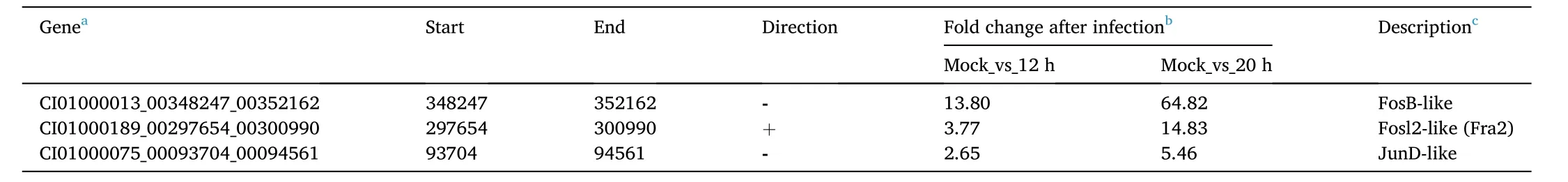
Table 2Transcriptome analysis of AP-1 transcription factors in CIK cells.
3.2.Conserved function domains of the transcription factors
The FosB protein possessed 4 conserved regions from the N to C terminals, which might be responsible for the transcription activity and dimerization of FosB (Fig. 1). There were more about 40 amino acids at the N terminal and the middle region of the carp FosB protein, which suggested that it might have potential unknown functions compared to the corresponding protein in mammals. The Fosl2 protein also contains a basic leucine zipper (b-ZIP) domain in its middle region, similarly to that in the other Fosl2 and FosB proteins (Figs. 1 and 2). The grass carp JunD protein also contains transactivation regions in the middle region and a b-ZIP domain at its C terminal (Fig. 3), which is different from the two Fos proteins (FosB and Fosl2). The FosB, Fosl2, and JunD proteins of grass carp shared slightly high homology with mammalian, avian, and elasmobranch proteins, and higher homology with teleost fish proteins,such as those ofDanio rerioandCyprinus carpio(Fig. 4).
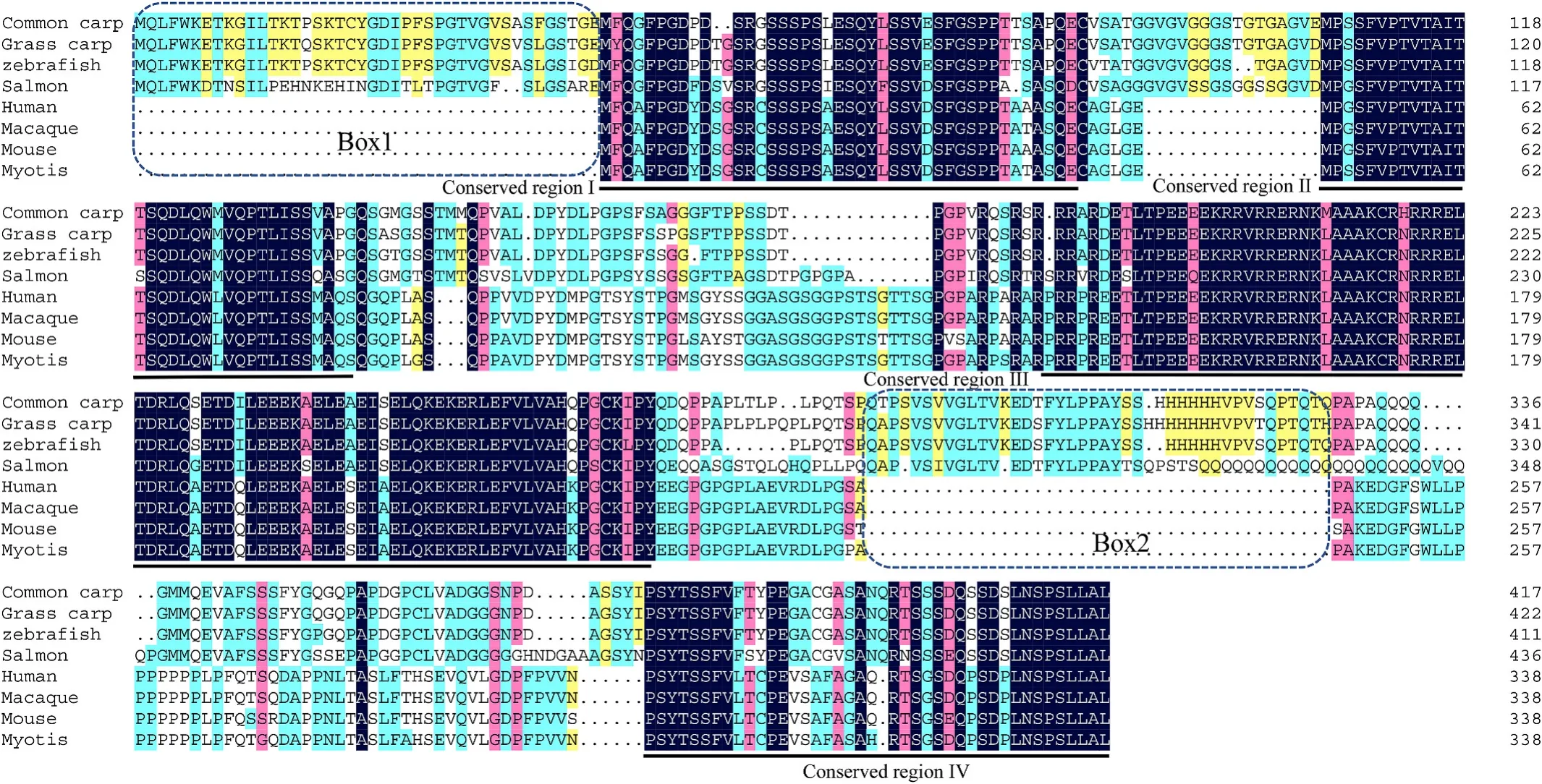
Fig. 1.Multiple alignments of amino acid sequences of FosB protein. Species and accession numbers are listed. The similarity at a specific position is indicated by blue (100%), pink (>75%), cyan (>50%), and yellow (>30%) bars, and the conserved domains, sequence gaps were marked by the underlines or boxes respectively.Species and accession numbers for FosB proteins: Homo sapiens [NP_006723.2], Macaca mulatta [XP_014979764.1], Myotis lucifugus [XP_006107289.1], Mus musculus[NP_032062.1], Chrysemys picta bellii [XP_023969385.1], Coturnix japonica [XP_015706635.1], Cyprinus carpio [XP_018978739.1], Danio rerio [XP_005173698.1],Ctenopharyngodon Idella [MH107063], Oryzias latipes [XP_011480826.1], Oncorhynchus mykiss [XP_021437622.1], Salmo salar [XP_014069147.1], Xenopus tropicalis[XP_002932681.2]. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)

