Integrated UHPLC-MS and network pharmacology to explore the active constituents and pharmacological mechanisms of Shenzao dripping pills against coronary heart disease
Tao Hu,Ke-Ning Zheng,Jia-Yin Liang,Dan Tang,Lu-Yong Zhang,Ming-Hua Xian*,Shu-Mei Wang
1Key Laboratory of Digital Quality Evaluation of Chinese Materia Medica of State Administration of Traditional Chinese Medicine,Guangdong Pharmaceutical University,Guangzhou 510006,China.2Engineering &Technology Research Center for Chinese Materia Medica Quality of the Universities of Guangdong Province,Guangzhou 510006,China.3School of Traditional Chinese Medicine,Guangdong Pharmaceutical University,Guangzhou 510006,China.4Guangzhou Key Laboratory of Construction and Application of New Drug Screening Model Systems,Guangdong Pharmaceutical University,Guangzhou 510006,China.5School of Pharmacy,Guangdong Pharmaceutical University,Guangzhou 510006,China.
Abstract Background:Shenzao dripping pills(SZDP)is an empirical prescription of traditional Chinese medicine that is mainly used to treat coronary heart disease.However,the chemical composition and pharmacological mechanisms of SZDP are unknown.Methods:In this study,ultra-high performance liquid chromatography-quadruple-Exactive Orbitrap mass spectrometry was used to identify the chemical components in extracts and medicated plasma of SZDP.Subsequently,we performed network pharmacology methods,including target prediction by Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform and Integrative Pharmacology-based Research Platform of Traditional Chinese Medicine,protein-protein interaction network via STRING database;further,the key targets and compounds were screened using Cytoscape.Finally,the key targets and compounds were validated by molecular docking.Results:72 chemical constituents were identified from SZDP by high performance liquid chromatography and mass spectrometry technology.Among the components absorbed into plasma by SZDP,24 prototype components and 9 metabolized components were identified.The network pharmacology analysis of the prototype components showed that there are 13 key compounds(including ginsenoside Rc,Rb1,Rb2,ferulic acid,etc.),90 proteins(including proto-oncogene tyrosine-protein kinase Src,nuclear receptor subfamily 3 group C member 1,caspase-3,etc.),and 10 pathways(including estrogen,IL-17 and VEGF signaling pathway,etc.)that play an essential role in the treatment of coronary heart disease with SZDP.In addition,the results of molecular docking revealed that ginsenosides Rc,Rb2 and Rb1 have strong binding activities to the caspase-3,as well as ginsenoside Rb2 to the nuclear receptor subfamily 3 group C member 1.Conclusion:This study showed that SZDP might act through multiple chemical constituents and targets against coronary heart disease.
Keywords:Shenzao dripping pills;coronary heart disease;chemical constituents;network pharmacology;molecular docking
Background
Coronary heart disease(CHD)is a multifactorial disease characterized by the rupture of atherosclerotic plaques,which results in myocardial ischemia,hypoxia,and necrosis.It has significant morbidity,recurrence,and mortality,and it poses a significant threat to human life,much like a ticking bomb[1].Even though chemically synthesized medications account for the majority of contemporary CHD therapy,adverse drug responses are a considerable hazard.Traditional Chinese medicine(TCM)therapeutic efficacy in the treatment of CHD has long been recognized.TCM differs from chemically produced drugs in that it contains various active ingredients and has a variety of pharmacological pathways[2].It offers many advantages when it comes to preventing,treating,and recovering from heart disease.However,the compounds of TCM are complicated because of the interaction between different compounds.It is hard to fully explain the active constituents and pharmacological mechanisms of TCM.
CHD belongs to the category of “Xiongbi” in the theory of TCM.The concept of “Xiongbi” was first proposed by the ancient Chinese medical classicsInner Canon of Huangdi ·Plain Questionsin the pre-Qin period(221 B.C.E.,author unknown).Their clinical symptoms were heart pain,which was unbearable,which is also the main representative symptom of CHD in modern Western medicine.Shenzao dripping pills(SZDP),as an empirical prescription of TCM,is made up of six herbs,including Guangzao(Choerospondias Axillaris),Danshen(Salvia Miltiorrhiza),Renshen(Panax Ginseng),Jiangxiang(Dalbergia Odorifera),Suhexiang(Liquidimibar Orientalis)and Bingpian(Cinnamomum Camphora)[3].It is an effective hospital preparation for CHD with a clinical dose of 8 g/d.The prescription composition of SZDP is derived from two classic Chinese formulas for CHD treatment,namely classic ancient prescription of Chinese medicine Suhexiang pills and Chinese patent medicine compound Danshen formula(approved number by State Food and Durg Adminnistration:Z44023372)[4,5].Suhexiang pill was first recorded in theSu Shen Liang Fangco-authored by Shi Su and Kuo Shen in C.E.1075,which is often used to treat Xiongbi.Suhexiang pills are made up of fifteen herbs,includingLiquidimibar Orientalis,Baizhu(Atractylodes Macrocephalae),Zhusha(Chinnabaris),Chenxiang(Aquilaria Sinensis),Hezi(Terminalia Chebula),Dingxiang(Eugenia Caryophyllata),Xiangfu(Cyperus Rotundus),Muxiang(Aucklandia Lappa),Tanxiang(Santalum Album),Ruxiang(Boswellia Carterii),Biba(Piper Longum),Wuxixie(Rhinoceros Unicornis),Anxixiang(Styrax Tonkinensis),Shexiang(Moschus Berezovskii),and Cinnamomum Camphora.Compound Danshen formula is a Chinese patent medicine based on the theory of TCM,consisting ofSalvia Miltiorrhiza,Sanqi(Panax Notoginseng),andCinnamomum Camphora.SZDP was integrated Suhexiang pills and compound Danshen formula,addedDalbergia Odoriferaand Mongolian medicineChoerospondias Axillaris.Our research team prepared it into dripping pills,which improved its bioavailability and was easy to carry and take.In addition,some active components in the six herbs of SZDP have been reported to treat CHD.Total flavonoids inChoerospondias Axillarishave been discovered to have anti-arrhythmic and anti-myocardial ischemia activities in experiments[6,7].The compound inSalvia Miltiorrhizacontains protocatechualdehyde,salvianic acid A,salvianolic acid C,lithospermic acid,caffeic acid and other substances,which have anti-inflammatory,immune regulation,and protective effects on the cardiovascular system.Panax Ginseng,Dalbergia Odorifera,Liquidimibar Orientalis,andCinnamomum Camphoraall have active components in treating myocardial ischemia[8-11].Furthermore,in a prior study,we detected volatile components in SZDP by liquid chromatography and mass spectrometry(LC-MS)[12].However,these studies can only reflect part of the chemical composition of SZDP,which is not comprehensive enough.The active components and pharmacological effects of SZDP are still unknown.
Ultra-high performance liquid chromatography-quadruple-Exactive Orbitrap mass spectrometry(UHPLC-Q-Exactive Orbitrap MS)combines ultra-high performance liquid chromatography with high-resolution Orbitrap mass spectrometry(UHPLC-MS)detection technology to produce a more accurate analysis for compounds identification of SZDP.Furthermore,network pharmacology analysis is complementary with TCM,which shifts drug development from a“one target,one drug” to a “multiple targets,multiple compounds”approach.This is unquestionably a more useful tool for studying SZDP’s pharmacological effects[12].By combining these two technologies,we can not only overcome the problem of rapid qualitative identification of SZDP,but we can also explain the intricate mechanisms of SZDP with multiple projected pathways,allowing for a more in-depth examination of its pharmacological mechanisms of action.These methods also provide accurate data for the following discussion.
