The artificial intelligence watcher predicts cancer risk by facial features
Hao-Ran Zhang,Guang-Yi Lv,Shuang Liu,Dan Liu,Xiong-Zhi Wu*
1National Clinical Research Center for Cancer,Key Laboratory of Cancer Prevention and Therapy,Tianjin Medical University Cancer Institute and Hospital,Tianjin 300060,China.2University of Science and Technology of China,Hefei 230026,China.
Abstract Background: Early diagnosis of malignant tumors effectively reduces the mortality rate.The special craniofacial structure serves as a diagnosis basis of early screening for many hereditary diseases.However, cancer is also considered a genetic disorder.Could facial images direct tumor screening? Methods: We developed an image recognition program, the artificial intelligence watcher, which could extract implicit knowledge from faces and distinguish cancer patients from normal persons.The artificial intelligence program used a convolution neural network with 6 layers.Then, we conducted a retrospective clinical study of 643 cancer patients and 219 local normal people at 20-80 ages from China to analyze their photos and medical history.By analyzing their facial features, disease and family gene sequencing, the relationship between appearance and cancer can be inferred. Results: We demonstrated that the accuracy of artificial intelligence watcher achieved up to about 90%.Statistical results showed lung and gastric cancer patients have more narrow-set eyes.Furthermore, we suspected the physiological basis for artificial intelligence watcher is craniofacial genes are closely related to cancer susceptibility genes.Through the analysis of the tumor database and craniofacial development gene database,we found many single nucleotide polymorphism mutations related to appearance are also related to the tumor,in particular,WW domain containing E3 ubiquitin protein ligase 2,SH3 and PX domains 2B, DNA-directed RNA polymerase III core subunit and coatomer subunit zeta. Conclusion: According to our gene sequencing results, there are some polymorphisms of the same locus in patients with similar facial features different from their healthy relatives, such as WW domain containing E3 ubiquitin protein ligase 2 and ATP binding cassette subfamily A member 4 genes.Our model provides a new efficient auxiliary diagnosis method for tumor screening.
Keywords: artificial intelligence; cancer genetics; tumor screening; facial features; facial recognition
Background
In the last 10-15 years, attempts to develop a general screening technique to reduce cancer mortality have uniformly been unsuccessful.Although there are many genetic tests and marker screening for tumors, they are costly and cumbersome.Recently, face recognition technology applied in all fields of clinical diagnosis, in particular, some genetic diseases screening [1-3].Many hereditary diseases are accompanied by abnormal facial features, such as Down syndrome [4], ankyloblepharon-ectodermal dysplasia-clefting syndrome [5], and phenylketonuria [6].It could be speculated that the links between facial features and disease are the gene mutations that determined some pathologic change and craniofacial growth.Interestingly, many ancient Chinese medical books also recorded the relationship between disease and facial features.For instance,Inner Canon of Yellow Emperor(unknown author, written around 26 C.E.) is probably the first book to document the relationship between appearance and disease..It said that an ancient physician can predict fetal disease by observing nasion and nasal bone.In normal people,the nasal root is high, the nasal bone is straight, facial skin color is normal and the five sense organs are well placed.If these features are abnormal, it indicates the occurrence of disease in the body.There are similar records in ancient Chinese Xiang Shu (a kind of book describing the relationship between appearance and fate).It is recorded inYu Guan Zhao Shen Juthat the features of the nose symbolize the length of a person’s life.This book was written by Qiu Qi in the Southern Tang Dynasty (937 C.E.-975 C.E.).Although these ancient texts do not have sufficient evidence, other modern studies have come to a similar conclusion.More and more studies have shown that the normal people with standard looks represent healthy bodies,on the contrary, abnormal facial features lead to some disease even if they are not ugly or deformed [7-9].
In recent years, artificial intelligence (AI) is fast becoming a powerful tool for imaging diagnosis and optimization treatment.Facial recognition and clinical imaging diagnosis have always been the hot fields of AI.The ability of automated facial recognition technology to detect the genetic disease phenotype from images was assessed by many research [10].In forensic studies, genes could be used to predict a person’s appearance and skin color.So, can we predict disease by portrait images? Recent pure data-driven neural network methods directly leverage the raw images to perform analytical tasks and achieved great success.In this paper, we designed an AI watcher to recognize patients based on various human facial photos.
The tumor can also be regarded as a hereditary disorder to some extent.Is it possible to speculate on cancer susceptibility from certain facial features? There is some indirect evidence.For example, patients with Down syndrome have a strikingly lower incidence of many solid tumors except germ cell tumors.Some researchers think some genetic abnormalities such as regulatorofcalcineurin 1 gene, microRNA-99a,microRNA-125b-2 and microRNA-let-7c are the main candidates for explaining this inverse comorbidity [11].Thus, the gene mutations causing the occurrence of tumors and craniofacial development might be inextricably linked.However, many tumor-related gene mutations and craniofacial developmental genes in different research areas are studied separately, such as fibroblast growth factor receptor mutation[12, 13], tumor protein 63 mutation [14, 15] and so on.
Do certain facial features reflect traits of genes that increase tumor susceptibility? With advances in technology, face recognition programs, gene sequencing and AI analysis and other new techniques provided the possibility that we could dive deep into the relationship between appearance-gene-tumor than ever before.Image analysis of relevant facial structures, which is based on applying mathematical functions to evaluate image texture and yield a binary classification result, is widely used in clinical imageology [3].But those AI programs could only improve the diagnosis rate on the basis of doctor awareness, which cannot find the new rules that doctors have never noticed.
For example, although genes have been mapped severally for various rare craniofacial syndromes and cancer susceptibility, the genetic basis of the connection between facial variation and tumorigenicity is still poorly understood.The characterization of gene mutations in dysmorphology and in animal models has identified many rare genetic variants with major effects on craniofacial development [16, 17].Facial appearance and cancer both have a strong genetic component.Here, we analyzed many facial images from different types of cancer patients using AI watcher.In addition,in the genetic analysis of the cancer family, we obtained quantitative measures related to the ordinal phenotypes examined.Combining the findings on both sides, AI watcher and gene sequencing allowed us to reveal the potential cancer-gene-appearance association and to identify some gene regions impacting both face morphology and cancer susceptibility.So, the new AI watcher could not only contribute to cancer screening but also discover the hidden link between diseases and appearance that human doctors will never find.Here, by using genetic analysis of real clinical data, our goal is to be able to further explore the genetic association between appearance and cancer.