Fig. 2.Multiple alignments of amino acid sequences of Fosl2 protein. Species and accession numbers are listed. The similarity at a specific position is indicated by blue (100%), pink (>75%), cyan (>50%), and yellow (>30%) bars, and the conserved domains, sequence gaps were marked by the underlines or boxes respectively.Species and accession numbers for Fosl2 proteins: Homo sapiens [NP_005244.1], Macaca mulatta [AFI33861.1], Pteropus vampyrus [XP_011369037.1], Mus musculus[NP_032063.2], Chelonia mydas [XP_007071748.1], Gallus gallus [NP_001039301.1], Parus major [XP_015478085.1], Xenopus tropicalis [XP_002938465.1], Callorhinchus milii [XP_007908258.1], Salmo salar [XP_014051373.1], Cyprinus carpio [XP_018976129.1], Sinocyclocheilus anshuiensis [XP_016300174.1],Ctenopharyngodon idella [MH107064], Danio rerio [NP_001076467.1 ]. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
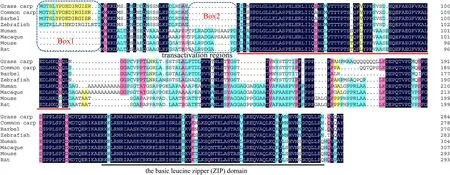
Fig. 3.Multiple alignments of amino acid sequences of JunD protein. Species and accession numbers are listed. The similarity at a specific position is indicated by blue (100%), pink (>75%), cyan (>50%), and yellow (>30%) bars, and the conserved domains, sequence gaps were marked by the underlines or boxes respectively.Species and accession numbers for JunD proteins: Mus musculus [NP_001273873.1], Rattus norvegicus [NP_001273895.1], Meriones unguiculatus [XP_021493699.1],Homo sapiens [NP_001273897.1], Macaca mulatta [XP_014978833.1], Pogona vitticeps [XP_020636717.1], Pseudopodoces humilis [XP_005531699.1], Xenopus tropicalis[XP_002937979.1], Salmo salar [XP_014004833.1], Ctenopharyngodon idella [MH107065], Danio rerio [NP_001121814.1], Cyprinus carpio [XP_018959779.1],Sinocyclocheilus rhinocerous [XP_016408475.1], Rhinolophus sinicus [XP_019592535.1]. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
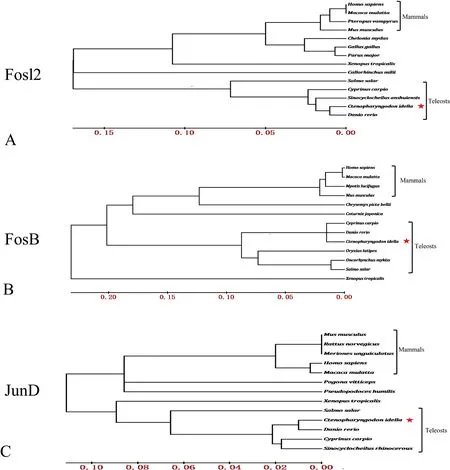
Fig. 4.Phylogenetic analysis of grass carp FosB, Fosl2, and JunD. The Phylogenetic trees were constructed by the neighbor-joining method based on amino acid sequences. Species and accession numbers are listed in Figs. 1–3.
