Characterization and expression analysis of gonad specific igf3 in the medaka ovary
Jiale Xie , Ying Zhong , Yuli Zhao , Wenjie Xie , Jing Guo , Lang Gui ,,Mingyou Li ,
a International Research Center for Marine Biosciences, Ministry of Science and Technology, Shanghai Ocean University, Shanghai, 201306, China
b Key Laboratory of Exploration and Utilization of Aquatic Genetic Resources, Ministry of Education, Shanghai Ocean University, Shanghai, 201306, China
Keywords:
igf3
igf1
Medaka
Ovary
A B S T R A C T
The gonad specific expression of insulin-like growth factor 3 (igf3) has suggested an important role of igf3 in fish reproduction. In this study, medaka igf3 was isolated and its expression patterns were compared with igf1, and germ cell gene vasa in adult medaka ovary. Molecular cloning and sequencing showed that the open reading frame (ORF) of medaka igf3 was comprised of 501 nucleotides and encoded 166 amino acid residues. Reverse transcription polymerase chain reaction (RT-PCR) analysis showed that igf3 RNA was specifically expressed in developing embryos and adult gonads of both sexes. Real time quantitative PCR (q-PCR) indicated that igf3 expression was gradually increased during oogenesis, and reached its highest level at stage V. Using Chromogenic and fluorescent in situ hybridization, igf3 was shown to be present in the germ cells of oogonia and in oocytes at stages I-III, as well as abundant in granulosa cells and theca cells of oocytes at stages IV–V. In addition,igf3 and igf1 were expressed mutually in the outer theca cells of stage V oocytes in ovary. Collectively, we demonstrated that gonad specific igf3 could mark medaka ovarian somatic cells and germ cells. These findings highlight the importance of igf3 during ovarian development.
1.Introduction
The insulin-like growth factors (igfs) are recognized as key factors in regulating growth and reproduction axis by triggering a series of physiological process in a variety of vertebrates (Giudice, 2001; Wood et al.,2005). The above responses process depend on interacting with the Igf signals, specific proteases, extracellular matrix proteins as well as intracellular signals transduction pathways (Duan, 2002; Ozaki et al.,2013).
The Igfs system contains three ligands: Igf1, Igf2 and Igf3. Among them, Igf1 and Igf2 were mainly produced in the liver in endocrine manner as well as most of extrahepatic sites such as the gonads of fish by autocrine or paracrine mechanisms (Reinecke, 2010). Igf1 RNA and protein have been well studied in the ovary of various fish species,including tilapia (Berishvili et al., 2006), sturgeon (Wuertz et al., 2007),common carp (Paul et al., 2009) and zebra fish (Xie et al., 2016). Igf2 RNA and protein were also reported in the ovary of rainbow trout(Perrot & Funkenstein, 1999), seabream (Perez et al., 2016) and zebra fish (Nelson & Van Der Kraak, 2010). Additionally, functional analyses have revealed physiological roles of Igf1 and Igf2 in oocyte maturation and steroidogenesis during ovarian development (Bobe et al., 2003; Campbell et al., 2006). However, different from the conventional Igf1 and Igf2 that are ubiquitously expressed in a majority of tissues,igf3is exclusively expressed in fish gonads, it is envisaged thatigf3may involve in fish reproduction (Wang et al., 2008).
Igf3 has been identified in several fish species in recent years,including tilapia (Li et al. 2012b, 2020; Reinecke, 2010; Wang et al.,2008) zebra fish (Li, Liu, et al., 2011; Zou et al., 2009), orange-spotted grouper (Yang et al., 2015) and common carp (Song et al., 2016). The medakaigf3sequence was also predicted (Wang et al., 2008). Furthermore, researches have demonstrated that zebra fishigf3has functions on oocyte maturation (Li et al., 2018) and gonad steroidogenesis (Li, Wu,et al., 2012). Apart from the specific presence in gonads,igf3has also been reported in zebra fish embryos (Li, Liu, et al., 2011; Zou et al.,2009). While there are several relevant reports onigf3, the expression patterns ofigf3among different fish species remains unclear. Much more studies are need.