Materials and methods
Retrospective analysis and patient family samples.We conducted a retrospective clinical study of 643 cancer patients and 219 local normal people (133 male and 86 female) aged 22-85 from Tianjin Medical University Cancer Institute and Hospital in China to analyze their photos and medical history.The study protocols were approved by the Cancer Foundation of China’s Institutional Review Board (the approved number: 2017 quick review C02).The cancer type of patient group included lung cancer, gastric cancer, hepatobiliary tumor,colorectal cancer, esophageal cancer, pancreatic cancer, brain tumor(meninges and glial), breast cancer, ovarian cancer, uterine tumor(cervical cancer, endometrium cancer, and uterine leiomyosarcoma),leukemia, lymphoma, throat cancer (oral cavity, gums, tongue,nasopharynx, and larynx), multiple myeloma, melanoma, thyroid cancer,small intestine tumor,bone tumor, soft tissue tumors(sarcoma,fibroma, and mesothelioma), prostate cancer, urinary tract tumor, and thymoma.The medical records of these patients are intact, and all participants are Northern Chinese.The normal people group were from the Tianjin Medical Center.The criterion was that all normal people were native residents in Tianjin and they had no tumors or other serious diseases.Family 1 includes father, mother, daughter, and two brothers.Family 2 includes daughter, father, and mother.All participants included in this study were of Chinese ancestry aged 22-85 and gave written informed consent.
Digital image acquisition
The photographs of the patient and normal people were captured by a machine (DS01-B, Shanghai Daosh Medical Technology, Shanghai,China).The spectrum of illuminant was close to natural light and color rendering index ≥92%.All photos were taken in the same pattern.More details of the instrument were in the Supplementary Material.
Data availability
We applied an artificial neural network-based integrative data mining approach to data from the photos of cancer patients and normal people.The data that support the findings of this study are available from the corresponding author upon reasonable requests, such as gene sequencing results and AI program code.Identifiable images of research participants cannot be released.
Whole exome sequencing
The quality of isolated genomic DNA was verified by using these two methods in combination.DNA degradation and suspected RNA/protein contamination were verified by electrophoresis on 1%agarose gels.The concentration and purity of DNA samples were further quantified precisely by the Qubit dsDNA hs assay kit in Qubit®3.0 Fluorometer (Life Technologies, Carlsbad, CA, USA).A total amount of 0.4 μg DNA per sample was required for library generation.The exome sequences were efficiently enriched from 0.4 μg genomic DNA using Agilent liquid capture system (Agilent SureSelect Human All Exon V6, Beijing, China) according to the manufacturer’s protocol.Firstly, qualified genomic DNA was randomly fragmented to an average size of 180-280 bp by the Covaris S220 sonicator.The remaining overhangs were converted into blunt ends via exonuclease/polymerase activities.Secondly, DNA fragments were end-repaired and phosphorylated, followed by A-tailing and ligation at the 3’ ends with paired-end adaptors (Illumina, San Diego,California, USA).DNA fragments with ligated adapter molecules on both ends were selectively enriched in a quantitative real-time polymerase chain reaction.After polymerase chain reaction, libraries hybridize with liquid phase with biotin-labeled probe, then use magnetic beads with streptomycin to capture the exons of genes.Captured libraries were enriched in a polymerase chain reaction to add index tags to prepare for sequencing.Products were purified using AMPure XP system (Beckman Coulter, Beverly, MA, USA) and quantified using the Agilent high sensitivity DNA assay on the Agilent bioanalyzer 2100 system.At last, DNA library was sequenced on Illumina for paired-end 150 bp reads.See Supplementary Materials for more details.The sequencing data have been uploaded to the National Center for Biotechnology Information Sequence Read Archive database(SUB4274592 and SUB4275181).
AI program
We utilize a convolution neural network with 6 layers as it is shown in Figure 1A.The task of “dominant feature” analysis uses a costumed convolutional neural network.To measure the eyes positions and size,we use the free software OpenCV 2.0.We perform the classification for eyes’ data through the open-source tool Weka 3.7.Here we give the details of the network.To reduce the impact of light or color, the input photo is first converted into a gray-scaled image.Then it is sent to 5 convolutional layers, where all the kernel size is 5 times 5.The numbers of the feature maps for the 5 layers are 8,16,32, 64 and 128.This process can be expressed as formulas(1) and (2).
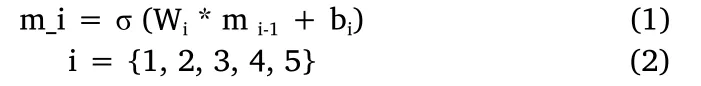
“*” is the convolution operation, “σ” is the activation function, and“m0” is the input image.After the 5thconvolutional layer, the output tensor is flattened into a vector and further mapped into a 4,096 dimensions vector through a fully connected layer.This process can be expressed as formula (3).
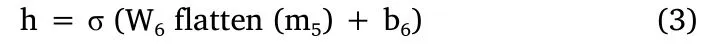
“h” is usually called the representation/embedding of the image.Finally, we can get the output label (which indicates whether the person is a tumor patient or not)based on“h”by formulas(4)and(5).
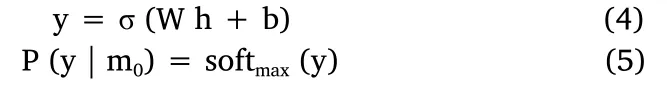
Here,“P(y |m0)”means the probability of the person being a tumor patient based on the image.
Please note that all the notations of “Wi” and “bi” above are our model parameters which need to learn for the training process.To train our model, mini-batch gradient descent is used, where the batch size in our case is 32.In detail, we choose RMSProp optimizer(http://www.cs.toronto.edu/~tijmen/csc321/slides/lecture_slides_lec 6.pdf) to learn the parameters with momentum, where the learning rate and momentum are separately set as 0.0001 and 0.9 respectively.Kernels(W1to W5) of the convolutional layers are initialized randomly using truncated normal distribution with mean = 0 and standard deviation = 0.001.Weights of dense layers (W6and W) are randomly set following the uniform distribution in the range between −(6/(nin+ nout))0.5and (6/(nin + nout))0.5, where “nin” and “nout” refer to the number of input and output neurons respectively.
Graphing and statistical analysis
All graphs were generated using WPS and SPSS 12.0 statistical software program.Data represent the mean ± standard deviation of the mean, except for in vivo data, which represent the mean ±standard error of the mean.U-tests were performed, andP<0.05 was considered significant.
Results
AI watcher distinguish between cancer patients and normal people
We have developed a new AI facial recognition program, AI watcher,to distinguish tumor patients from normal groups based on facial photos.The overall sample included high-definition facial photographs of 643 patients involving different types of tumors.First,AI watcher could find out which site has the greatest impact on the predictive value of cancer according to different combinations of indicators in the rectangular coordinate system (Figure 1A).Second,AI watcher could get the probability of each person, and then judge the risk of the tumor based on the difference in probability.The results show that it could clearly distinguish between cancer patients and normal people (Figure 1B).We also found that the cancer probability increases with the age, which is in line with the pathogenesis of tumors.Although other areas may also play a part, the most dominant area is obviously around the eyes (Figure 1C).These suggested genes which affect the development of areas around the eyes may be involved in tumors progression.
