小RNA测序揭示miRNAs在油棕合子胚发育中的作用
张安妮 曹红星 陈萍 金龙飞




摘 要:油棕是世界上產油效率最高的热带木本油料作物,其棕榈油产量与合子胚发育密切相关。油棕的合子胚发育是一个系统且复杂的生物过程,其中涉及许多基因的精确调控。MicroRNAs(miRNAs)是重要的信号分子,调节植物的各种发育过程。一些miRNAs已经被整合到基因调控网络中协调植物胚发育的可塑性,但对于其在油棕合子胚发育过程中的作用知之甚少。本研究采用小RNA测序对油棕S1(早期)、S2(中期)和S3(晚期)的合子胚进行高通量测序分析,鉴定了6个已存在的和334个已知的miRNAs,并预测到376个新的miRNAs。其中97、102和19个miRNAs在S1与S2、S1与S3、S2与S3之间差异表达;从S1到S3共有7个miRNAs持续差异表达。另外,miRNAs在调控胚胎早期和晚期的发育差异明显,即与S1相比,S2有36个miRNAs上调和61个miRNAs下调,但与S2相比,S3只有12个miRNAs上调和7个miRNAs下调。依据表达量的变化将135个miRNAs划分为4种趋势,并将对应的938个靶基因与转录组数据进行关联分析,检测到71个miRNA靶基因对。GO富集分析显示7个miRNAs的9个靶基因富集到152个生物过程,且与生长发育相关的基因被鉴定为miRNAs的靶标,表明miRNAs可能在调控油棕胚发育的激素信号、生殖生长等生物学过程中发挥作用。另外,KEGG分析表明miRNAs通过调控次生代谢途径相关基因影响合子胚的成熟。进一步筛选出4个miRNAs家族参与植物激素的合成和信号传导调控:miR159-MYB调控赤霉素和脱落酸来维持胚的发生潜力以及诱导胚成熟,miR164-NAC调控乙烯和生长素参与胚细胞扩增,miR172-AP2调控乙烯和脱落酸诱导胚成熟,novel-m004-SPL调控赤霉素来诱导胚的形态建成。本研究初步鉴定了参与油棕合子胚发育相关的miRNAs,为后续研究miRNAs调控合子胚发育的分子机制奠定基础。
关键词:油棕;胚发育;miRNAs;激素
中图分类号:S565.9 文献标识码:A
sRNA Sequencing Reveals the Role of miRNAs During Zygotic Embryo Development in Oil Palm (Elaeis guineensis Jacq.)
ZHANG Anni1, CAO Hongxing2, CHEN Ping1*, JIN Longfei2*
1. College of Horticulture, Hainan University / Key Laboratory for Quality Regulation of Tropical Horticultural Crops of Hainan Province, Haikou, Hainan 570228, China; 2. Coconut Research Institute, Chinese Academy of Tropical Agricultural Sciences / Hainan Key Laboratory of Tropical Oil Crops Biology, Wenchang, Hainan 571339, China
Abstract: Oil palm is the most efficient oil-producing tropical woody oil crop in the world. There is a direct correlation between palm oil production and the development of zygotic embryos. The development of zygotic embryos of oil palm is a systematic and complex biological process, which involves the precise regulation of substantial genes. MicroRNAs (miRNAs) are important signal molecules in regulating various developmental processes of plants. Some miRNAs have been integrated into gene regulatory networks to coordinate the plasticity of plant embryo development, but little is known about the roles in the development of zygotic embryos of oil palm. In this study, the high-throughput sequencing analysis was performed from embryos at S1 (early stage), S2 (middle stage), and S3 (late stage) by small RNA sequencing, and 6 exist miRNAs and 334 known miRNAs were identified, and 376 new miRNAs were predicted. There were 97, 102, and 19 miRNAs differentially expressed in S1 vs S2, S1 vs S3, and S2 vs S3, and a total of 7 miRNAs were continuously differentially expressed from S1 to S3. In addition, miRNAs have significant differences in regulating early and late embryonic development. Compared with S1, S2 has 36 miRNAs up-regulated and 61 miRNAs down-regulated. However, compared with S2, S3 has only 12 miRNAs up-regulated and 7 miRNAs down-regulated. According to changes in expression levels, 135 miRNAs were divided into 4 trends. The correlation analysis between corresponding 938 target genes and transcriptome data detected 71 miRNA target gene pairs. GO enrichment analysis showed that 9 target genes of 7 miRNAs were enriched to 152 biological processes, and genes related to growth and development were identified as miRNAs targets, indicating that miRNAs may play a role in regulating hormone signaling and reproductive growth of oil palm embryos. Moreover, KEGG analysis showed that miRNAs affected the maturation of zygotic embryos by regulating genes related to secondary metabolic pathways. A total of 4 miRNAs families were further screened to be involved in the synthesis and signal transduction regulation of plant hormones: miR159-MYB regulates gibberellin and abscisic acid to maintain embryogenic potential and induce embryonic maturation, miR164-NAC regulates ethylene and auxin to participate in embryonic cell expansion, miR172-AP2 regulates ethylene and abscisic acid to induce embryo maturation, and novel-m004-SPL regulates gibberellin to induce embryonic morphogenesis. This study preliminarily identified miRNAs involved in the development of oil palm zygotic embryos, which laid the foundation for further study on the molecular mechanism of miRNAs regulating the development of zygotic embryos.