In this study,the chemical components of SZDP were swiftly identified by LC-MS,and the material basis of SZDP was clarified.It is believed that components absorbed into the blood are more likely to exert therapeutic effects in vivo[13,14].Based on this,we conducted a network pharmacology assessment of the prototype components of SZDP in plasma and predicted the critical chemical components and targets for CHD intervention.To validate our predictions,we performed molecular docking experiments to show the relationship between drugs and targets.In summary,using LC-MS,network pharmacology,and molecular docking techniques,the chemical composition and potential pharmacological effects of SZDP were thoroughly investigated in this study,resulting in a reliable analytical strategy for identifying TCM substances and assessing their pharmacological effects.
Methods
Chemical and reagents
Gallic acid was purchased from National Institutes for Food and Drug Control(Beijing,China).Chengdu Croma Biotechnology Co.,Ltd.(Chengdu,China)provided all reference compounds(purities >98%),including ginsenoside Rb2,ginsenoside Rc,ginsenoside Rd,ginsenoside Rf,dihydrotanshinone I,cryptotanshinone,tanshinone I,tanshinone IIA,liquiritigenin,isoliquiritigenin,butin and medicarpin.Acetonitrile was bought from Shanghai Macleans Biotech Co.,Ltd.(Shanghai,China)and methanol was acquired from Oceanpak Co.(Gothenburg,Sweden),both of chromatographic grade.Ultra-pure water is provided by Watsons Group(Hangzhou,China).SZDP was made by the laboratory according to the prescription and technology.
The preparation process of SZDP is as follows:the four herbs ofChoerospondiasAxillaris(voucherspecimennumber:GDPUTCM-CA2019-001),Salvia Miltiorrhiza(voucher specimen number:GDPUTCM-SM2020-601),Panax Ginseng(voucher specimen number:GDPUTCM-PG2020-501)andDalbergia Odorifera(voucher specimen number:GDPUTCM-DO2020-601)are decoction and alcohol precipitation,concentrated and dried to obtain extract powder.It was mixed with PEG6000 and addedLiquidimibar Orientalis(voucher specimen number:GDPUTCM-LO2019-040)andCinnamomum Camphora(voucher specimen number:GDPUTCM-CC2020-907).Voucher specimens were deposited at the School of Traditional Chinese Medicine,Guangdong Pharmaceutical University.After melting together at 80 °C,and prepared into dropping pills.These herbs were identified as genuine by Professor Hong-Yan Ma from the TraditionalChineseMedicineAuthentication,Guangdong Pharmaceutical University;and they were stored in a display cabinet of medicinal materials in the School of Traditional Chinese Medicine,Guangdong Pharmaceutical University,protected from light.
Animal experiment
Specific pathogen free male Sprague-Dawley rats(120-150 g)were provided by the Guangdong Medical Laboratory Animal Center(approval number:SCXK-Guangdong-2018-0002).Rats were housed in a specific pathogen free grade laboratory(25 ± 1 °C,48%-50%relative humidity)with a 12 hours dark/light cycle.Animal feed and drinking water were freely given.The experiment started after 7 days of adaptive feeding.The animal protocol was approved by the Animal Ethics Committee of Guangdong Pharmaceutical University(approved number:gdpulacspf2017353).
Before this experiment,we set a series of administration doses through the pre-experiment.We selected the administration dose with the most compounds in the plasma as the dose of this experiment.Sprague-Dawley rats were divided into control group,SZDP group(n= 6 each)and different blood collection time point SZDP group(including 0.5,1,2,3,4 hours)(n=8).SZDP was administered orally at a dose of 30 g/kg/d for 5 days[15],and twice a day.The control group was given an equal volume of pure water.The dose of SZDP administration group was 42 times the clinical dose.The formula was based on this dose conversion:logS = 0.8762 + 0.698 logW(S:body surface area,cm2;W:weight,g)[16].The body weight of humans is calculated as 70 kg,and the body weight of rats is calculated as 0.2 kg.
Preparation method of samples
Preparation of SZDP samples.The pulverized powder of SZDP(1 g)was extracted by sonicating with 20 mL of 80% methanol for 30 min.After filtration,it was prepared into a test solution with a concentration of 10 mg/mL,and filtered with a 0.22 μm organic phase microporous membrane before use.
Preparation of mixture solution of the reference standards compounds.A certain amount of 13 reference compounds were dissolved in methanol to form a mixed standard(about 8 μg/mL for each compound),then stored at 4 °C.The solution was centrifuged at 13,000 rpm for 10 min before analysis.
Preparation of plasma samples.Before the last day of dosing,all animals were fasted for 12 hours,but drinking water was allowed.On the 5thday,the optimal blood collection time was determined by different blood collection time points(0.5,1,2,3,4 hours)SZDP group.Next,the control group and SZDP group were anesthetized with isoflurane.Under the conditions of the above optimal blood collection time point,the retro-orbital venous plexus was used to collect blood.Then the hepatic portal vein and abdominal aorta were used to collect blood.All blood was centrifuged at 4,000 rpm for 10 min at 4 °C and the supernatant was taken to obtain plasma.The plasma was stored in the refrigerator at-80°C for later use.
LC-MS analysis conditions
The type of column is ACQUITY UPLCBEH C18column(2.1 mm×100 mm,1.7 μm).Gradient elution was carried out with water with 0.1 %(volume ratio)formic acid in water(solvent A)and acetonitrile(solvent B).Gradient elution was as follow:0-1 min,5% B;1-10 min,5%-20% B;10-20 min,20%-26% B;20-21 min,26%-32% B;21-22 min,32%-50% B;22-24 min,50%-60% B;24-32 min,60%-80% B;32-34 min,80%-86% B;34-36 min,86%-90% B;36-37 min,90% B.The flow rate was 0.3 mL/min,and the sample injection volume was 5 μL.
LC-MS analysis was performed with a U3000 UHPLC-Q-Exactive Orbitrap MS(Thermo Fisher Scientific Corp.,Waltham,Massachusetts,USA).The ion source is the electrospray ionization source,which is 350 °C.Parameters were set as follows:positive and negative ion switching mode in the range of mass-to-charge ratio(m/z)100-1,500 was set for the MS analysis;the collision energies were 20,30 and 60 eV;the sheath gas flow rate was 10 arbitrary units and its temperature was 300 °C;the auxiliary gas flow rate was 45 arbitrary units.
Data analysis and manual verification
The mass spectrum data was imported into XCalibur(version 4.0)for qualitative analysis of the chemical components of SZDP.First,the chemical formula was calculated according to the exact mass of the molecular ion peak of the compounds,and compared the literature and reference materials by secondary mass spectrometry(MS/MS)fragments.Then,the LC-MS data of SZDP were analyzed by comparing the PubChem(https://pubchem.ncbi.nlm.nih.gov/)[17]and Chemspider(http://www.chemspider.com/)[18]databases.Next,the cleavage method was further compared with MS/MS data to identify the compounds.