Since quantitative traits are expected to provide higher power for discrimination than categorical traits, we assessed facial features on an ordered categorical scale reflecting the distinctiveness of each trait.The samples were divided into the training set and test set for cross test analysis.Then, we set up a convolutional neural network to classify the two groups and found them to reliably distinguish and to also show extensive traits in the cancer sample.The results demonstrated that classification based on eyes’ photos outperformed those using nose and forehead in distinguishing between tumor patients and normal groups.
Therefore, to further refine the extensive facial traits and remove individual differences in the cancer sample, we performed a detailed statistical analysis of the relative position of eyes in different groups through programming and localization.We just focus on the eye’s size and position on the coordinate system (Figure 1D).A number of statistical classifiers were performed based on the coordinates,including decision trees, logistic regression, and neural network analysis (Table 1).The results of the classification experiment with 10 characteristic values (x1, x2, y1, y2, w1, w2, h1, h2, Ed, Fw) show that most of the tumor patients could be discriminated from the normal group (Table 1).x1 and y1 represent coordinates of the right eye in the plane system; y2 and x2 represent coordinates of the left eye in the plane system; w1 and w2 represent width of left and right eyes;h1 and h2 represent the height of two eyes; Fw represents the distance between the edges of a face in a flat photograph; Ed represents the distance between the inner corners of the eyes.Correctly classified rate = (True positive + True negative)/Total.For example, the classification accuracy of the decision tree is up to 91%.The calculation method of mean square error (MSE) is as formula(6).
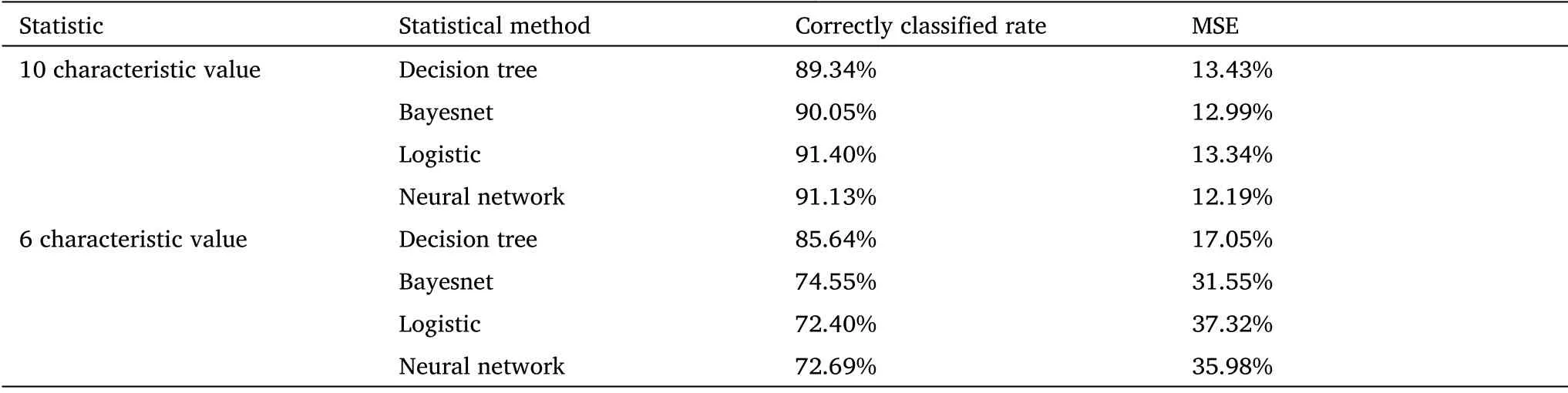
Table 1 Results of tumor-normal dichotomy in ocular localization in coordinates.
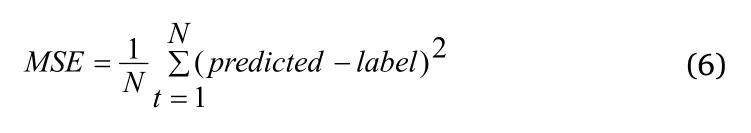
In order to further simplify the statistical results, we used six characteristic values (w1, w2, h1, h2, Ed, Fw) to observe the difference in eye characteristics of different groups.The results showed 10 indicators were more obvious than 6 indicators in differentiating patients with tumors from normal people.However, in order to further remove the individualized differences between people’s faces and test the hypothesis, we turn from a holistic approach to a single index.
We selected the ratio of binocular spacing (Eye-d) and face width(Fw) as an indicator (the ratio of Eye-d and Fw) to analyze the difference between cancer patients and the normal (Figure 1E).The ratio of Eye-d and Fw could represent the characteristics of the distance between the inner canthus, eliminating the inter-individual error and reducing the measurement error as much as possible.As a result, it could more accurately reflect the differences of real facial features between different patients.The results showed that the relative eye distance distribution and median of the normal and the patient were different,P< 0.001 (Figure 2A), and the eyelid ratio of the normal was 0.5% wider than that of the patient.As shown in the typical image, the eye distance between the male and female subjects was wider than that of the tumor patient.In addition, the features of different types of tumors are not identical, so we calculated the relationship between relative eye distance of tumor patients and patient gender, age, local location, tumor type and metastasis through retrospective clinical studies(Table 2).
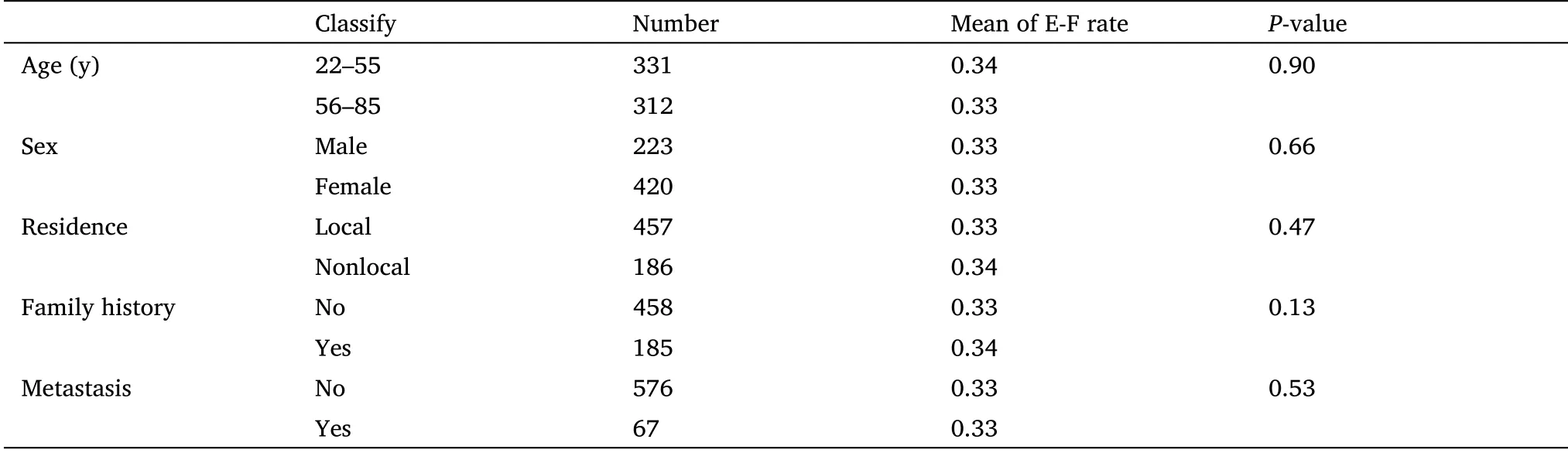
Table 2 The influence of different factors on eye distance (U-test).