Keywords: Elaeis guineensis; embryonic development; miRNAs; phytohormone
DOI: 10.3969/j.issn.1000-2561.2022.01.003
油棕(Elaeis guineensis Jacq.)是世界上产油效率最高的热带木本油料作物之一,产生的棕榈油占全球食用油的33%[1]。随着全球对棕榈油需求的不断加大,油棕种植面积持续增加,威胁着热带雨林的生物多样性[2]。提高油棕单产是维持棕榈油的需求与油棕种植园扩张之间的动态平衡的重要途径。目前生产上通常采用厚壳种和薄壳种杂交培育高产的薄壳种油棕。果实发育决定油棕的产油量,而果实的生长与合子胚的发育密切相关。
MicroRNAs(miRNAs)是一类小的(19~24 nt)非编码RNAs,通过剪切或抑制靶基因的翻译来调节基因的表达[3]。在拟南芥的早期胚发育过程中,miR156通过负调控Squamosa启动子结合类蛋白(squamosa promoter-binding-like protein 10/ 11, SPL10/11)防止种子过早成熟,保证合子胚的正常发育[4]。而在胚发育后期,这些miRNAs的表达量降低从而促进合子胚的成熟[5]。植物激素响应途径在合子胚形态发育和成熟过程中起着一定作用[6]。例如干扰miR160的表达导致靶基因生长素响应因子17(Auxin response factor17, ARF17)的表达量增加,进而导致生长素偶联蛋白的大量积累,产生畸形胚[7]。起剪切加工miRNAs前体功能的Dicer-like1(DCL1)核酸酶基因突变会改变生长素的转录反应,产生形状不规则的胚[8]。在大麦中的研究发现,过表达miR393导致靶基因TIR1/AFBs生长素受体的表达降低,增加了种子的长宽比[9]。目前油棕miRNAs的研究主要集中在花发育和脂肪酸合成等方面[10-11],而在调控合子胚发育中的研究还少见报道。随着油棕基因组测序的完成[12],利用高通量测序挖掘重要的miRNAs成为可能。本研究使用高通量测序对油棕合子胚发育前、中和后3个时期的miRNAs进行表达谱分析,并结合mRNAs测序数据预测miRNAs靶基因对在胚发育过程中的作用,初步鉴定了4个可能参与合子胚发育的miRNAs家族,为后续研究miRNAs调控油棕合子胚发育的分子机制提供基础理论。
1 材料与方法
1.1 材料
本实验的油棕(Elaeis guineensis Jacq.)品种为‘热油4号’。取油棕花后120 d(S1)、140 d(S2)和160 d(S3)的果实的合子胚,液氮速冻后,–80℃保存。
1.2 方法
1.2.1 sRNA文库构建和深度测序 总RNA采用Trizol试剂(美国英杰生命技术有限公司)抽提,每个样品设置3个生物学重复。小RNA文库构建由广州基迪奥生物技术公司完成,并采用Illumina HiSeqTM 2500测序仪进行高通量测序,原始数据上传到SRA数据库(登录号:SRP265708)。