The laboratory fish medaka is a useful model for studying reproductive biology, it is the first fish whose sex-determining genes have been identified (Matsuda et al., 2002; Nishimura et al., 2015). Medaka has well established germline stem cells cultures (Hong et al., 2004; Li et al., 2015a; Nakamura et al., 2010), and a lot of transgenic fish for germ cells visualization (Hong et al., 2016; Li et al. 2009, 2012a).Igf1andigf2have been reported in our previous reports (Yuan et al., 2018;Yuan & Hong, 2017). The present study was aimed to elucidate the temporal and spatial expression patterns ofigf3during ovarian development. Medakaigf3sequence was obtained and its RNA expression was analyzed in adult tissues and embryogenesis by RT-PCR. Furthermore,expression patterns ofigf3withigf1andvasawere compared on ovarian sections by chromogenic and fluorescentin situhybridization. We demonstrated thatigf3was expressed in somatic cells and germ cells in mature ovary.
2.Materials and methods
2.1.Fish and embryos
Medaka fish was raised under a constant temperature at 26 ℃ and an artificial photoperiod of 14 h light to 10 h dark cycle. Embryo developmental stage was determined as described previously (Iwamatsu,2004). All experiments carried out with medaka were conformed with the stipulations of the Committee for Laboratory Animal research of Shanghai Ocean University Committee.
2.2.Cloning of igf3
A previous work has predicted the medakaigf3sequence (Wang et al., 2008). However, the 5′end ofigf3cDNA was incorrect after our verification. Therefore,igf3sequence was re-cloned from medaka by RACE approach. Total RNA was extracted from medaka ovary by using TRizol® (Invitrogen, Carlsbad, CA) reagent, cDNA was synthesized by using RACE cDNA Amplification Kit (Clontech, USA) according to the manufacturer’s instructions. The medakaigf3EST sequences was blasted and a cloning called BJ711729, which containing a partial sequence including stop code ofigf3was finally obtained. Based on this fragment,the gene-specific primer 5′R & 5′NR (CTCCCACAACCACCGCAAGCAGCACGA and ATGTACTGTGCCAAAGCCAAGAGCCCG) were designed and 5′RACE was performed. Finally,igf3cDNA was obtained by combining 5′RACE sequence with BJ711729 sequence. Protein sequence alignment was conducted by using the Vector NTI Advance® 11.0. Neighbor-joining (NJ) Phylogenetic tree was construct by using MEGA 7.0 software.
2.3.RT-PCR and q-PCR
The RNA of adult medaka organs (kidney, liver, gut, eye, brain, ovary and testis) and embryos were extracted as described above, 1 μg total RNA was used to synthesized cDNA with oligo (dT)18by using M-MLV reverse transcription (Takara, Shiga, Japan), then theigf3expression was detected using reaction transcription-polymerase chain reaction(RT-PCR). The reaction system was conducted in 25 μl volume containing 20 ng cDNA templates, dNTPs, EX-taq enzyme (Takara, Shiga,Japan), EX-taq buffer andigf3primers (F1&R2: ATGGAGAGAGGGGAAGGGCTG and TTAGAGTCCGCCCCTCTATGG) orβ-actinprimers (F&R: TTCAACAGCCCTGCCATGTA and CCTCCAATCCAGACAGTAT), the reaction procedure was conducted for 35 cycles (10 s at 95 ℃, 10 s at 56 ℃ and 30 s at 72 ℃),β-actinwas used as internal control. The RT-PCR amplification products were documented on 1%agarose gel as described previously (Yuan et al., 2018; Zhu et al., 2018).3–5 mature ovaries were collected and 2.5 g/L trypases and 0.02% EDTA mixture were added at 37 ℃ for 5 min. Oocytes were washed 3–5 times with PBST and blowed gently. Different stages oocytes were isolated.The different stages of oocytes were identified according to morphology and collected into different EP tube for preparing RNA sample. For real time quantitative RT-PCR (q-PCR) analysis, the specific primers ofigf3F2&R2 was used (GCACGACCACCTCTCACAT and CTGAACAGAAAGCGGCACTT), andβ-actinwas used as the internal reference gene.The q-PCR process was performed on Bio Rad system using SYBR Green PCR Kit (TliRNaseH Plus). The 20 ng cDNA was conducted for q-PCR reaction. The relative expression level ofigf3at stages I–V was calculated using 2−ΔΔCtmethod as described in our previous research (Qiu et al., 2018).