Target prediction
Studies have proved that the compounds absorbed into the plasma play an essential role in the treatment of diseases.Therefore,this study focused on the compounds identified from SZDP by LC-MS.Firstly,we input the chemical names of the prototype components in the blood component into the Traditional Chinese Medicine Systems PharmacologyDatabaseandAnalysisPlatform(https://tcmspw.com/tcmsp.php)[19]database and the Integrative Pharmacology-based Research Platform of Traditional Chinese Medicine(TCMIP)(http://www.tcmip.cn/TCMIP/index.php/Home/)[20]to obtain the targets of the prototype components in the blood component.The parameters are set to default values and all targets are collected.These targets were calibrated against Uniprot(https://www.uniprot.org/)[17]database.Secondly,we searched the GeneCards(https://www.genecards.org/)[21],PharmGKB(https://www.pharmgkb.org/)[17]and TCMIP databases with“coronary heart disease” as the keyword,and selected targets with a score of more than 10.We used the Uniprot database to correct the obtained targets.Finally,the intersection of known compound targets and disease targets are used as common targets for drug and disease by Venn diagrams.
Protein-protein interaction(PPI)
PPI can reflect the interaction relationship between proteins and predict the potential functions of proteins and potential disease genes.The PPI network of common targets was established through STRING(https://string-db.org)with parameters set to“homo sapiens”[17].
Key targets and compounds
We imported the PPI network into Cytoscape software(version 3.7.1)for visual analysis,and screened common targets based on the six most important topological parameters of betweenness centrality,closeness centrality,degree centrality,eigenvector centrality,local average centrality,and network centrality point of the core targets.These six parameters could reflect the importance of the topological attributes of nodes in the network.We choose the median value that satisfies more than six parameter values at the same time as key targets.The prototype components in plasma corresponding to these targets are found by the correspondence between compounds and targets,and key compounds are obtained.
Enrichment of Gene Ontology(GO)and Kyoto Encyclopedia of Genes and Genomes(KEGG)pathways
By the “BiocManager” package of R software(version 4.1.0),we performed GO and KEGG enrichment analysis on common targets.The common targets were input into the R software,and adopted gene ID conversion.The GO information and KEGG pathway information of the common targets are enriched by “ClusterProfile”,“Enrichplot” and“Pathview” packages.Analysis and visualization were performed using the R software,with parameters at default values,and statistical significance thresholds for GO and KEGG enrichment analyses were set atP<0.05.