1.2.2 miRNAs的鉴定和分组间差异表达分析 原始数据先去掉接头和低质量序列得到纯净序列,然后与GenBank、Rfam数据库进行比对,去除rRNA、scRNA、snoRNA、snRNA、tRNA。与油棕参考基因组比对,去除外显子、内含子和重复序列片段。将剩下的序列与miRBase数据库进行比对,鉴定已存在的和已知的miRNAs。基于miRNA前体序列,利用MiRdeep2软件预测新的miRNAs。miRNAs的表达量采用TPM(tags per million)法进行计算。对差异表达的miRNAs(differentially expressed miRNAs, DEMs)采用edgeR软件进行鉴定,筛选标准为表达量变化2倍以上(Fold change≥2,FC≥2),且P值小于0.05。3个发育时期合子胚的miRNAs表达趋势采用ShortTime-series Expression Miner软件分析。
1.2.3 miRNAs的靶基因预测和分析 miRNAs的靶基因采用patmatch_v1.2软件预测,以miRNA的TPM值大于20為标准并与前期的转录组测序结果(SRA数据库,登录号:SRP265717)进行关联分析,找出与miRNAs表达趋势相反的靶基
因。将靶基因映射到GO(gene ontology)数据库(http://www.geneontology.org/)的各术语,并计算每个术语的基因数目。应用超几何检验,找出与整个基因组背景相比显著富集的GO条目。以KEGG(kyoto encyclopedia of genes and genomes)(http://www.kegg.jp/kegg/pathway.html)通路为单位,应用超几何检验,确定与整个基因组背景相比,在基因中显著富集的途径。
2 结果与分析
2.1 油棕miRNAs测序的标准数据分析
原始数据过滤后得到9 185 842~15 458 765个纯净序列(表1)。与GenBank、Rfam数据库以及参考基因组比对后,47.15%~59.49%的序列被成功定位到参考基因组上。外显子比率变化范围为0.5%~1.0%。比对miRBase数据库后,从3个sRNA文库中共鉴定到438个已存在的miRNAs、7 773 939个已知的miRNAs和129 689个新的miRNAs序列。miRNA序列的长度范围为18~25 nt,其中,24 nt的序列丰度最高,其次是21 nt的序列(图1)。
2.2 DEMs的鉴定
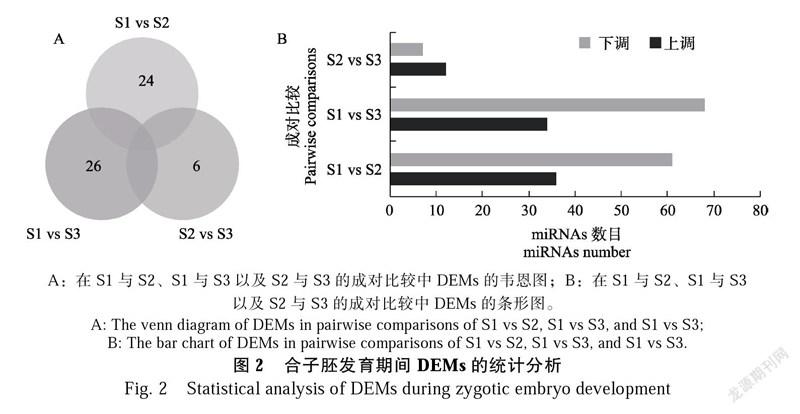
根据与miRBase数据库的比对结果,本研究在3个文库中共鉴定到6个已存在的miRNAs和334个已知的miRNAs,分别属于276个miRNAs家族。另外,还预测到376个新的miRNAs。以FC≥2且P<0.05为标准,在3对成对比较中各筛选出97(S1 vs S2)、102(S1 vs S3)和19(S2 vs S3)个DEMs(图2)。其中,从S1到S3共有7个DEMs,包含4个在S1 vs S2、S2 vs S3和S1 vs S3中均差异表达的miRNAs(图2A)。与S1相比,S2有36个miRNAs上调和61个miRNAs下调,但S3与S2相比只有12个miRNAs上调和7个miRNAs下调,表明在胚胎发育的早期和晚期miRNAs调控的差异明显(图2B)。利用Short Time-series Expression Miner软件将筛选出的135个DEMs的表达趋势划分为4种:从S1到S3,有49个miRNAs的表达量呈上升趋势,76个miRNAs的表达量呈下降趋势,10个miRNAs的表达量在S2达到峰值或最小值。
2.3 DEMs靶基因的预测与功能分析