2.4.Chromogenic and fluorescent in situ hybridization
In situhybridization (SISH) on ovarian sections by chemical staining with BCIP/NBT and fluorescentin situhybridization (FISH) with TSA™Plus Fluorescence Systems (NEN Life Science) were conducted as described previously with minor modifications (Li, Shen, et al., 2011;Yuan et al., 2018). For chromogenic or fluorescentin situhybridization,2–3 frozen sections was selected to verifyigf3expression patterns. The experiment was repeated at least three times. Brie fly, pGEM-T vectors containing the 501 ntigf3ORF, 558 ntigf1ORF and 1.8 kbvasaORF were linearized for the synthesis of sense and anti-sense RNA probes from Sp6 or T7 promoter by using the digoxigenin (DIG) RNA (Roche,Basel, Switzerland) forigf1andvasa, or FITC RNA Labelling Kit forigf3(Roche, Basel, Switzerland). Nucleus was stained with DAPI for blue fluorescence and embedded in the Gold Antifade reagent (Invitrogen,Carlsbad, CA) (Yuan et al., 2018).
2.5.Microscopy
Observation and photography on Leica TCS SP8 Laser Scanning Confocal Microscope (Leica, Germany) and Nikon Ds-Ri2 camera(Nikon, Tokyo, Japan) were as described (Qiu et al., 2018; Yuan et al.,2018).
3.Results
3.1.Cloning and characterization analysis of medaka igf3
The medakaigf3ORF was 501 nucleotides, and encoded 166 amino acid residues (GenBank accession no. MK183116) (Fig. S1). The signal peptide prediction was consistent with the database annotation. The output for thioredoxin domain contained protein 4 precursor (endoplasmic reticulum protein ERp44 (Fig. S1). Alignment analysis with other species indicated that medaka Igf3 was 50%, 48%, 47%, 29% and 29% identical to killifish, sea bass, tilapia, common carp and zebra fish(Fig. 1A). Besides, medaka Igf3 also possessed the conserved characteristics of Igf family, including conserved cysteine residues and A, B, C and D domains (Fig. 1A). Phylogenetic tree analysis revealed that medaka Igf3 was clustered to the Igf3 clade that was independent of Igf1 and Igf2 clades (Fig. 1B).

Fig. 1.Align ment and Phylogenetic tree of Igf3. (A) Alignment of Igf3. Domains A-D of the mature Igf3 are highlighted. Asterisks depict the six conserved cysteine residues within domains A-D. Sequence identity values are given at the end of the alignment. Species abbreviations are as follows: Ola, Oryzias latipes(medaka); Om, Oncorhynchus mykiss (rainbow trout); Nf, Nothobranchius furzeri (killifish); Dl, Dicentrarchus labrax (sea bass); On, Oreochromis niloticus (tilapia); Cc,Cyprinus carpio (common carp); Dr, Danio rerio (zebra fish). (B) Phylogenetic tree of Igf3. Insulin serves as the outgroup. Bootstrap values are given. Accession numbers follow the organism.
3.2.Medaka igf3 was expressed in adult gonads and embryos
The expression of medakaigf3in adult tissues and embryos was analyzed by RT-PCR. In embryos,igf3RNA was persistently expressed in all embryogenesis ranging from 2-cell stage to hatched fry (Fig. 2A),which suggested thatigf3RNA is maternally deposited. In adult tissues,igf3RNA was specifically detected in gonads of both sexes but was absent in other somatic tissues (Fig. 2B). Interesting, theigf3expression level in testis was higher than that in ovary (Fig. S3). To further verify the relative expression level ofigf3in different stages of oocytes, q-PCR was conducted. The expression ofigf3was low at the early stages of oocytes at I-III, and increased at stage IV, finally reached highest level at the stage V (Fig. 2E).
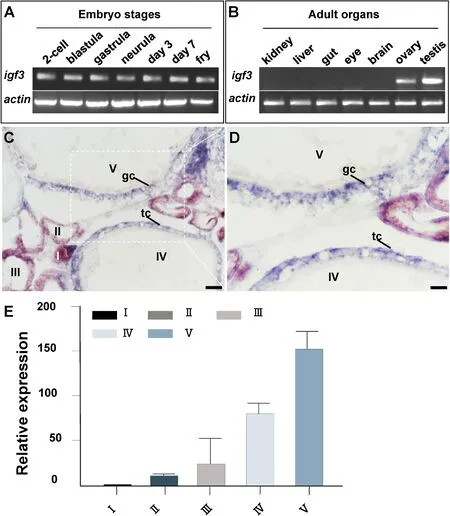
Fig. 2.RNA Expression of igf3. (A–B) Embryonic and adult expression by RT-PCR. (C–D) Adult ovarian cryosections were hybridized to antisense probe and the signal was visualized by chromogenic staining, igf3 signal was expressed throughout oogenesis, igf3 was expressed in the oocytes at I– III and high expression was detectable in granulosa cells and theca cells at stages IV–V. (E) q-PCR analysis of medaka igf3 RNA in oocytes from stages I–V. I–V, stages of oocytes; gc, granulosc cell; tc, theca cells; Scale bars, 100 μm.