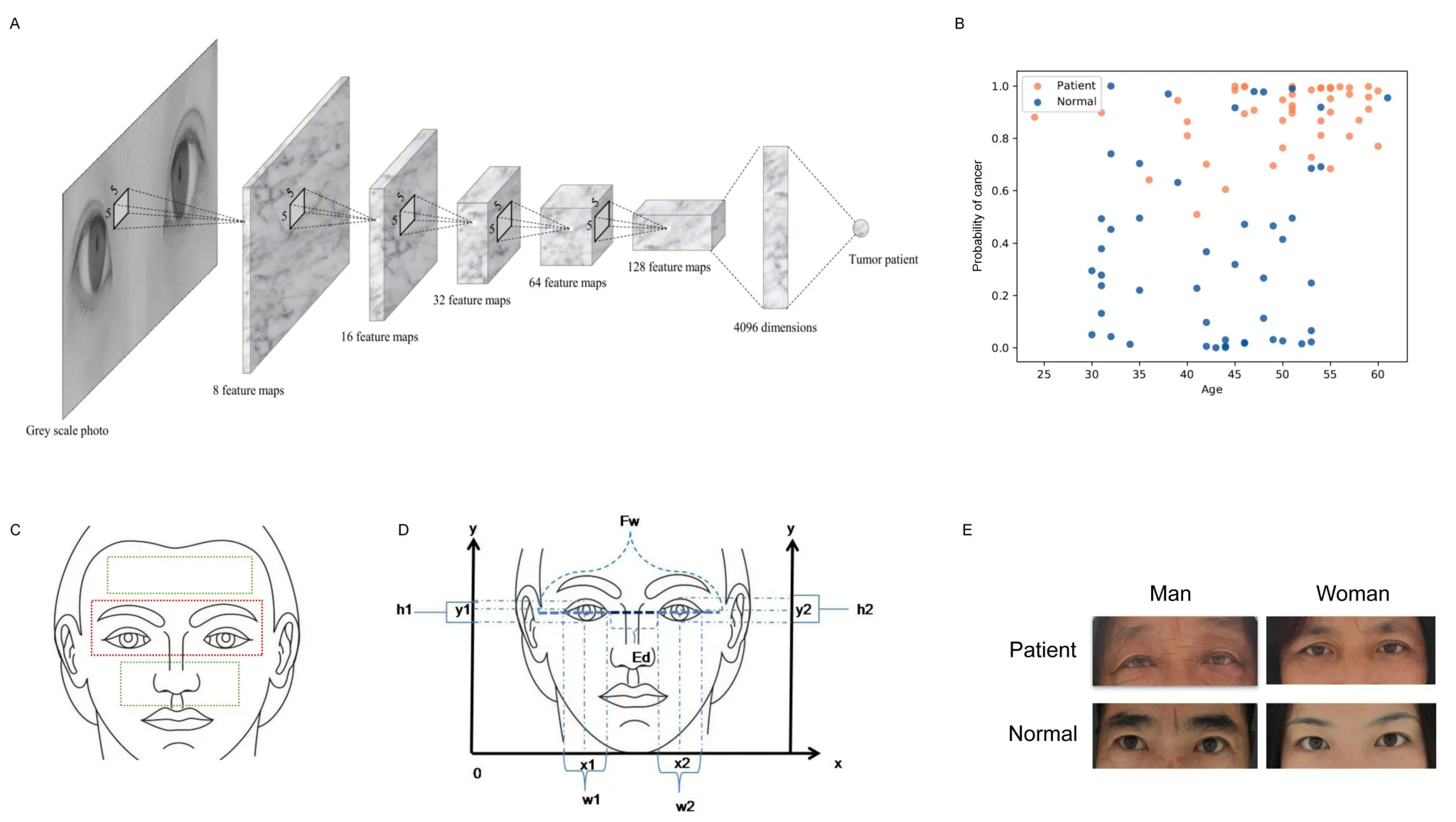
Figure 1 AI watcher finds features in facial image to distinguish patients from normal.(A) The model shows the AI watcher analyzing process of the facial image.(B) AI predicts the probability of each sample in the test set.Green is normal, red is tumor patients.X-axis represents the probability of tumor risk, and Y-axis represents age.(C) The eyes were statistically more heavily weighted than the forehead and nose.(D) Convert the photo to a diagram locating in rectangular coordinate system.x1 and y1, coordinates of the right eye in the plane system; y2 and x2,coordinates of the left eye in the plane system;w1 and w2,width of left and right eyes;h1 and h2, the height of two eyes;Fw,the distance between the edges of a face in a flat photograph; Ed, the distance between the inner corners of the eyes.(E) Eye image screenshots of patients and normal people’s faces.AI, artificial intelligence.
There was no statistically significant difference in relative eye distance between the groups based on gender, age, residence and family history.Tumor type, whether metastasis is related to relative eye distance (Figure 2B).The relative eye distance of patients with lung cancer and gastric cancer is narrower than other types, and one possible reason is that tumor protein 63 is more closely related to lung cancer, colorectal cancer and breast cancer.The distance between the inner canthus in the eyes of tumor patients is narrower than normal suggesting that cancer patients could be recognized by facial features,especially lung cancer, colorectal cancer and breast (Figure 2B).
According to ancient Chinese physiogn my,the collapse of the nasal base can also mean serious disease.Considering that area around the eyes had better differentiation, we then analyzed the degree of nasal root collapse.We simulated the shape of the nasion by using the brightness of the pixel of the nasal root position in the plane photo(Figure 2C).According to the comparison between the histogram results and the real results, the simulated results using pixel gray values could basically represent the real face collapse situation (Figure 2D).The results showed that the shape of the nose root of the patients was statistically different from that of the normal people (P< 0.001).The nasion condition of the patient is lower and thinner in shape,while the normal group is plumper (Figure 2E).The accuracy rate of the two classifications is 78.14% and the mean absolute error is 26.05% by logistic analysis.There was no statistically significant difference in relative eye distance between the groups based on gender,age, residence, family history and metastasis (Table 3).So, the nasion condition could be used as one of the criteria to identify cancer patients, and the collapse of the nasion is also a facial marker of many genetic diseases.The genes that determine the development of the nasal roots are likely to be closely related to genetic diseases, which deserve more sequencing studies.