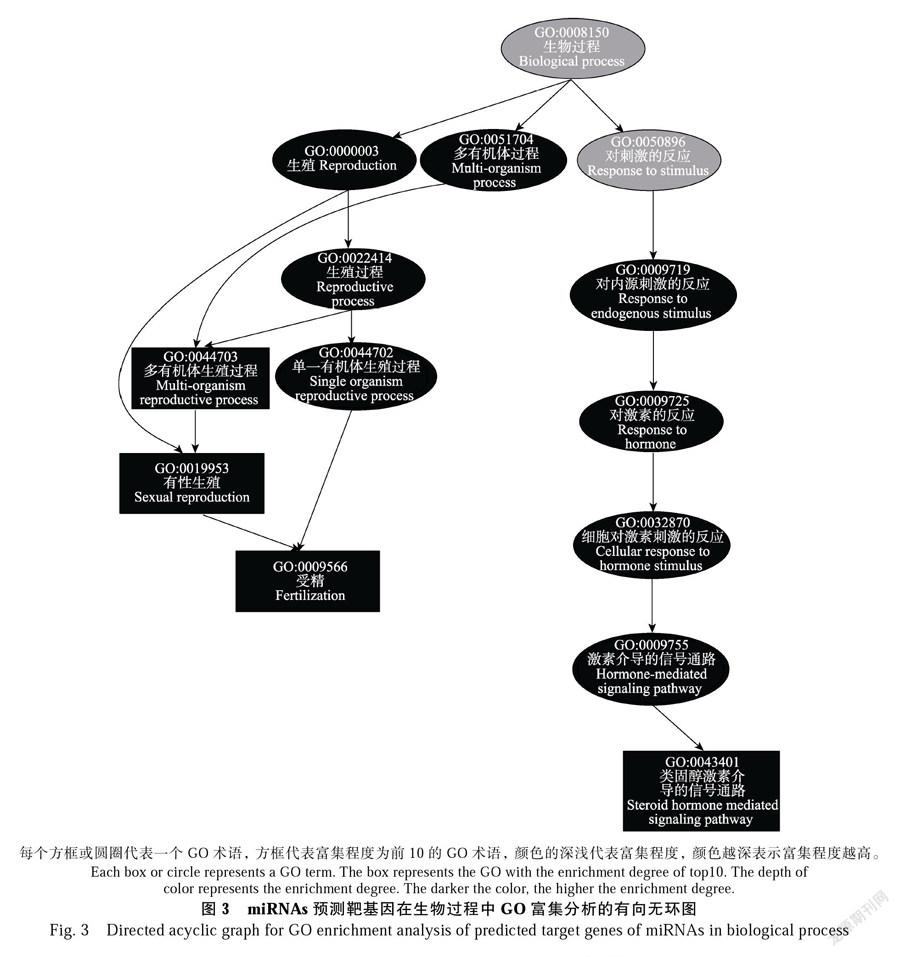
将135个DEMs的938个靶基因与之前的转录组测序数据联合分析。以miRNAs的TPM值大于20为标准筛选出71个miRNA靶基因对,其中包含4个表达量升高的miRNAs和14个表达量下降的miRNAs(表2)。在GO富集分析中,7个miRNAs的9个靶基因富集到152个生物过程。将GO分析用有向无环图展示并屏蔽无意义的节点(图3),结果表明:在生物过程中,靶基因注释到“对内源性刺激的反应”“对激素
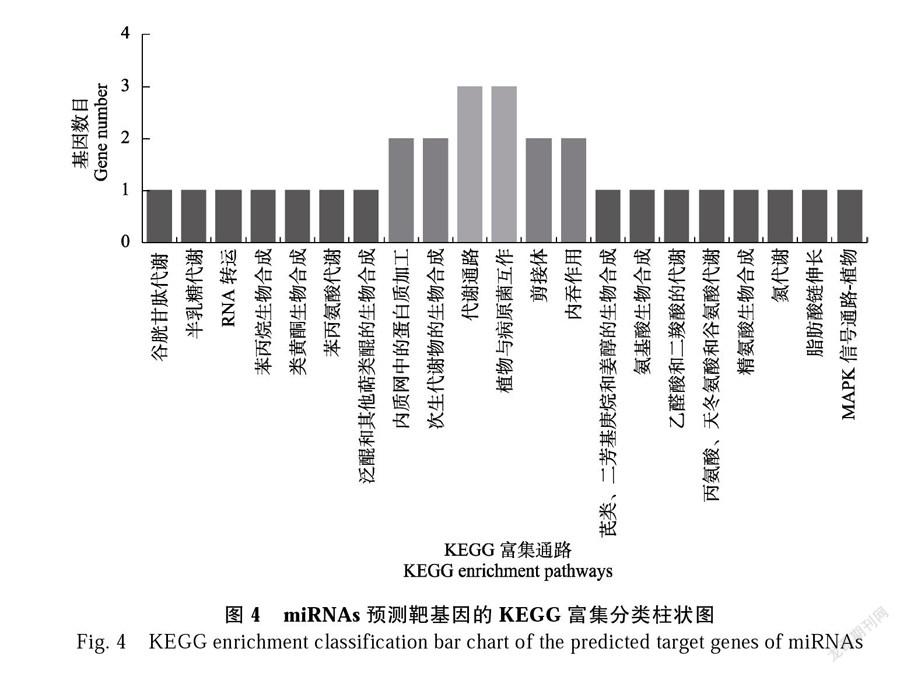
的反应”“细胞对激素刺激的反应”“激素介导的信号通路”“类固醇激素介导的信号通路”“单一有机体生殖过程”“有性生殖”和“受精”等GO术语。12个靶基因通过KEGG分析富集在“植物病原菌互作”“内吞作用”“苯丙烷生物合成途径”和“类黄酮生物合成途径”等代谢途径中(图4)。
3 讨论
对影响miRNAs生物发生的等位基因的突变体表型进行研究发现,miRNAs促进胚的形态建成和成熟进程[8, 13]。与胚发育相关的miRNAs及其预测的靶基因已经在拟南芥[4]、水稻[14]、菊花[15]、花生[3]等植物中得到了系统的鉴定。miRNAs调控油棕合子胚发育的分子机制还不明晰。本研究从3个sRNA文库中鉴定到6个已存在的miRNAs、334个已知的miRNAs和376个新的miRNAs。在GO富集分析中,“植物激素信号传导”“类固醇激素介导的信号通路”“有性生殖”“受精”等与生长发育相关的基因也被鉴定为miRNAs的靶标,表明miRNAs可能在调控油棕胚发育的激素信号、生殖生长等生物学过程中发挥作用。另外,基于KEGG分析,与苯丙烷、类黄酮、芪类、泛醌等次级代谢产物相关的生物合成途径也被富集,暗示了miRNAs通过调控次生代谢途径相关基因影响油棕胚的成熟。
本研究共鉴定到4个上调的miRNAs。例如,novel-m0004-5p的靶基因属于SPL家族。在胚的形态发生阶段,SPL10和SPL11的转录水平降低会抑制早熟相关基因的表达[4]。另外,赤霉素通过间接激活SPL促进开花[16]。在油棕胚发育过程中,novel-m0004-5p的表达量上升,而SPL的表达量下降。结果表明,SPL的低表达可能诱导胚的形态建成,该过程依赖低水平的赤霉素。在水稻中也发现在胚成熟期间有低水平的赤霉素合成[17]。
表达量下调的miRNAs有14个。靶基因为MYB101的miR159-y和miR159-z属于miRNA159家族。miR159通过影响MYB33的活性来调节赤霉素介导的花发育过程[18]。另外,脱落酸也可以诱导miR159控制拟南芥种子萌发过程中MYB101和MYB33的转录水平[19]。在本研究中,miR159-y和miR159-z的表达量下降,而MYB101的表达量增加。与LI等[20]的发现一致,即胚发生潜力的维持以及体胚的成熟与miR159对MYB33转录后的调控有关。这可能是响应赤霉素或者脱落酸信号的结果,因为单个miRNA可能对不同的激素信号有反应[19]。miR164-x的靶基因NAC098编码的CUP-SHAPED COTYLEDON 2(CUC2)蛋白属于NAC转录因子家族NAM亚家族。在本研究中,与miR164-x的表达降低相比,NAC098特异性mRNA水平有所增加。以往研究表明,CUC1/ CUC2/CUC3在胚发生过程中参与茎尖分生组织的形成和子叶的分离[21]。