Network and analysis of “TCM-components-targets-pathway-disease”
The compounds entering the plasma,common targets,top ten KEGG pathways,disease and TCM were imported into Cytoscape software to construct a “TCM-components-targets-pathway-disease”network.
Molecular docking technology validation
Key targets and compounds were molecular docked through the SYSAL(version 2.1.1)to verify the interaction between the compounds and the targets.The structure of compounds can be downloaded from the PubChem database and optimized by ChemDraw software(version 20.0).The three-dimensional structure of the key target proteins can be obtained from Protein Data Bank(http://www.rcsb.org/)[22]for docking with core targets.The results of the docking were visualized and analyzed by Pymol software(version 1.0.0).
Results
Identification of the prototype components of SZDP
The positive and negative base peak intensity current diagram of SZDP was shown in Figure 1.The composition of SZDP has 72 chemical components inferred,whose attributions and the identification results were listed in Table 1.In addition,we display several typical identification methods of compounds,including tanshinones,phenolic acids,ginsenosides and organic acids(Figure 2).In addition,the mass spectra of the 13 standard substances are shown in Supplementary Material Figure 2.
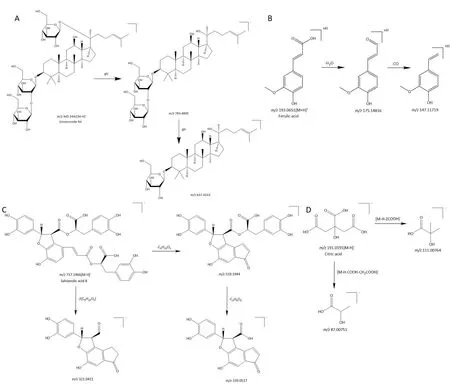
Figure 2 Analysis of cleavage pathways of typical compounds in SZDP.(A)Analysis of the cleavage pathway of ginsenoside Rd in ginsenosides;(B)analysis of the cleavage pathway of ferulic acid in phenolic acids;(C)analysis of the cleavage pathway of salvianolic acid B in phenolic acids;(D)analysis of the cleavage pathway of citric acid in organic acids.SZDP,Shenzao dripping pills; m/z,mass-to-charge ratio.
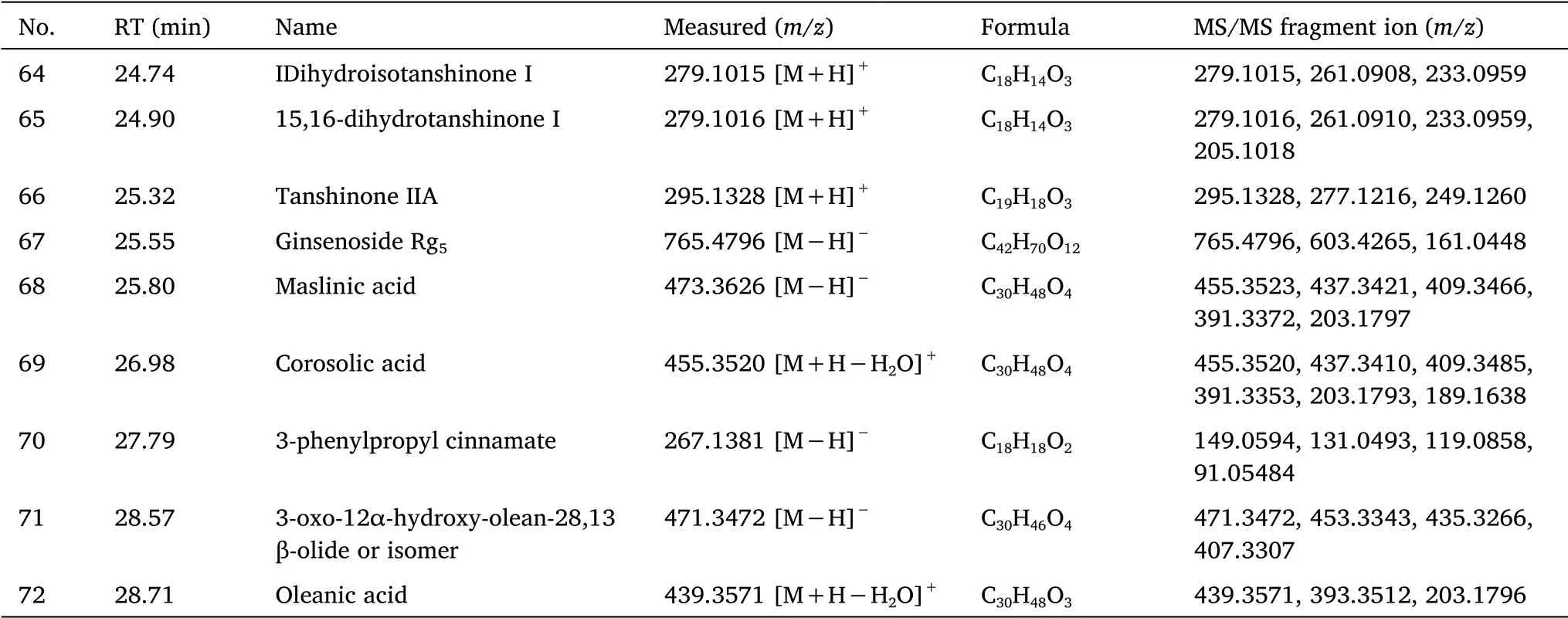
Table 1 Identification of the chemical components of SZDP by UHPLC-Q-Exactive Orbitrap MS(Continued)
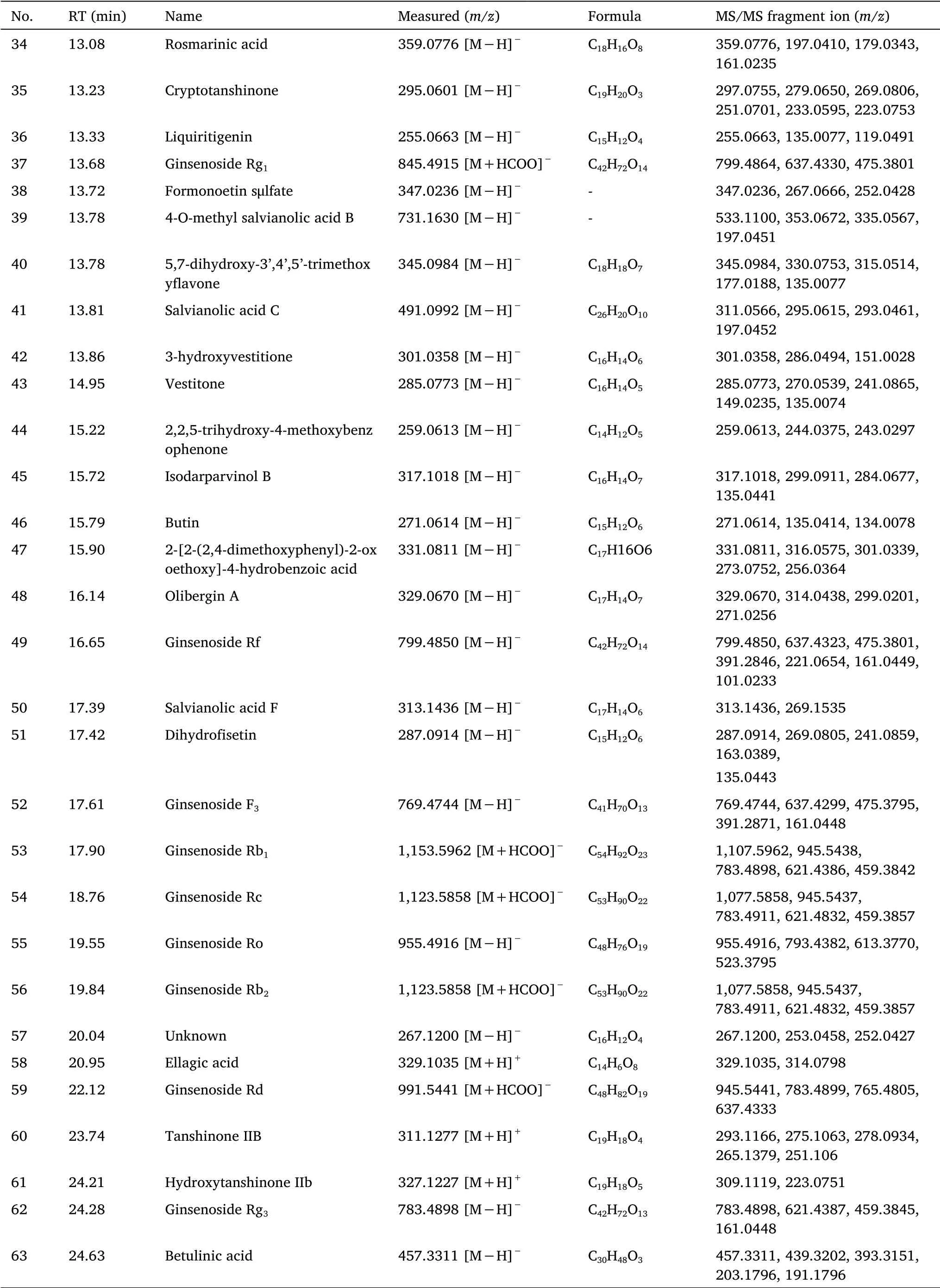
Table 1 Identification of the chemical components of SZDP by UHPLC-Q-Exactive Orbitrap MS(Continued)
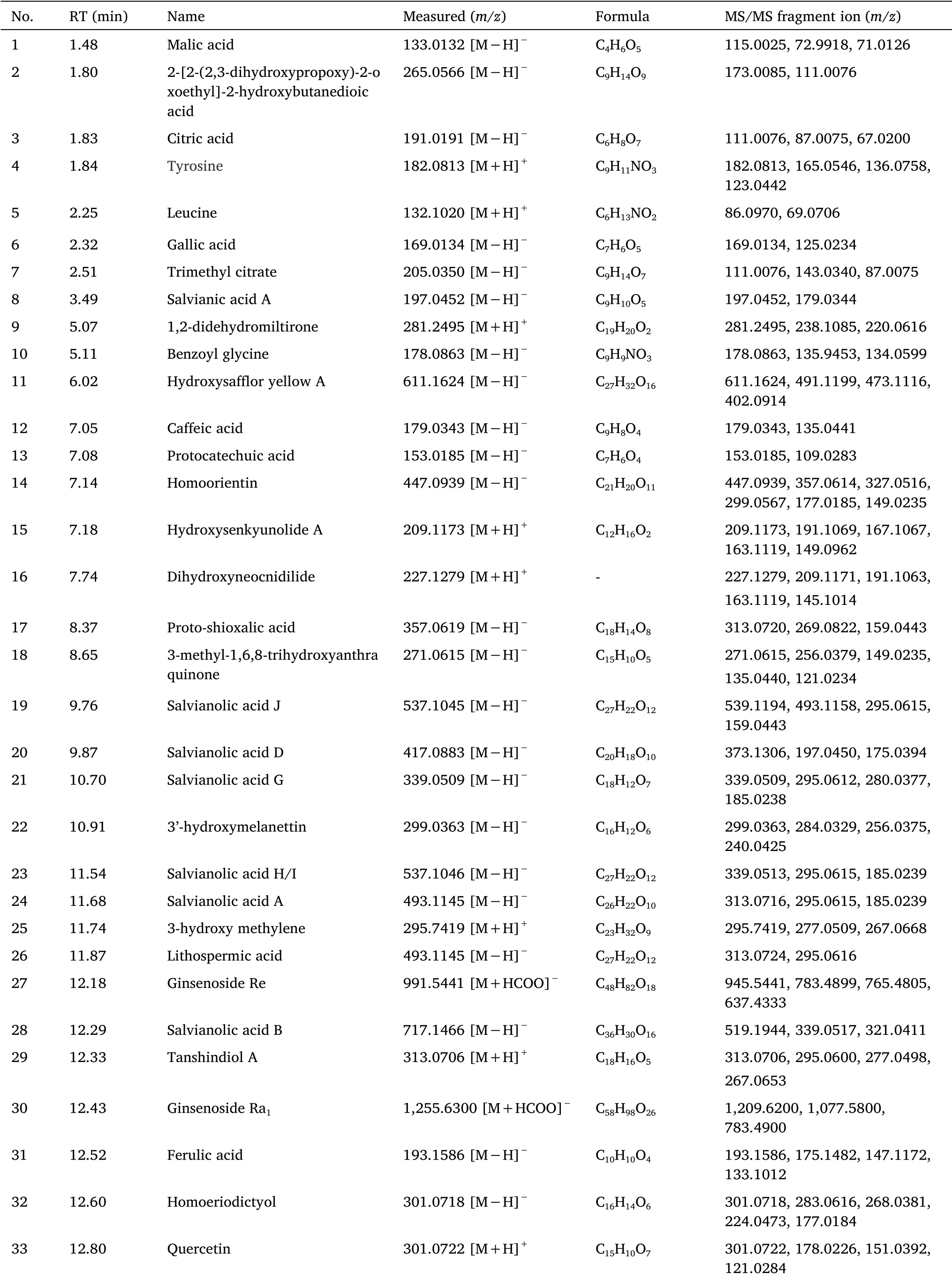
Table 1 Identification of the chemical components of SZDP by UHPLC-Q-Exactive Orbitrap MS
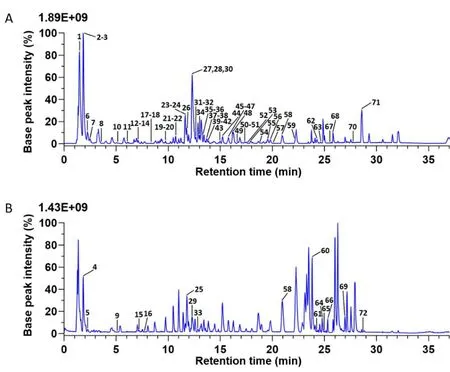
Figure 1 The base peak intensity chromatograms of SZDP by UHPLC-Q-Exactive Orbitrap MS.Negative ion mode(A)and positive ion mode(B);the numbers marked in the figure correspond to the compound numbers in Table 1.SZDP,Shenzao dripping pills;UHPLC-Q-Exactive Orbitrap MS,ultra-high performance liquid chromatography-quadruple-Exactive Orbitrap mass spectrometry.