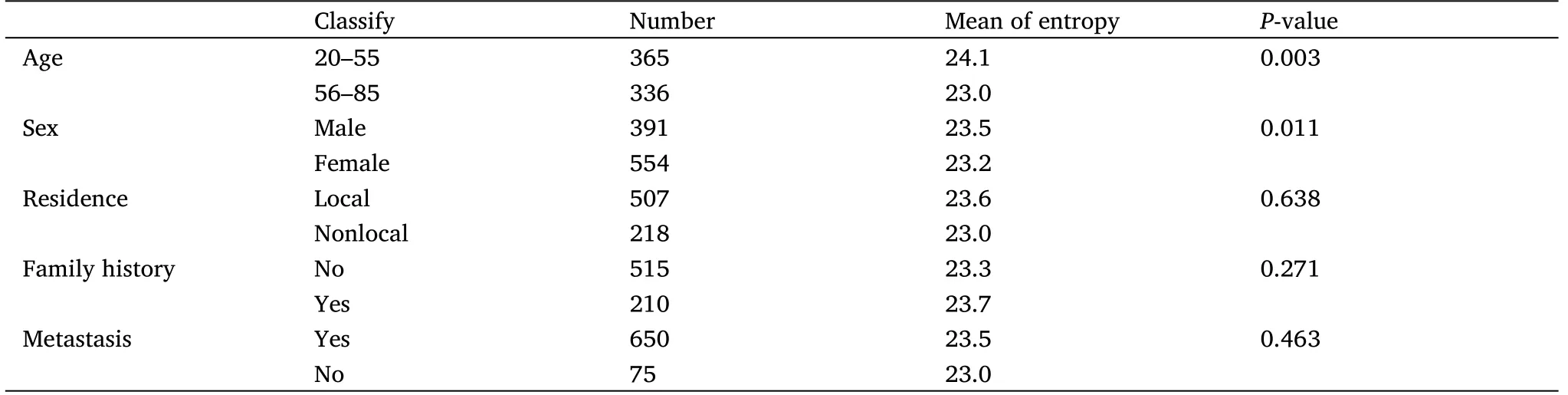
Table 3 The degree of sag in the nasal root position (U-test).
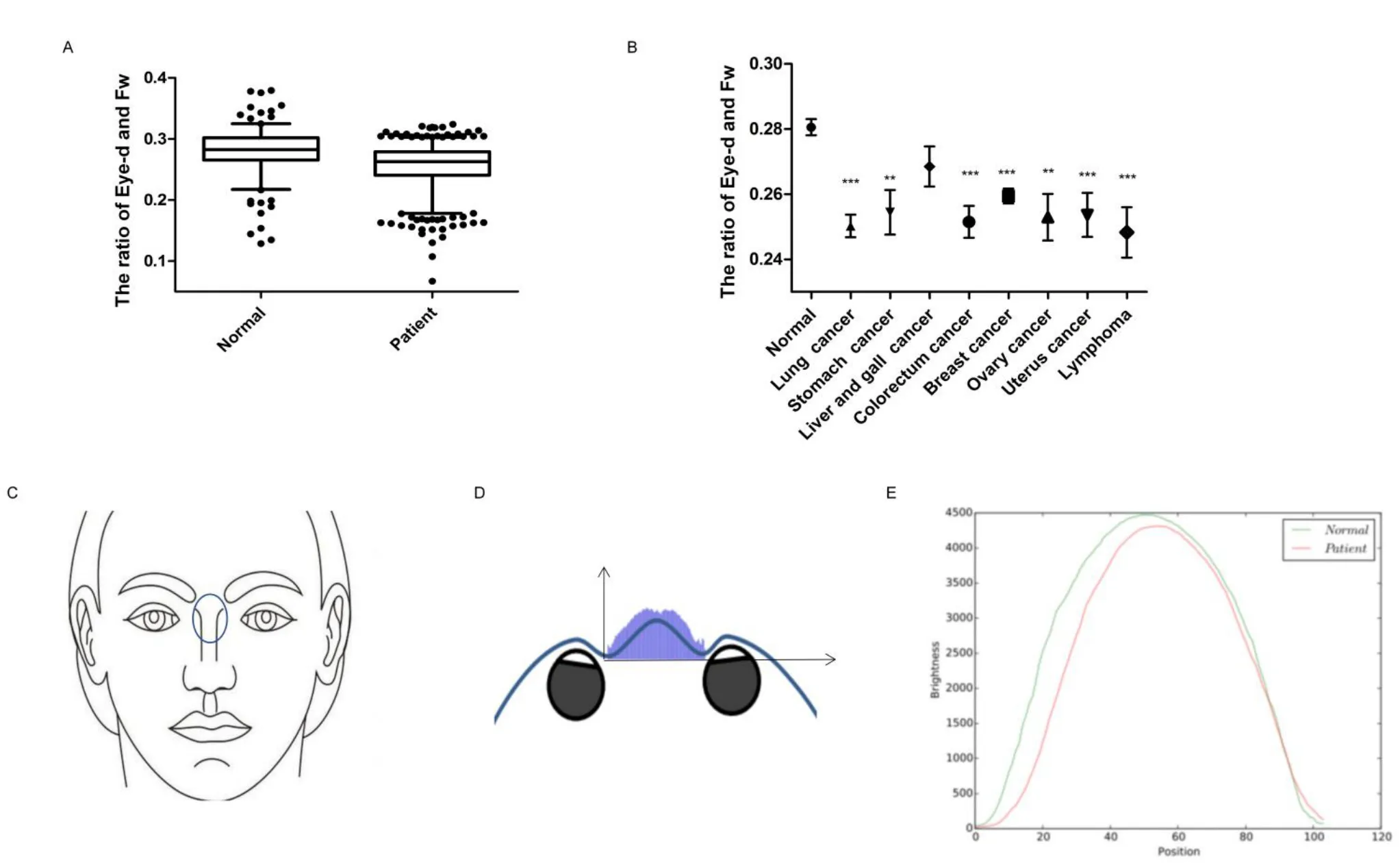
Figure 2 Facial characteristics of patients are different from normal people.(A) Distributional difference of the ratio of Eye-d and Fw between normal and tumor groups.(B)The ratio of Eye-d and Fw of different tumor groups in which the number of patients is greater than 20(*P<0.05;**P< 0.01;***P < 0.001 versus control).(C) The nose root we focused is between the eyes, below the eyebrows line and above the nose.(D) The simulated results using pixel gray values could basically represent the real face collapse situation.(E) Brightness value (gray value) to location curve for modeling of the nasal root surface condition.Eye-d,binocular spacing; Fw,face width.
Gene sequencing explain the biological foundation of AI watcher prediction
According to our conjecture, AI watcher could predict tumor risk through facial features, because some genes that determine facial features are closely related to those tumor development genes.So, we made efforts to identify genes for facial features with quantitative assessments.We collected facial images and blood samples of two tumor families, analyzed their facial features in detail and sequenced 10X exome in order to the analysis of the hereditary basis of cancer and appearance in the family.By candidate gene study carried out in two independent cancer family samples, we investigated a potential association between cancer risk alleles and facial features in cancer family analysis.We have compared the gene results with the human reference genome to filtrate and define the mutation sites which may determine both the inheritance of facial features and tumor susceptibility.