在拟南芥中,乙烯通过抑制miR164调控NAM亚家族成员RhNAC100的表达进而控制花瓣的细胞扩增[22]。miR164也可以通過剪切NAC1的mRNA来负调控侧根发育过程中生长素信号的传递[23]。结果表明,NAC098可能在介导乙烯信号调节油棕胚细胞扩增的过程中起重要作用。另外,本研究中egu-miR172a靶定APETALA2(AP2)转录因子。在油棕中,EgAP2-1的转录本在合子胚的分生组织中积累最多[24]。在番茄和水稻中,AP2通过乙烯或脱落酸的介导参与调节果实成熟和植物衰老[25-26]。在本研究中,AP2的表达量上升,这可能是egu- miR172a通过乙烯或脱落酸途径介导的诱导胚成熟的机制之一。
综上所述,本研究共筛选出4个miRNAs家族,即novel-m0004、miR172、miR159和miR164均可能通过植物激素调控途径参与油棕合子胚的发育(图5),为进一步探索miRNAs调控油棕胚发育的分子机制奠定基础。
参考文献
[1] LEE F, ONG-ABDULLAH M, OOI S, HO C, NAMASIVAYAM P. Cloning and characterization of somatic embryogenesis receptor Kinase I (EgSERK I) and its association with callus initiation in oil palm[J]. In Vitro Cellular & Developmental Biology-Plant, 2019, 55(2): 153-164.
[2] CARLSON K M, HEILMAYR R, GIBBS H K, NOOJIPADY P, BURNS D N, MORTON D C, WALKER N F, PAOLI G D, KREMEN C. Effect of oil palm sustainability certification on deforestation and fire in Indonesia[J]. Proceedings of the National Academy of Sciences, 2018, 115(1): 121-126.
[3] MA X, ZHANG X, ZHAO K, LI F, LI K, NING L, HE J, XIN Z, YIN D. Small RNA and degradome deep sequencing reveals the roles of microRNAs in seed expansion in peanut (Arachis hypogaea L.)[J]. Frontiers in Plant Science, 2018, 9: 1-15.
[4] NODINE M D, BARTEL D P. MicroRNAs prevent precocious gene expression and enable pattern formation during plant embryogenesis[J]. Genes & Development, 2010, 24(23): 2678-2692.