3.3.Expression of FosB, Fosl2, and JunD in healthy grass carp
The transcription levels ofFosB, Fosl2, and JunDin the following healthy grass carp tissues were determined: the head kidney, spleen,muscle, brain, heart, intestine, trunk kidney, gill, skin, and liver (Fig. 5).The three genes were found to be expressed in all the examined tissues,which suggested the three proteins functioned widely in grass carp tissues. Furthermore,FosBwas highly expressed in the spleen, muscle,trunk kidney, gill, and skin;Fosl2was highly expressed in the head kidney, spleen, heart, trunk kidney, and gill; andJunDwas highly expressed in the spleen, muscle, heart, and gill. The kidney, spleen, and gill were the common tissues of high abundance of the three gene transcripts.
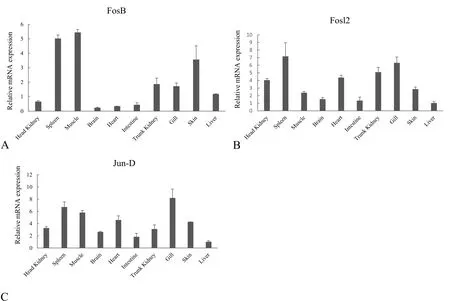
Fig. 5.The relative abundance of FosB, Fosl2, and JunD transcripts in healthy grass carp tissues. The mRNA abundance of FosB (A), Fosl2 (B), and JunD (C) was determined by real-time RT-qPCR. The relative mRNA levels were normalized to those of 18 S rRNA and determined by comparison with the expression in the liver by using the 2 –ΔΔCT method. The tissues involved in this assay are indicated.
3.4.Co-expression of FosB, Fosl2, and JunD in response to GCRV infection
The transcriptome data of CIK cells uninfected or infected with GCRV suggested thatFos-B,Fosl2, andJun-Dwere expressed in response to viral infection at 12 and 20 h after infection of GCRV. Fold changes inFos-B,Fosl2, andJun-Dexpression at 12 and 20 h after infection were>2.65 or even higher compared to those in the mock group (Table 1). To better understand the response of the three genes during GCRV infection, their relative expression levels were determined by real-time RT-qPCR (Fig. 6). The results showed thatFos-B,Fosl2, andJun-Dexpression was induced 8 h after GCRV infection in CIK cells, which preceded the expression of the GCRV capsid protein VP7. The results indicated thatFos-B,Fosl2, andJun-Dcould be up-regulated and co-expressed in response to GCRV infection.