The first family includes two colorectal cancer patients and three normal relatives (Figure 3A).Using the cancer gene database to routinely analyze the tumor susceptibility gene of the family members(Figure 3B).In terms of these candidate tumor susceptibility genes,the individuals with similar appearance have the more similar cancer gene.The patient-common cancer genes include pericentriolar material 1 (PCM1), abl interactor 1, transformation/transcription domain associated protein, exostosin glycosyltransferase 1 and tumor necrosis factor receptor superfamily member 17.The mutated genes shared only by the patients may be the key to revealing the genetic nature of appearance and tumor (Figure 3C).Through the analysis of the tumor database and craniofacial development gene database together,we found many genes single nucleotide polymorphisms(SNP)mutations related to appearance are also closely related to the tumor,in particular, WW domain containing E3 ubiquitin protein ligase 2(WWP2), sperm hammerhead 3 (SH3) and PX domains 2B(SH3PXD2B), DNA-directed RNA polymerase III core subunit and coatomer subunit zeta (Table 4).Protein three-dimensional structure analysis results predicted the changes as a result of genetic mutations(Figure 3D).Through the analysis using the tumor database and facial database,there are many genes related to facial appearance and tumor(Table 4).Three-dimensional protein structure analysis results predicted the changes as a result of genetic mutations(Figure 3D).The WWP2 was shown to be the gene that determines eye distance in mice,which is also a key tumor gene [18, 19].The gene-face pattern was observed in patients’ appearance photos (Figure 3A).There are many similar facial characteristics between the two patients.For example,we focused on the relative position of the eyes to the nose.The angles of three angles in a triangle formed by the eye outer corner and the nose tip were similar in two patients (> 90°), but different from normal relatives (< 90°).Path enrichment result prompted that the Notch pathway is mainly enriched, which was also closely related to appearance, such as skull development.

Table 4 Gene SNP mutations associated with the craniofacial development and cancer genetic only in two patients in the first family
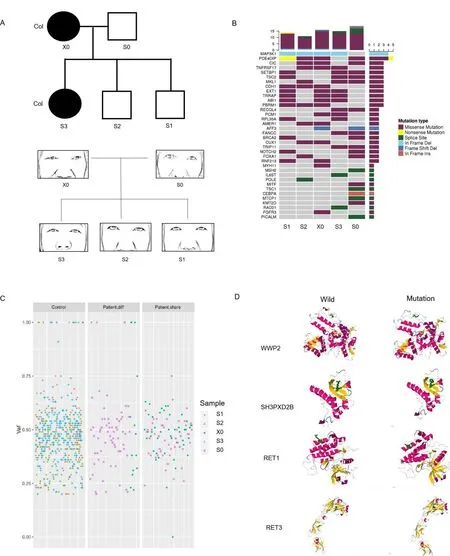
Figure 3 The pedigrees of the five people cancer families showing co-dominance inheritance of facial appearance and cancer susceptibility.(A) The cancer patients are indicated with shading.Squares and circles denoted males and females respectively.The figure below corresponds to each family member’s appearance around the eyes and nose.X0 and S0 refer to parents; S1, S2, and S3 refer to three offspring samples(2 brothers and 1 sister).Col, colorectal cancer.(B)Comparative analysis of tumor susceptibility genes.(C) Comparative analysis of genetic differences between different samples except the genes that all people share.Control, the genes of the two healthy individuals in the cancer family;patient.diff, the specific genes to different patients; patient.share, the same genes between patients; vaf, variant allele fraction.(D) Protein simulation of genetic mutations common to cancer genetics and craniofacial development.WWP2, WW domain containing E3 ubiquitin protein ligase 2; SH3PXD2B, SH3 and PX domains 2B; RET1, DNA-directed RNA polymerase III core subunit;RET3, coatomer subunit zeta.
In the second cancer genetic family, we observed the inheritance of different tumor types and faces in a family using the same sequencing and analysis methods (Figure 4).The second family includes two cancer patients (one got lymphoma and hepatoma, one got thyroid cancer) and one normal relative (Figure 4A).The list of tumor susceptibility genes showed the patient inherited oncogene Gprotein signalling modulator 2, Rho GTPase activating protein 29,erythropoietin producing hepato-cyte receptor A (Figure 4B).The mutated genes shared only by the patients were extracted and collated for analysis (Figure 4C).Through the analysis of tumor database and craniofacial development gene database, we found many genes SNP mutations related to appearance are also closely related to the tumor such as ATP binding cassette subfamily A member 4 (ABCA4),melanocortin 1 receptor (MC1R) and dachsous cadherin-related 2(DCHS2) (Table 5).Three-dimensional protein structure analysis results predicted the changes as a result of genetic mutations (Figure 4D).It was obvious that the daughter’s facial features were similar to the father’s but significantly different from the mother’s.Patients Z0(father with the tumor) and Z1 (daughter with the tumor) have many similar facial features such as the ratio of Eye-d and Fw or the relative position of the eyes to the nose(Figure 4A).In conclusion,our analysis of the genetic database found that patients who inherit the same facial features implied they inherit the same facial development genes,which are very likely to be involved in cancer development.Besides,we also found some mutations are the enhancers (Table 6).The gene regulation of facial development and tumorigenesis is very complex and not limited to exons.Our study as a preliminary exploration could reveal the genetic correlation between tumor-related genes and specific tumor appearance.