3.3.Ovarian expression of igf3 by chromogenic ISH
To investigate the temporospatial expression ofigf3RNA, SISH was carried out on ovarian sections. Chromogenic SISH results showed thatigf3was detected in the oocytes from stages I–III (Fig. 2C). Besides, theigf3RNA was also expressed in granulosa cells and theca cells surrounding oocytes at stages IV-V,. (Fig. 2D). No signal was detected above background in the gonads by sense probe (date not shown).
3.4.Ovarian expression of igf3 and vasa by FISH
To precisely identify the medakaigf3expression patterns, RNA expression ofigf3andvasawere further performed by FISH.Vasais a well-studied germ cell marker in various organisms including medaka(Li et al., 2009; Yuan et al., 2014; Li et al., 2015b). In ovary, the expression ofvasawas high in oogonia and early stages of oocytes, then decreased at advanced stages of oocytes and finally disappeared at mature oocytes (Fig. 3B, D, 3F). In contrast, except for the expression of theigf3was present in the germ cells of early stages (I-III) of oogenesis including oogonia, its strong signal was also detected in numerous somatic cells, such as granulosa cells and theca cells of oocytes at advanced development stages IV-V (Fig. 3A, C, 3E).

Fig. 3.Expression of igf3 and vasa in the ovary. Adult ovarian cryosections were hybridized to antisense probes by fluorescence staining. Nuclei were stained by DAPI. (A–D) Micrographs at low magnification, igf3 signal was expressed throughout oogenesis. (E–F) Micrographs of the area framed in (C–D) at high magnification,high expression was expressed in granulosa cells and theca cells as well as in oogonia. I–V, stages of oocytes. og, oogonia; gc, granulosa cells; tc, theca cells; Scale bars, 100 μm.
3.5.Ovarian differential expression of igf3 and igf1 by FISH
To further elucidate the expression patterns ofigf3during ovarian development, a test for co-localization ofigf3andigf1was performed by FISH. Results showed that the expression patterns ofigf3andigf1were different.Igf3was present in oogonia (Fig. 4A and E), whileigf1was absent in oogonia (Fig. 4B and F). Interestingly, bothigf3andigf1were expressed in granulosa cells in oocytes from stages IV-V (Fig. 4C and D),but they were expressed in two distinct types of theca cells in ovary(Fig. 4G and H).
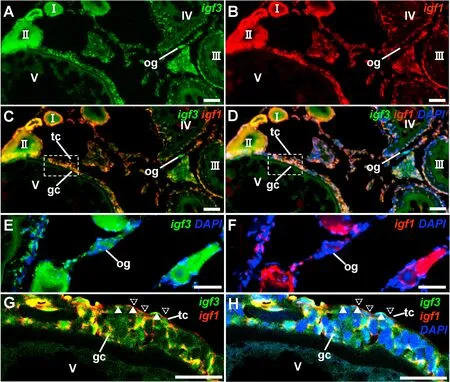
Fig. 4.Expression of igf3 and igf1 in the ovary. Adult ovarian cryosections were hybridized to antisense probes by fluorescence staining. Nuclei were stained by DAPI. (A–D) Micrographs at low magnification. (E–F) igf3 was present in oogonia but igf1 was absent. (G–H) Micrographs of the area framed in (C and D) at high magnification, igf3 and igf1 were expressed in distinct theca cells in ovary, theca cells showing igf3 signal and igf1 signal are indicated. I–V, stages of oocytes. og,oogonia; gc, granulosa cells; tc, theca cells; Scale bars, 100 μm.
4.Discussion
The Igfs are key factors in regulating growth and reproduction in a variety of vertebrates (Giudice, 2001; Wood et al., 2005). Among theigfs,igf3has been studied in several fish species, such as tilapia (Wang et al., 2008; Berishvili et al., 2010; Li et al. 2012b, 2020), zebra fish (Li,Liu, et al., 2011; Zou et al., 2009), orange-spotted grouper (Yang et al.,2015). To elucidate the potential roles ofigf3in medaka ovary development, medakaigf3was cloned in the present study. We found that medakaigf3RNA was expressed in adult gonads and developing embryos. Additionally, expression ofigf3was also found in somatic cells and germ cells during medaka ovarian development, which was different from medakavasathat only expressed in germ cells origf1expressed in oocytes.