[5] WILLMANN M R, MEHALICK A J, PACKER R L, JENIK P D. MicroRNAs regulate the timing of embryo maturation in Arabidopsis[J]. Plant Physiology, 2011, 155(4): 1871- 1884.
[6] MERINO I, ABRAHAMSSON M, STERCK L, CRAVEN- BARTLE B, CANOVAS F, ARNOLD S. Transcript profiling for early stages during embryo development in Scots pine[J]. BMC Plant Biology, 2016, 16(1): 255.
[7] MALLORY A C, BARTEL D P, BARTEL B. MicroRNA- directed regulation of Arabidopsis auxin response factor17 is essential for proper development and modulates expression of early auxin response genes[J]. The Plant Cell, 2005, 17(5): 1360-1375.
[8] ARMENTA-MEDINA A, LEPE-SOLTERO D, XIANG D, DATLA R, ABREU-GOODGER C, GILLMOR C S. Arabidopsis thaliana miRNAs promote embryo pattern formation beginning in the zygote[J]. Developmental Biology, 2017, 431(2): 145-151.
[9] BAI B, SHI B, HOU N, CAO Y, MENG Y, BIAN H, ZHU M, HAN N. microRNAs participate in gene expression regulation and phytohormone cross-talk in barley embryo during seed development and germination[J]. BMC Plant Biology, 2017, 17(1): 150.
[10] GAO L, WANG Y, ZHU Z, CHEN H, SUN H, ZHENG Y, LI D. EgmiR5179 from the mesocarp of oil palm (Elaeis guineensis Jacq.) regulates oil accumulation by targeting NAD transporter 1[J]. Industrial Crops and Products, 2019, 137: 126-136.
[11] HO H, GUDIMELLA R, ONG-ABDULLAH M, HARIKRISHNA J A. Expression of microRNAs during female inflorescence development in African oil palm (Elaeis guineensis Jacq.)[J]. Tree Genetics & Genomes, 2017, 13(2): 35.