Fig. 6.mRNA expression levels of FosB, Fosl2, and JunD in CIK cells infected with GCRV JX01. The mRNA abundance of FosB (A), Fosl2 (B), and JunD (C) was determined by RT-qPCR. The relative mRNA levels were normalized to those of 18 S rRNA and determined by comparison with the expression in the Mock group by using the 2 –ΔΔCT method. Error bars indicate the standard deviation of the mean for experiments performed in triplicate, and *P <0.05 and **P <0.01 are indicated. (D) The expression of the outer capsid protein, VP7, was determined by Western blot to re flect the viral replication level at the indicated time points of GCRV infection, and the cellular GAPDH protein was used as an internal control.
3.5.Association of FosB, Fosl2, and JunD proteins
The expression of the three genes has been characterized during GCRV infection. It is interesting that they all have a transcriptional regulatory function on member of AP-1 proteins and were uniformly induced by virus infection. To better understand protein function and the internal relationships, the protein-protein association and coexpression network were analyzed in mammals and fish (Fig. 7). The results also showed thatFosB, Fosl2, andJunDcan interact with each other and are co-expressed in mammals. Almost the same results were obtained in fish, however, there was no evidence of an association betweenFosBandFosl2in fish. These findings suggested thatFosB,Fosl2,andJunDhad similar functions and are co-expressed in mammals and fish.
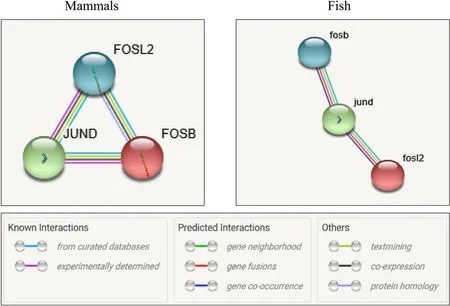
Fig. 7.Protein-protein association and co-expression network analysis of FosB, Fosl2, and JunD. The edges between the proteins are indicated at the bottom of the picture and represent protein-protein associations, which are meant to jointly contribute to a shared function. Co-expression of proteins is marked as black edges. The data analyzed for mammals and fish are from Homo sapiens and Danio rerio respectively.
4.Discussion
In the present study, we aimed to determine the association between the AP-1 transcription factors and GCRV infection. Our findings suggested that grass carp FosB, Fosl2, and Jun-D proteins shared high homology with those of other species and were commonly expressed in immune-organ tissues, and that FosB, Fosl2, Jun-D could be coinduced or co-expressed in response to GCRV infection in CIK cells.
So far, a variety of AP-1 transcription factors have been studied in mammals due to their important effects on pathology and inflammation;however, not much attention has been focused on AP-1 proteins in fish.As a result, there are limited reports on AP-1 protein function during pathogen invasion or even sequence characterization of these proteins in fish. Here, the sequences ofFosB,Fosl2, andJunDand their putative proteins were characterized, and their function domains were conserved. The kidney, spleen, and gill of healthy grass carp were common tissues showing high abundance ofFosB,Fosl2, andJunDtranscripts. These tissues form the main immune organs in fish (Austbo et al., 2014; Valenzuela-Miranda et al., 2015). However, we also found that theFosBwas also highly expressed in the muscle of the healthy grass carp. Although there is no evidence to explain whyFosBhighly expressed in this immune-irrelated tissue, one preliminary hypothesis is that theFosBmay relate to its immune-related function of regulating cell cycle and proliferation, bone and fat formation and so on (Madrigal &Alasoo, 2018; Renthal et al., 2008; Renthal; Nestler, 2008).
The high expression of theFosB,Fosl2, andJunDgenes in the immune organs might be associated with host defense in grass carp. In primary ATL cells, the JunB, Fosl2, and JunD proteins are highly expressed, and the Fosl2/JunD and/or Fosl2/JunB heterodimers can jointly activate the promoter of CCR4, a chemokine receptor known to be selectively expressed by various T cells (Nakayama et al., 2008). We also found that the FosB, Fosl2, and JunD proteins could interact with each other and are co-expressed in mammals and fish (Fig. 7). The many interactions of AP-1 proteins with each other suggest they jointly participate in a series of life processes in mammals and fish.