Identification of tanshinones.The fragmentation of compound 66 was similar to that of the reference substance tanshinone ⅡA.The molecular ion peak[M+H]+wasm/z295.1328(C19H18O3+,retention time(tR)= 25.32 min),and the two-stage mass spectrometry(MS2)fragmentm/z277.1216 was[M+H-H2O]+andm/z249.1260 were[M+H-H2O-CO]+.Thus,compound 66 was inferred to be tanshinone IIA.
The molecular ion peak[M-H]-of compound 29 wasm/z313.0706(C18H16O5-,tR= 12.33 min),and the MS2fragmentm/z295.0600 was[M+H-H2O]-,m/z277.0498 It was[M+H-2H2O]-andm/z267.0653 was[M+H-H2O-CO]-.Thus,compound 29 was inferred to be tanshinol A.
The molecular ion peak[M+H]+of compound 60 wasm/z311.1277(C19H18O4-,tR= 23.74 min),the MS2fragmentm/z293.1166,m/z278.0934,m/z265.1379 andm/z251.1060 were[M+H-H2O]+,[M+H-H2O-CH3]+,[M+H-H2O-CO]+and[M+H-H2O-CO-CH2]+.Therefore,compound 60 was inferred to be tanshinone IIB.
The fragmentation of compound 64 was similar to that of the reference substance dihydrotanshinone I.The molecular ion peak[M+H]+wasm/z279.10132(C18H14O3+,tR= 24.74 min),and the MS2fragmentsm/z261.0908 andm/z233.0959 was[M+H-H2O]+and[M+H-H2O-CO]+.By comparing with the reference peak,compound 64 was inferred to be dihydrotanshinone I.
Identification of ginsenosides.The fragmentation of compound 54 was similar to that of the reference ginsenoside Rc,the molecular ion peak[M+HCOO]-wasm/z1,123.5858(C53H90O22+,tR=18.76 min),and the MS2fragmentsm/z945.5437 andm/z621.4832 were[M-glc-H]-and[M-ara-2glc-H]-.By comparing with the reference peak,compound 54 was inferred to be ginsenoside Rc.
The fragmentation of compound 56 was similar to that of reference ginsenoside Rb2.The molecular ion peak[M+COOH]-wasm/z1,123.5858(C53H90O22-,tR= 19.84 min),and the MS2fragmentm/z1,077.5858,m/z945.5437,m/z621.4832 andm/z459.3857 were[M-H]-,[M-ara-H]-,[M-ara-2glc-H]-and[M-ara-3glc-H]-.Thus,compound 56 was inferred to be ginsenoside Rb2.
The molecular ion peak[M+COOH]-of compound 27 wasm/z991.5441(C48H82O19-,tR= 12.18 min),and the MS2fragmentsm/z945.5441,m/z783.4899 andm/z637.4333 were respectively[M-H]-,[M-glc-H]-and[M-glc-rha-H]-.Thus,compound 27 was inferred to be ginsenoside Re.
The fragmentation of compound 59 was similar to that of the reference ginsenoside Rd.The molecular ion peak[M+COOH]-was 991.5441(C48H82O19-,tR= 22.12 min),and the MS2fragmentsm/z945.5441 andm/z783.4899.It was[M-H]-and[M-glc-H]-.Therefore,compound 59 was inferred to be ginsenoside Rd.
Identification of Phenolic acids.The molecular ion peak[M-H]-of compound 28 wasm/z717.1466(C36H30O16-,tR= 12.29 min),the MS2fragmentm/z519.1944 was[M-H-C9H10O5]-,m/z321.0411 was[M-2C9H10O5]-andm/z339.0517were[M-HC9H10O5-C9H8O4]-.Thus,compound 28 was speculated to be salvianolic acid B.
The molecular ion peak[M-H]-of compound 50 wasm/z313.1436(C17H14O6-,tR= 17.39 min),and the MS2fragmentm/z269.1535 was[M+H-CO2]-.Thus,compound 50 was speculated to be salvianolic acid F.
The molecular ion peak[M-H]-of compound 21 wasm/z339.0509(C18H12O7-,tR= 10.70 min),and the MS2fragmentm/z295.0612 was[M+H-CO2]-andm/z280.0377 It was[M+H-CO2-CH3]-.Therefore,compound 21 was speculated to be salvianolic acid G.
The molecular ion peak[M-H]-of compound 23 wasm/z537.1046(C27H20O12-,tR= 11.54 min),the MS2fragmentm/z339.0564 was[M-H-C9H10O5]-andm/z295.06146 was[M-H-C9H10O5-CO2]-.Thus,compound 23 was speculated to be salvianolic acid H/I.
The molecular ion peak[M-H]-of compound 41 wasm/z491.0992(C26H20O10-,tR= 13.81 min),and the MS2fragmentm/z295.0615 was[M-H-C9H10O5]-.Therefore,compound 41 was speculated to be salvianolic acid C.
Identification of Flavonoids.The fragmentation of compound 36 was similar to that of the reference glycyrrhizin.The molecular ion peak[M-H]-wasm/z255.0663(C15H12O4-,tR= 13.33 min),and the MS2fragments werem/z135.0077 andm/z119.0491 was consistent with the secondary fragments of the reference substance.By comparing with the reference substance,compound 36 was predicted to be liquiritigenin.
The molecular ion peak[M-H]-of compound 51 wasm/z287.0914(C15H12O6-,tR= 17.42 min),and the MS2fragmentm/z269.0805 was[M-H-H2O]-.Thus,compound 51 was predicted to be dihydrofisetin.
The molecular ion peak[M-H]-of compound 22 wasm/z299.0363(C16H12O6-,tR= 10.91 min),the MS2fragmentm/z284.0329 was[M-H-CH3]-,Thus,compound 22 was predicted to be 3’-hydroxymelanettin.
The molecular ion peak[M-H]-of compound 45 wasm/z317.1018(C16H14O7-,tR= 15.72 min),and the MS2fragmentm/z299.0911 was[M-H-H2O]-,Thus compound 45 was predicted to be isodarparvinol B.