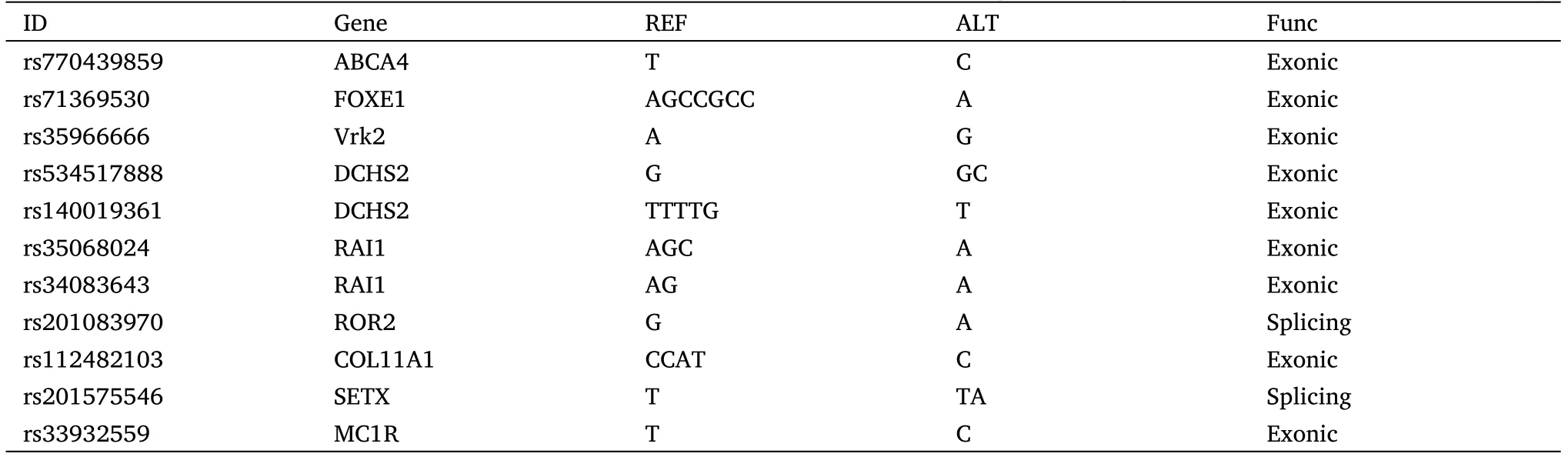
Table 5 Gene SNP mutations associated with the craniofacial development and cancer genetic only in two patients in the second family
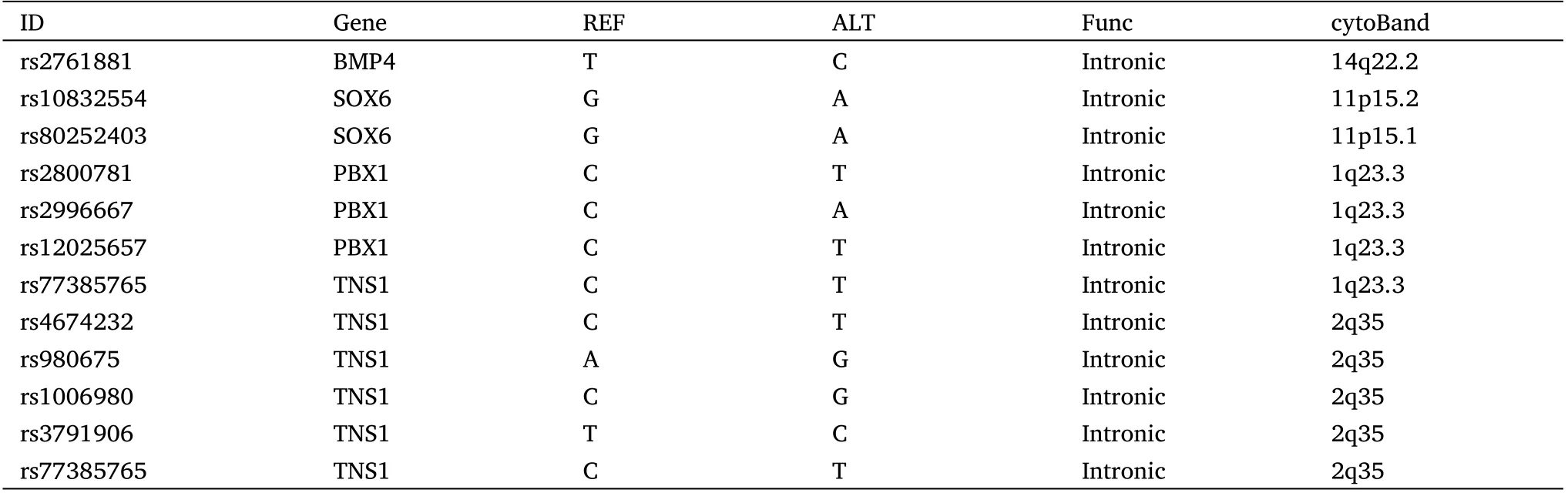
Table 6 The enhancer mutations of cancer patients in tumor families
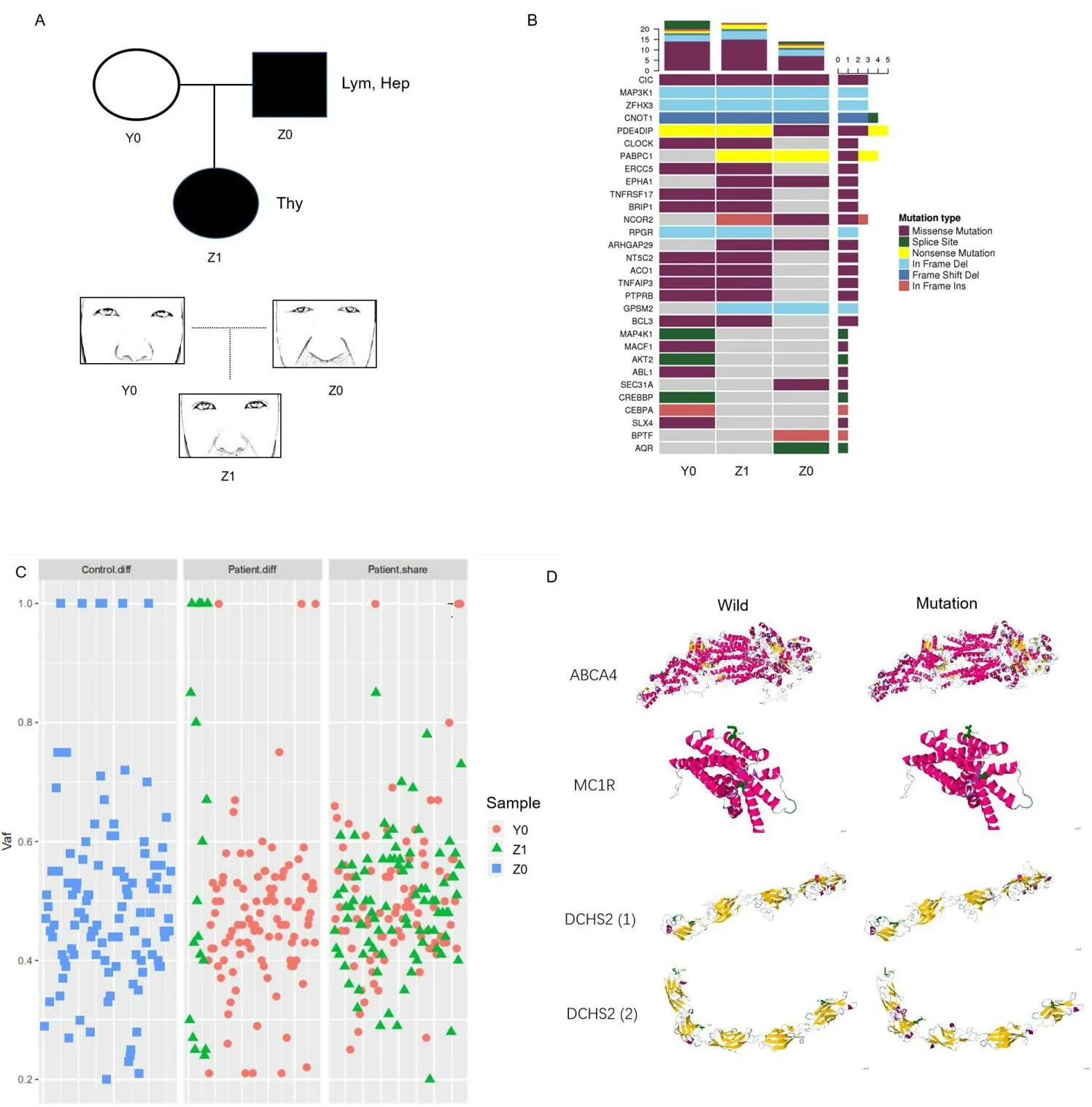
Figure 4 Sequencing analysis of facial genes and tumor genes of the three people family with different cancer.(A) The cancer patients are indicated with shading.Thy, thyroid cancer; Hep, hepatocellular carcinoma; Lym, lymphoma.Y0 refers to normal mother; Z0 refers to father with the tumor; Z1 refers to daughter with the tumor.The figure below corresponds to each family member’s appearance around the eyes and nose.(B)Comparative analysis of tumor susceptibility genes.(C) Comparative analysis of genetic differences between different samples except the genes that all people share.Control, the genes of the healthy individual in the cancer family; patient.diff,the specific genes to different patients; patient.share,the same genes between patients; vaf, variant allele fraction.(D) Protein simulation of genetic mutations common to cancer genetics and craniofacial development.ABCA4,ATP binding cassette subfamily A member 4;MC1R,melanocortin 1 receptor.DCHS2,dachsous cadherin-related 2.