Medaka Igf3 was also comprises of four domains as the other members ofigfsfamily (A, B, C and D) and conserved cysteine residues which shares a similar tertiary protein structure with Igf1 and Igf2, as similar to orange-spotted grouper (Yang et al., 2015). The four domains play a key role during the signal peptide yields and mature Igf peptides. (Wood et al., 2005). Sequence analysis showed that medaka Igf3 exhibits low similarity to other teleost Igf3, further indicates that the low conservation of Igf3 in different fish species. According to the Phylogenetic tree,medaka Igf3 was in an independent group, different from Igf1 and Igf2 clades, further proves that Igf3 is a novel number of Igfs family. So far,twoigf3transcripts have been found in zebra fish and common carp (Li,Liu, et al., 2011; Song et al., 2016). However, only one subtype ofigf3was present in tilapia (Wang et al., 2008), orange-spotted grouper (Yang et al., 2015) and our study in medaka. This might imply that evolutionary diversity of Igf3 was caused by distinct genome duplications in teleost fishes.
Medakaigf3RNA was highly present in gonads and expression level of testis is higher than in ovary. However, it was absent in other somatic tissues. The gonad specific expression ofigf3was similar to the previous findings in zebra fish (Li, Liu, et al., 2011) and tilapia (Wang et al., 2008),indicating thatigf3may play important role during gonadal development in teleost fish. Sinceigf3was detectable throughout embryogenesis, we suggested thatigf3may exert specific roes during embryogenesis. For example, zebra fishigf3can affect midline formation and notochord development (Zou et al., 2009).
Theigf3expression during oogenesis proceeds were analyzed, its expression increased from stages I–V, and high expression was also present in granulosa cells and theca cells at later stage of oocytes.Consistent with this result, q-PCR also showed that oocytes at stage V exhibited the strongestigf3expression level. The exactly expression of Igf3 through antibody staining might be needed in the future study. The increasing expression ofigf3in oocytes from stages I–V suggested the role in vitellogenic growth, and its highest expression at stage V suggested thatigf3may play important roles in oocyte maturation. Similar results were observed in zebra fish, which also exhibited progressive increases ofigf3during follicle growth, besides, recombinant zebra fish Igf3 could enhance oocyte maturation, suggesting the important role of Igf3 in oocyte matruation (Li, Liu, et al., 2011). In the following study,researchers found that the Igf3 plays a crucial role in mediating luteinizing hormon (LH) action on oocyte maturation through phosphorylate Igf1 receptors, it was known that the LH is involved in oocyte maturation and ovulation (Li et al., 2015b). Further findings also showed that theigf3transcript level is increased in hCG-induced ovulation in vivo,inhibited the Igf3 signaling by its receptor inhibitors could significantly attenuate the hCG-induced ovulation, implying that Igf3 serves as a mediator of LH action in zebra fish ovulation (Li et al., 2018). Considering the functions of zebra fish Igf3 in oocyte maturation and ovulation regulation, we supposed that medaka Igf3 may exert its function through interact with its receptors to induce oocyte maturation and ovulation,but further research was needed.
The mature medaka ovary consisted of a small number of oogonia and a large number of oocytes (Sakai et al., 1988). During the course of ovarian development, estrogens are produced by granulosa cells in the inner layer of ovarian follicles, while steroidal precursors of estrogen are supplied by theca cells in outer layer of follicles. These estrogens and estrogens precursors produced by granulosa cells and theca cells are important for oocytes growth and maturation. We found that bothigf3andigf1were expressed in granulosa cells, indicating they might involve in ovary development. Interestingly, they were expressed in distinct types of theca cells in medaka ovary. As described in the previous reports, two distinct types of theca cells, the steroidogenic gene-expressing cells and aromatase gene-expressing cells, which derived from the same precursor were identified in medaka ovary (Nakamura et al., 2009;Nishimura & Tanaka, 2014). But the exact cell types thatigf3andigf1locate need further study. To better distinguish the differential expression patterns ofigf3andigf1in ovary, a schematic illustration has been proposed (Fig. 5). The different expression patterns ofigf3andigf1in ovarian section may indicate their different roles in oocytes functions.These findings could provide fundamental information to further analysis cellular level in ovarian development.