The molecular ion peak[M-H]-of compound 48 wasm/z329.0670(C17H14O7-,tR= 16.14 min),and the MS2fragmentsm/z314.0438,m/z99.0201 andm/z271.0256 were respectively[M-H-CH3]-,[M-H-2CH3]-and[M-H-2CH3-H2O]-,Therefore,compound 48 was predicted to be olibergin A.
Identification of organic acids and ester compounds.The molecular ion peak of compound 3[M-H]-wasm/z191.0191(C6H8O7
-,tR= 1.83 min),the MS2fragmentm/z111.0076 was[M-H-2COOH]-andm/z87.0075was[M-H-COOH-CH2COOH]-.Thus,compound 3 was speculated to be citric acid.
The molecular ion peak of compound 1[M-H]-wasm/z133.0132(C4H6O5
-,tR= 1.48 min),the MS2m/z115.0025 was[M-H-H2O]-andm/z71.0126 was[M-H-H2O-CO2]-.Thus,compound 1 was speculated to be malic acid.
The molecular ion peak[M-H]-of compound 6 wasm/z169.0134(C7H6O5,tR= 2.32 min),and the MS2fragmentm/z125.0234 was[M-COOH]-.Compound 6 was determined by comparison with the reference substance.It was gallic acid.
Compounds 2 and 7 have the same ion fragmentsm/z111.0076 andm/z87.0075 as citric acid.It was speculated that compounds 2 and 7 were derivatives of citric acid,and the molecular ion peak of compound 7[M-H]-wasm/z205.0350(C9H14O7-,tR= 2.51 min),the molecular weight was 14 Dalton more than citric acid,it was inferred that compound 7 was methyl citrate,and the molecular ion peak of compound 2[M-H]-wasm/z265.0566(C9H14O9-,tR=1.80 min),after consulting the HMDB library comparison.Compound 2 was citrate monoglyceride.
The molecular ion peak[M+H]+of compound 12 wasm/z179.0343(C9H8O4+,tR= 7.05 min),and the MS2fragmentm/z135.0441 was[M+H-H2O-CO]+.Thus,compound 12 was speculated to be caffeic acid.
The molecular ion peak[M+H]+of compound 31 wasm/z193.1586(C10H10O4+,tR= 12.52 min),and the MS2fragmentm/z175.1482was[M+H-H2O]+,m/z147.1172was[M+H-H2O-CO]+.Thus,compound 31 was speculated to be ferulic acid.
The molecular ion peak[M-H]-of compound 13 wasm/z153.0185(C7H6O4-,tR= 7.08 min),and the MS2fragmentm/z109.0283 was[M+H-CO]+.Therefore,compound 13 was speculated to be original catechin.
Identification of amino acids.The quasi-molecular ion peak of amino acid compounds was[M+H]+peak,secondary fragments were easy to produce characteristic fragment ions without HCOOH and NH3,and the molecular ion peak of compound 5 was[M+H]+It wasm/z132.1020(C6H13NO2+,tR= 2.25 min),the MS2fragmentm/z86.0970 was[M+H-HCOOH]+andm/z69.0706was[M+H-HCOOH-NH3]+Therefore,compound 5 was speculated to be leucine.
The molecular ion peak[M+H]+of compound 4 wasm/z182.0813(C9H11NO3+,tR= 1.84 min),and the MS2fragmentm/z165.054 was[M+H-NH3]+,m/z136.0758 was[M+H-HCOOH]+andm/z123.0442 was[M+H-HCOOH-NH3]+.Thus,compound 4 was speculated to be tyrosine.
Identification of chemical components of SZDP entering the plasma
Through multiple time points investigation of the blood in the SZDP group,we determined that the substance that appeared in the plasma was greatest after 1 hour after administration of the TCM.As a result,the time frame for collecting blood was established at 1 hour.
The sample was analysed using LC-MS after the rat’s plasma was collected,and a positive and negative ion base peak intensity chart was constructed(Supplementary Material Figure 1).The plasma components of the SZDP group were analysed by comparing the chemical components of SZDP to a reference substance and validating them with a database.As stated in Supplementary Material Table 1,a total of 33 compounds were discovered.There were 24 metabolites and 9 prototype components among all the components.
Identification of metabolic components of SZDP in plasma
The molecular ion peak[M-H]-of compound 24 wasm/z343.1405(C15H12O6-,tR= 17.42 min),and its MS2fragments werem/z175.0242,m/z167.1071,m/z113.0234,m/z85.0283,MS2fragmentsm/z175.0242 andm/z113.0234 were characteristic ion fragments of the uronic acid group of glucose.It was speculated thatm/z167.1071 was vanillic acid,which was the methyl group of protocatechuic acid.Thus,compound 24 was speculated to be the metabolite vanillic acid-glucuronic acid.
The molecular ion peak of metabolite 6[M-H]-was 246.9920(C8H8O7S-,tR= 4.15min),and its MS2fragments werem/z203.0112 andm/z167.0343,and MS2fragments werem/z167.0343.It was obtained by removing a sulfuric acid group from the parent ion,m/z167.0343 should be the methylation product of protocatechuic acid,the ion peak of vanillic acid.Thus,compound 6 was speculated to be the metabolite vanillic acid-sulfonic acid.
The molecular ion peak[M-H]-of compound 13 wasm/z477.1049,the MS2fragment wasm/z301.0723(C24H30O10-,tR= 9.19 min),and it was identified as the metabolite quercetin-gluconic acid by consulting the database.
Compounds 12,14,15,16,17 were glucuronate conjugates because their[M-H]-exhibit the characteristic neutral loss of glucuronic acid and produce[Aglycon-H]-corresponding to their flavonoid skeleton.
Network pharmacology analysis
Matching of prototype components in plasma and disease targets from two databases.The Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform and TCMIP databases were used to look for targets for the 24 prototype components found in the plasma,and the UniProt protein database was used to rectify the results.A total of 150 targets were acquired in the end.A total of 3,592 CHD targets were retrieved from GeneCards,PharmGKB,and TCMIP databases,with 90 overlapping targets produced by matching the CHD targets with the SZDP compound targets(Figure 3A).