[12] SINGH R, ONG-ABDULLAH M, LOW E L, MANAF M A A, ROSLI R, NOOKIAH R, OOI L C, OOI S, CHAN K, HALIM M A, AZIZI N, NAGAPPAN J, BACHER B, LAKEY N, SMITH S W, HE D, HOGAN M, BUDIMAN M A, LEE E K, DESALLE R, KUDRNA D, GOICOECHEA J L, WING R A, WILSON R K, FULTON R S, ORDWAY J M, MARTIENSSEN R A, SAMBANTHAMURTHI R. Oil palm genome sequence reveals divergence of interfertile species in old and new worlds[J]. Nature, 2013, 500(7462): 335-339.
[13] SEEFRIED W F, WILLMANN M R, CLAUSEN R L, JENIK P D. Global regulation of embryonic patterning in Arabidopsis by microRNAs[J]. Plant Physiology, 2014, 165(2): 670-687.
[14] XUE L, ZHANG J, XUE H. Characterization and expression profiles of miRNAs in rice seeds[J]. Nucleic Acids Research, 2009, 37(3): 916-930.
[15] YANG Y, GUO J, CHENG J, JIANG Z, XU N, AN X, CHEN Z, HAO J, YANG S, XU Z, SHEN C, XU M. Identification of UV-B radiation responsive microRNAs and their target genes in chrysanthemum (Chrysanthemum morifolium Ramat) using high-throughput sequencing[J]. Industrial Crops and Products, 2020, 151: 112484.
[16] YU S, GALVÃO V C, ZHANG Y, HORRER D, ZHANG T, HAO Y, FENG Y, WANG S, SCHMID M, WANG J. Gibberellin regulates the Arabidopsis floral transition through miR156-targeted SQUAMOSA PROMOTER BINDING– LIKE transcription factors[J]. The Plant Cell, 2012, 24(8): 3320-3332.
[17] HU Y, SONG S, WENG X, YOU A, XING Y. The heading-date gene Ghd7 inhibits seed germination by modulating the balance between abscisic acid and gibberellins[J]. The Crop Journal, 2021, 9(2): 297-304.
[18] ACHARD P, HERR A, BAULCOMBE D C, HARBERD N P. Modulation of floral development by a gibberellin-regu¬lated microRNA[J]. Development, 2004, 131(14): 3357- 3365.
[19] REYES J L, CHUA N. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination[J]. The Plant Journal, 2007, 49(4): 592- 606.
[20] LI W, ZHANG S, HAN S, WU T, ZHANG J, QI L. Regulation of LaMYB33 by miR159 during maintenance of embryogenic potential and somatic embryo maturation in Larix kaempferi (Lamb.) Carr.[J]. Plant Cell, Tissue and Organ Culture (PCTOC), 2013, 113(1): 131-136.
[21] OLSEN A N, ERNST H A, LEGGIO L L, SKRIVER K. NAC transcription factors: structurally distinct, functionally diverse[J]. Trends in Plant Science, 2005, 10(2): 79-87.
[22] PEI H, MA N, TIAN J, LUO J, CHEN J, LI J, ZHENG Y, CHEN X, FEI Z, GAO J. An NAC transcription factor controls ethylene-regulated cell expansion in flower petals[J]. Plant Physiology, 2013, 163(2): 775-791.
[23] GUO H, XIE Q, FEI J, CHUA N. MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for Arabidopsis lateral root development[J]. The Plant Cell, 2005, 17(5): 1376-1386.
[24] MORCILLO F, GALLARD A, PILLOT M, JOUANNIC S, ABERLENC-BERTOSSI F, COLLIN M, VERDEIL J L, TREGEAR J W. EgAP2-1, an AINTEGUMENTA-like (AIL) gene expressed in meristematic and proliferating tissues of embryos in oil palm[J]. Planta, 2007, 226(6): 1353-1362.
[25] KARLOVA R, ROSIN F M, BUSSCHER-LANGE J, PARAPUNOVA V, DO P T, FERNIE A R, FRASER P D, BAXTER C, ANGENENT G C, MAAGD A. Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening[J]. The Plant Cell, 2011, 23(3): 923-941.
[26] XU X, BAI H, LIU C, CHEN E, CHEN Q, ZHUANG J, SHEN B. Genome-wide analysis of microRNAs and their target genes related to leaf senescence of rice[J]. PLoS One, 2014, 9(12): e114313.