Discussion
In the whole experiment, we first used the AI watcher to conduct facial analysis training and identify the difference between tumor patients and the normal.Although 2D image recognition has its limitations, it still has a high accuracy rate.To some extent, the AI watcher simulates the human visual imaging and diagnosis by observation.The program we used to find features in the eye area that had more advantages in distinguishing cancer patients.This program may not only apply to diseases such as tumors, but also other serious genetic diseases.Our AI design is still efficient in analytical processing complex image details when the sample size is not very big.Of course,a larger sample size would allow the program to predict more accurately and identify the characteristics of each disease more clearly.AI watcher is capable of finding out the facial changes caused by many diseases that cannot be found by human beings or making the preliminary diagnosis through facial features differences, such as lupus erythematosus, jaundice, anemia, hyperthyroidism, facial paralysis and so on.It may even predict the probability of certain diseases through the regularity of facial features.
The heritability of craniofacial morphology is high in families [19].Some craniofacial traits, such as facial shape and position of the eyes and nose, appear to be more heritable than others [20].The general morphology of craniofacial development and tumor formation both is largely genetically determined and partly attributable to environmental factors [21].An appreciation of the genetic basis of facial feature differences in the cancer family has far-reaching implications for understanding the etiology of facial pathologies, the origin of the proto-oncogene, and even the evolution of tumor tissue.In this candidate gene study carried out in two independent cancer family samples, we investigated a potential association between risk alleles for tumorigenesis and facial characteristic in genetic genealogy.The genes that we found in our study and that cancer patients share are probably the intersection of tumor inheritance and facial inheritance [19].
In the present study, we focus on the gene polymorphism for capturing facial morphology since previous facial development studies have demonstrated that SNP represents a crucial factor in appearance[22].It is feasible to speculate that once the major genetic determinants of facial morphology are related to tumor susceptibility genes, predicting detrimental mutation heredity from facial appearance found in cancer screening will become useful as an investigative strategy in tumor prevention.The Genome-Wide Association Study found a significant number of genetic variants associated with phenotypes.One study showed that common variants below the Genome-Wide Association Study threshold can also contribute a considerable part of heritability [23].For instance, the possibility that variation in specific craniofacial structures could result from the action of different genes might also contribute to explaining why quantitative analyses using whole-face shape summaries from 3D images have had limited success in detecting significant genetic effects[24].
In addition, senataxin and MC1R are mutated genes in two family cancer patients.MC1R is the gene that affects facial color.Although we haven’t analyzed the facial color here, the observation of complexion is a diagnostic method for inspecting many diseases.The color and luster of the face may also hint at doctors with disease probabilities [25].Moreover, our study found that a lot of introns and enhancers mutations.Other studies in theSciencejournal found that enhancers determine facial features [26-28].Further research may build on our findings and reveal more connections between enhancers and faces.From the above, faces could reflect the sequences of specific genes that affect not only appearance but also the development of tumors and possibly development [29].Therefore, the ancient Chinese physiognomy may prove that the ancient physicians had found the connection between appearance, gene and disease only by observation the appearance to predict health or disease.
In spite of some limitations, we have been able to demonstrate that a phenotype as complex as human facial morphology may be successfully investigated via the AI program with moderate sample size.The associated DNA variants may also affect neighboring genes.Some identified genes such as PCM1 and COL24A1 are potential new players in the molecular regulation of facial patterning and cancer genetics.Overall, we have uncovered 17 genetic loci that contribute to normal differences in facial shape, representing a significant advance in our view of the genetic relevance of facial morphology and tumor genetics.
The AI watcher provides a new way to predict or diagnose various diseases not only by analyzing face photographs but also by other imaging data such as computed tomography and magnetic resonance imaging.Most noteworthy is the use of AI watcher may support image acquisition systems to screen for multiple diseases with physiognomic changes in the future.There are a number of studies that look at physical features to predict disease, such as earlobe folds, which have been observed to be associated with cardiovascular disease [30].Other studies have used a combination of 3D imaging and machine learning to analyze and identify facial features of acromegaly patients,and found that AI can be used to detect patients with acromegaly earlier [31].In the future, we will use 3D imaging and AI to further analyze the association between appearance and genes.
Conclusion
The AI watcher provided an innovation for cancer auxiliary diagnosis,which may guide the potential tumor genetic testing and treatment.Moreover, our data also highlight that the high heritability of facial shape phenotypes is associated with cancer genetic susceptibility,involving many ignored DNA variants with a degree of cancer risk.Future large samples on cancer group phenotype should help to identify more genes for the susceptibility of many other complex hereditary diseases.
 Traditional Medicine Research2022年1期
Traditional Medicine Research2022年1期
- Traditional Medicine Research的其它文章
- Evaluation of bioactive flavonoids in Citri Reticulatae Pericarpium from different regions and its association with antioxidant and α-glucosidase inhibitory activities
- Exploring the anti-diabetic effects and the underlying mechanisms of ethyl acetate extract from Sophora flavescens by integrating network pharmacology and pharmacological evaluation
- Effects of Shenling Baizhu powder on endoplasmic reticulum stress related signaling pathway in liver tissues of nonalcoholic fatty liver disease rats
- Artificial neural network techniques to predict the moisture ratio content during hot air drying and vacuum drying of Radix isatidis extract
- Safety of Lycium barbarum L.:more information needed
- Mechanisms and status of research on the protective effects of traditional Chinese medicine against ischemic brain injury