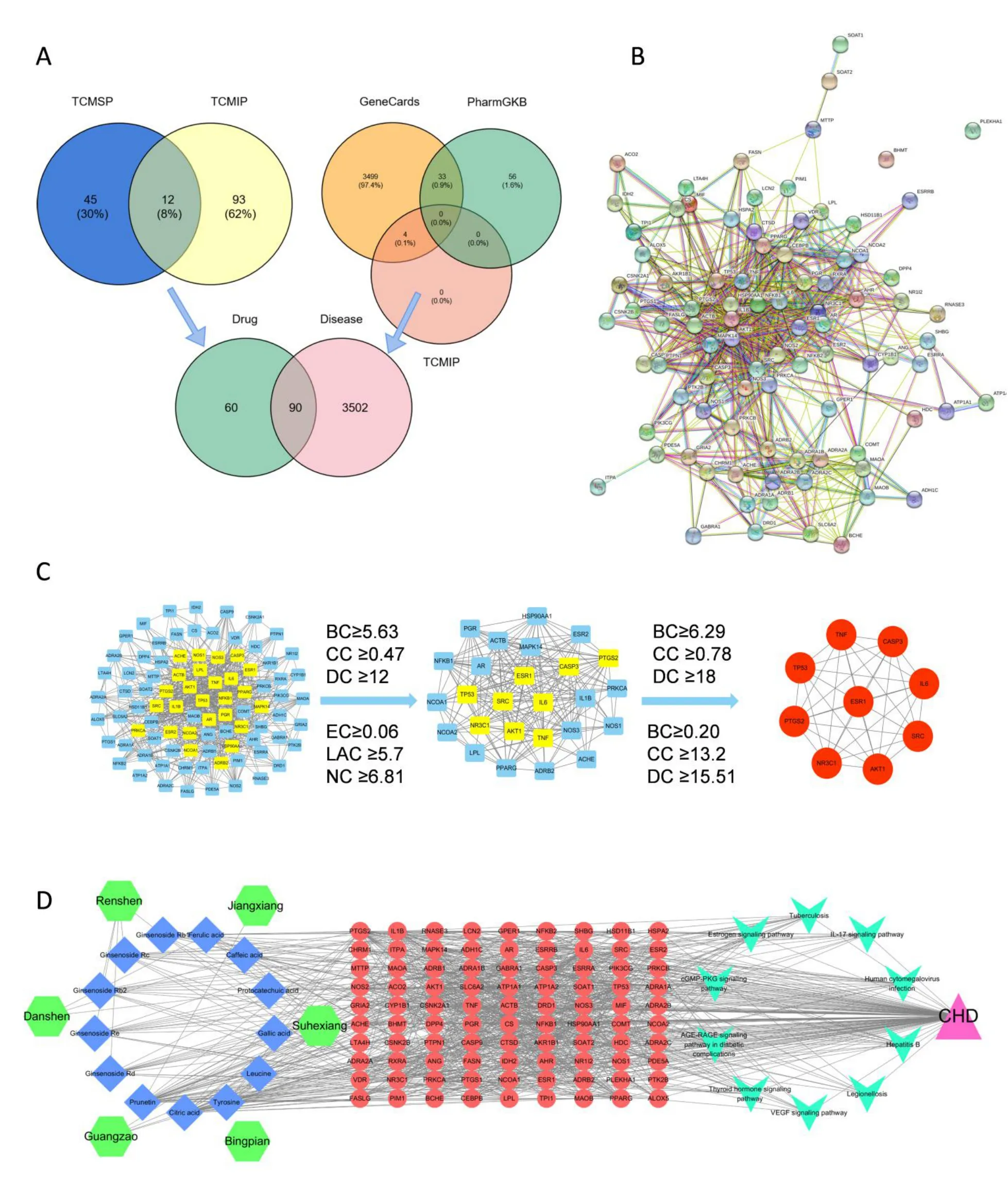
Figure 3 Network pharmacology analysis diagram of SZDP entry into plasma components.(A)The common target for drug and disease by Venn diagrams.Each circle represents the marked database,the sum of the numbers in them is the total number of targets in the database,and the number at the intersection of the two circles represents the number of targets shared by the two parts.(B)The common target PPI network for drug and disease.Each small circle represents a protein,and their interconnected lines represent interactions between them.(C)Network diagram after filtering by topological parameters.The blue squares represent the targets below the parameter value,the yellow squares represent the targets above the parameter value,which are selected for secondary screening,and the red circles represent the important targets left after the final screening.(D)The network of “TCM-components-targets-pathway-disease”.Green regular hexagons represent different Chinese medicines in SZDP;blue diamonds represent key compounds in SZDP;red circles represent common targets of drug and disease;cyan arrow shapes represent higher-scoring pathways;pink triangle represents coronary heart disease.SZDP,Shenzao dripping pills;PPI,protein-protein interaction;TCMSP,Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform;TCMIP,Integrative Pharmacology-based Research Platform of Traditional Chinese Medicine;BC,betweenness centrality;CC,closeness centrality;DC,degree centrality;EC,eigenvector centrality;LAC,local average centrality;NC,network centrality;CHD,coronary heart disease.
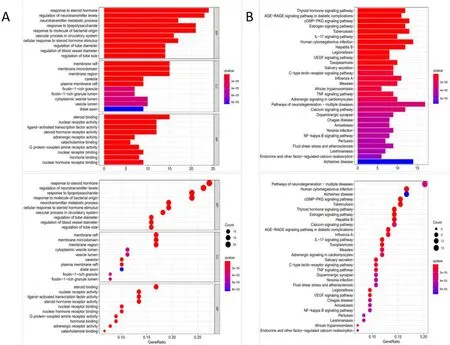
Figure 4 Enrichment analysis of GO and KEGG pathways.(A)GO enrichment analysis(including biological process,molecular function and cellular component).(B)Enrichment analysis of GO pathways.The abscissa represents the proportion of the target,and the ordinate represents the ID of the enriched word.The ordinate refers to the number of genes involved in each pixel.Colors ranging from red to blue represent Q-values from small to large,which means that the depth of the color is proportional to the degree of enrichment.GO,Gene Ontology;KEGG,Kyoto Encyclopedia of Genes and Genomes.
STRING analysis and PPI network of the overlapping targets.STRING analysis was used to compare 90 overlapping targets and produce a PPI network(Figure 3B).We then created a visualized network using the Cytoscape software.The PPI network comprises 656 edges and 88 nodes.After analysing the topological properties of the PPI network with the CytoNCA plug-in,26 significant targets were discovered(Figure 3C).After the second screening phase,nine promising targets were discovered.Through the correlation of compounds and targets,we found 13 essential compounds,including ginsenoside Rb1,ginsenoside Rb2,ginsenoside Rc,ginsenoside Rd,ginsenoside Re,protocatechuic acid,caffeic acid,ferulic acid,citric acid,tyrosine,gallic acid,and prunetin.
GO and KEGG pathways analysis.We used the GO and KEGG pathways enrichment analyses to look at the common targets indicated above.There were overall 1,601 biological function annotations in GO,including 1,349 biological processes(BP),61 cell components(CC),and 101 molecular functions(MF).According to BP enrichment analysis,the SZDP compound interacts with steroid hormones,alters neurotransmitter regulation,neurotransmitter metabolism,lipopolysaccharide response,bacteria-derived molecule reaction,and,most intriguingly,vascular processes in the circulatory system.The 101 co-identified MF encompass steroid receptor activity,nuclear receptor activity,ligand-activated transcription factor activity,steroid hormone receptor activity,adrenergic receptor activity,catecholamine binding,G protein-coupled amine receptor activity,nuclear receptor binding,and hormone binding.CC is linked to membrane raft,membrane microdomain,membrane region,caveola,and plasma membrane raft.Based on the adjustedP-value,we then choose the top 10 BP,CC,and MF biological function data for visual examination.KGGG pathways analysis showed 149 pathways including thyroid hormone signaling pathway,cGMP-PKG signaling pathway,estrogen signaling pathway,IL-17 signaling pathway,VEGF signaling pathway,and so on.Based on theirP-value ranking,the top 30 are picked for visual presentation with bar plots.Using the results of the KEGG enrichment analysis,the top 30 pathways are picked for the visual presentation of examples(Figure 4).
Establishmentandanalysisofthenetworkof“TCM-components-targets-pathway-disease”.We imported the compounds entering the plasma,common targets,top ten KEGG pathways,disease and TCM into Cytoscape software to construct a“TCM-components-targets-pathway-disease” network(Figure 3D).The synergistic effect of “multi-components,multi-targets,multi-pathways” can be seen in this figure,indicating that SZDP compounds are interfering with CHD.