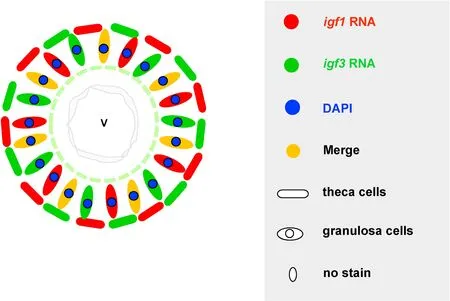
Fig. 5.Schematic illustration of igf3 and igf1 in the later stage of oocytes. The expression of igf3 in oogonia but igf1 was absent in oogonia. Both igf3 and igf1 were expressed in granulosa cells of ovary, igf3 and igf1 were detectable in two types of theca cells in ovary. V, fully grown oocytes.
Although the mechanisms ofigf3on oocyte maturation is elusive, we speculated thatigf3may exert its function by combining withigf1receptor to trigger signal transduction cascade (Li, Liu, et al., 2011). So far,there is no document about the cognate reporter forigf3. However, theigf3has similar structure to that of otherigfs(Wang et al., 2008). It has been reported thatigf3can trigger the Akt pathway which is a major downstream effector of theigf1receptor (Zou et al., 2009). Furthermore,another evidence also demonstrated thatigf1mediates oocyte maturation by activating phosphoinositide 3 (PI3) kinase signal pathways(Weber & Sullivan, 2001). However, the mechanism ofigf3actions on fish reproduction need further study. Additionally, evidences also shown thatigf3can stimulate spermatogonia proliferation and differentiation in zebra fish testis (Morais et al., 2017; Nobrega et al., 2015; Sa fian et al.,2018). Both oogonia and spermatogonia are germ cells,igf3might be involved in germ cells development. Since medakaigf3was also detected in testis, and the expression was relatively higher than that in ovary, its expression and function in testis would be analyzed in our further research.
In summary, medakaigf3was highly expressed in the germ cells of oogonia and early stages of oocytes, as well as in somatic cells surrounding oocytes at advanced stages, including granulosa cells and theca cells. The expression ofigf3in somatic cells and germ cells during ovarian development means that it may play unique and important roles on ovarian maturation in medaka.
Author contribution
Jiale Xie: Conceived and designed the experiments, Performed the experiments, Formal analysis Analyzed the Data curation data,Contributed to reagents/materials/analysis tools, Writing - original draft Wrote the paper. Ying Zhong: Conceived and designed the experiments, Performed the experiments, Writing - original draft Wrote the paper, Formal analysis Analyzed the Data curation data. Yuli Zhao:Conceived and designed the experiments, Performed the experiments,Writing - original draft Wrote the paper, Formal analysis Analyzed the Data curation data. Jing Guo: Contributed to reagents/materials/Formal analysis analysis tools. Lang Gui: Conceived and designed the experiments, Formal analysis Analyzed the Data curation data,Contributed to reagents/materials/analysis tools, Writing - original draft Wrote the paper. Mingyou Li: Conceived and designed the experiments, Formal analysis Analyzed the Data curation data Writing -original draft Wrote the paper
Declaration of competing interest
The authors declare that there is no conflicts of interest.
Acknowledgement
This work was supported by the National Natural Science Foundation of China (31672700) and National Key R&D Program of China(2018YFD0901205).
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aaf.2020.07.018.
 Aquaculture and Fisheries2022年3期
Aquaculture and Fisheries2022年3期
- Aquaculture and Fisheries的其它文章
- Anthropogenic temperature fluctuations and their effect on aquaculture: A comprehensive review
- Recent advances in application of moving bed bioreactors for wastewater treatment from recirculating aquaculture systems: A review
- Liver DNA methylation and transcriptome between 1- and 3-year-old grass carp
- Screening and validation of reference genes for qPCR analysis in gonads and embryos of Takifugu bimaculatus
- Assessment of genetic diversity, detection of strain-specific single nucleotide polymorphisms and identification of the Bangladesh and Vietnam strain of Channa striata by PCR-RFLP analysis of the mitochondrial COI gene fragment
- Assessment of dietary chaste tree (Vitex agnus-castus) fruit extract on growth performance, hemato-biochemical parameters, and mRNA levels of growth and appetite-related genes in gold fish (Carassius auratus)