The effects of AP-1 proteins on host defense and viral replication are complex and multifaceted. In this study,Fos-B,Fosl2, andJun-Dcould be co-induced or co-expressed in response to GCRV infection in CIK cells(Fig. 6 and Table 1), and we have previously reported that TNF-α expression in CIK cells was upregulated by GCRV infection, which could also trigger apoptosis via TNF-α-induced caspase-8 pathways (Lu et al.,2016). Since TNF-α expression is modulated by AP-1 transcription factors (Wagner, 2010), GCRV infection may trigger apoptosis via induction of AP-1 proteins to activate TNF-α-induced caspase-8 pathways. In grass carp, many putative AP-1 binding sites have been found in the regulatory regions of fork-head box 3 (foxp3) (Yang et al., 2012),calmodulin (CaM) gene (Huo & Wong, 2009), the TNF-related apoptosis-inducing ligand (TRAIL) (Chang et al., 2006), and the TRAF2-binding protein (T2BP) gene (Chang et al., 2005), suggesting that the transcriptional levels of these genes are regulated by AP-1 proteins. The alterations in the expression of numerous immune genes and inflammatory cytokines in GCRV-infected grass carp cells might be associated with the activation of AP-1 transcription factors to inhibit a certain stage of the viral replication cycle.
In contrast, GCRV may exploit the function of AP-1 transcription factors to present a response to host defense or to promote its own replication cycle, just as other reoviruses alter viral secretion via different pathways leading to AP-1 protein activation (Berard et al.,2015). In addition, reoviruses can induce G2/M cell cycle arrest to inhibit cell proliferation (Boehme et al., 2013; Poggioli et al., 2000), and some AP-1 transcription factors, such as JunD, may negatively regulate cell proliferation (Jia et al., 2007). Therefore, it is reasonable to assume that GCRV, a reovirus, might inhibit the cellular proliferation induced by JunD to fully utilize the limited cytosol for its own replication.
However, at present, there is no solid evidence to support these hypotheses: I) the transcriptions of many immune genes and inflammatory cytokines in response to GCRV infection were modulated by activation of AP-1 transcription factors; II) the co-expression of AP-1 transcription factors in response to GCRV infection is a host-defense way or viral replication strategy, which might be the focus of a future study on the aim of the up-regulated expression of the three AP-1 proteins in the host or the virus. In general, the co-expression of the AP-1 transcription factor family in response to a viral infection may provide insight into the pathogenesis of GCRV infection in grass carp.
CRediT authorship contribution statement
Fei Yu: Writing, Software, Preparation. Wanjuan Li: Validation,Verification, Methodology, Investigation, Preparation. Longlong Wang: Visualization. Shunzheng Que: Investigation. Liqun Lu:Conceptualization, Writing, Supervision.
Declaration of competing interest
The authors declare that there is no conflicts of interest.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (grant number 31672690) and the Earmarked Fund for China Agriculture Research System (grant number CARS-45-19).
Glossary
AP-1 Activator protein-1
FosB Transcription factor fosB
Fosl2 Fosrelated antigen 2
JunD Transcription factor junD
JunB Transcription factor junB
GCRV Grass carp reovirus
 Aquaculture and Fisheries2022年3期
Aquaculture and Fisheries2022年3期
- Aquaculture and Fisheries的其它文章
- Anthropogenic temperature fluctuations and their effect on aquaculture: A comprehensive review
- Recent advances in application of moving bed bioreactors for wastewater treatment from recirculating aquaculture systems: A review
- Characterization and expression analysis of gonad specific igf3 in the medaka ovary
- Liver DNA methylation and transcriptome between 1- and 3-year-old grass carp
- Screening and validation of reference genes for qPCR analysis in gonads and embryos of Takifugu bimaculatus
- Assessment of genetic diversity, detection of strain-specific single nucleotide polymorphisms and identification of the Bangladesh and Vietnam strain of Channa striata by PCR-RFLP analysis of the mitochondrial COI gene fragment