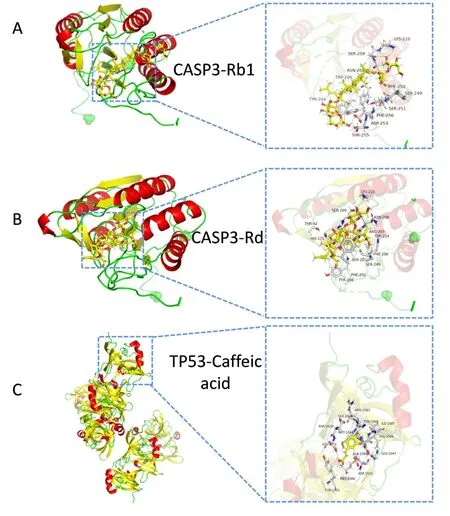
Figure 5 Results of molecular docking.(A)Docking diagram of CASP3 target and ginsenoside Rb1;(B)Docking diagram of CASP3 target and ginsenoside Rd;(C)Docking diagram of TP53 target and caffeic acid.On the left is the overall graph of the docking results;on the right,some details of the docking result are shown.TP53,cellular tumor antigen p53;CASP3,caspase-3.
Verification by molecular docking
Molecular docking technology was used to investigate the 9 key targets and the 13 essential compounds in order to determine the degree of interaction between the molecule and the target.The overall score for docking affinity was computed using the SYBYL software(version 2.1.1).The higher the score,the stronger the docking affinity(Supplementary Material Table 2).Higher docking scores for proto-oncogene tyrosine-protein kinase Src(SRC),prostaglandin G/H synthase 2(PTGS2),ethylene-responsive transcription factor ESR1(ESR1),nuclear receptor subfamily 3 group C member 1(NR3C1),potassium channel AKT1(AKT1),tumor necrosis factor(TNF),and caspase-3(CASP3)with their respective molecules indicate that the compound may function through these targets.Using the PyMOL program(version 1.0),we provide molecular docking data of the above molecular(Figure 5).
Discussion
In this study,the chemical compositions of SZDP were analyzed by UHPLC-Q-Exactive Orbitrap MS,and it was established that a reliable SZDP chemical compositions analysis method.A total of 72 compounds were discovered.This provides a research basis for studying the material basis and medicinal effects of SZDP and clarifying the mechanisms of these effects.In addition,we have also identified the chemical components of SZDP in plasma.On the one hand,it can provide evidence for the research on the material basis of SZDP.On the other hand,it can also correlate the important chemical components of SZDP with pharmacological effects.This has crucial implications for our understanding of SZDP’s mechanisms of action and identifying the pathway linked to CHD therapy.
The majority of research implies that the compounds that enter the plasma are active compounds[13,23].As a result,we are focusing on the SZDP components found in plasma.The possible active components of SZDP were discovered to be tanshinones,flavonoids,saponins,organic acid,and phenolic acid.The majority of these components have been shown to lower myocardial inflammation,oxidative stress,and platelet aggregation[24-28].Ginsenosides and their degradation products have been shown to significantlyreduceinfarctsizeaftermyocardial ischemia-reperfusion,reduce plasma viscosity and platelet aggregation levels,increase glutathione peroxidase and the activity of superoxide dismutase levels,and protected cardiomyocytes from myocardial ischemia-reperfusion injury[9].Tanshinone IIA can effectively reduce the levels of total cholesterol,triglycerides,and low-density lipoprotein in patients with CHD,further reduce body fat levels,and prevent CHD[29].Salvianolic acid B and ferulic acid act synergistically to promote angiogenesis by regulating the VEGF signaling pathway in zebrafish[30].In addition,we investigated the 9 SZDP metabolites and discovered that various metabolic pathways,such as glucuronidation,demethylation,and sulfonation,had changed.We observed that phase II metabolites were more frequently detected than phase I.Most of the phase I metabolites are lower than the detection limit in our MS.This might imply that phase I metabolites have minimal influence on disease.
Based on the results of network pharmacology and molecular docking investigations,we presented our unique finding of SZDP’s mechanisms of action in CHD.We determined that the drug and the disease had 90 target proteins in common.These target proteins are mostly involved in signaling networks such as the estrogen,IL-17,and VEGF signaling pathways.The 9 key target proteins found in our follow-up analysis include SRC,AKT1,and TNF.The 13 prototype components of SZDP that matched these key target proteins were ginsenoside Rb1,ginsenoside Rb2,tanshinone IIA,citric acid,and gallic acid.Studies have shown that ginsenoside Rb1can reduce the damage of myocardial ischemia-reperfusion by reducing the expression levels of CASP3 and TNF-α[31].Another study has also demonstrated that ginsenoside Rd inhibits the activities of CASP3 and caspase-9,increases phosphorylated AKT and glycogen synthase kinase-3 beta,and reduces plasma creatine kinase/lactate dehydrogenase levels after myocardial ischemia-reperfusion[32].Furthermore,the targets we examined have a significant role in the development of CHD.For example,AKT1 promotes inflammation in the early stages of hypertensive heart disease,improving cardiac fibrosis and protecting the cardiovascular system[33].SRC had been used as a biomarker to diagnose CHD[34].When compared to the control group,the SRC of the patients has significantly changed,indicating that CHD is closely correlated to the SRC protein.TNF is a protein that contributes to inflammation.According to the research,circulating TNF is linked to endothelial damage and thickening of the endothelium basement membrane in patients with CHD[35].Increases in TNF level might worsen endothelial dysfunction.TP53,CASP3,ESR1,NR3C1,and IL6 are among the major proteins identified in this investigation,and they are all linked to the cardiovascular system[36-39].These studies provide a worthy reference for the study of the pharmacological mechanisms of SZDP.
Our study had some limitations.Firstly,due to the complexity of TCM components,it is impossible to obtain all molecules using mass spectrometry.Therefore,some compounds of SZDP need to be identified by other technical methods.Secondly,we detected compounds in plasma by the chromatograph-mass spectrometer in the early stage and found that the compounds obtained were few,and the reliability of the data was not high.Therefore,no network pharmacology studies were performed in conjunction with volatile components in plasma.Furthermore,our network pharmacology and molecular docking studies were not validated in vitro or in vivo.Therefore,the pharmacological mechanisms of SZDP need further investigation.
Finally,using UHPLC-MS and network pharmacology analysis,the active compounds of SZDP were found,and the mechanisms of multi-component,multi-target,multi-pathway synergistic activity by SZDP to treat CHD was revealed.This also provides an idea for the rapid detection of TCM components and the treatment of CHD.
Conclusion
This study combines three techniques of LC-MS,network pharmacology and molecular docking to reveal the main active components of SZDP and make a prediction for the main pharmacological effects of SZDP in the treatment of CHD.SZDP mainly affects estrogen,IL-17 and VEGF signaling pathways through ginsenosides and ferulic acid,and then has a certain therapeutic effect on CHD.
 Traditional Medicine Research2022年3期
Traditional Medicine Research2022年3期
- Traditional Medicine Research的其它文章
- Traditional Chinese medicine:an important broad-spectrum anti-coronavirus treatment strategy on COVID-19 background?
- Anti-asthmatic mechanism of the Huashanshen dripping pill via suppressing contraction of the airway smooth muscle
- Application of network pharmacology in the prevention and treatment of COVID-19 by traditional Chinese medicine
- Pharmacological efficacy of the traditional Chinese medicinal formula Kun-Tai-1A in the treatment of letrozole-induced polycystic ovary syndrome
- Recent advances in research on natural product inhibitors of SREBPs
- Effects of Qingwen Baidu decoction on coagulation and multiple organ injury in rat models of sepsis
