Identification of QTLs Associated with Resistance to Pseudomonas syringae pv. Glycinea in Soybean (Glycine max (L.) Merr)
Mei Hong-yao, Liu Yang, Pan Xiao-cheng, Su An-yu, and Wu Xiao-xia,
1 College of Agriculture, Northeast Agricultural University, Harbin 150030, China
2 65301 Farm and Sideline Bases of Army, Wudalianchi 164100, Heilongjiang, China
3 College of Resources and Environmental Sciences, Northeast Agricultural University, Harbin 150030, China
4 Institute of Green Food in Heilongjiang, Harbin 150030, China
Abstract: Soybean bacterial spot disease caused by Pseudomonas syringae pv. Glycinea which is a bacterial disease seriously affects soybean yield. Ten soybean germplasms and recombinant inbred lines (RILs) population were used to identify the resistant trait after inoculated with P. sg (P. sgneau001) in this study. High-density genetic mapping was obtained by specific length amplified fragment sequencing (SLAF-seq) of 149 RILs population which was derived from the crossing between Charleston and Dongnong594. The results indicated that 10 germplasm resources had four resistant germplasms included highly resistant cultivar Charleston, four susceptible varieties included Dongnong594 and two moderately resistant cultivars. Five quantitative trait locus (QTLs) were detected in RILs population by the composite interval mapping (CIM) method, and located on Linkage Group (LG) D1b (chromosome two),LG C2 (chromosome six) and LG H (chromosome 12), respectively. LOD scores ranged from 2.68 to 4.95 and the phenotypic variation percentage was from 6% to 11%. Six candidate genes were detected, according to the result of gene annotation information.Four of them had relationship with protein kinase activity, protein phosphorylation and leucine rich repeat (LRR) transmembrane protein, which had high expression after inoculated with P. sg by qRT-PCR.
Key words: soybean, QTL mapping, Pseudomonas syringae pv. Glycinea, bacterial spot disease, candidate gene
Introduction
Soybean is an important crop globally that provides not only oil and protein for human consumption but also as feed for animals. However, the yield has decreased rapidly in recent years which is due to the occurrence of various diseases (Dallaet al., 2015;Fallet al., 2018). Soybean bacterial spot disease is one of the most economically damaging disease affecting soybean-producing area worldwide. This disease is caused by the bacteriaPseudomonas syringaepv. glycinea (P. sg) (Duncanet al., 2011;Duncanet al., 2014; Gonzalezet al., 2006). Soybean plant can be infected withP. sgin its whole growth period, but the incidence on the leaves is heavier.Susceptible infection can lead to yellow spots on leaves, surrounded by chlorotic spot halo and eventual death of the plant as well as seed staining (Almeidaet al., 2009; Liet al., 2017). Studies have shown that susceptible varieties under conducive condition, the loss caused by the disease can be as high as 30%-40%(Zhanget al., 2011).
Since 1980, many soybean germplasm resources have been screened using different methods for inoculation ofP. sg(Wilsonet al., 2001). At present,the inoculation ofP. sgincludes mainly high pressure spray method (Staskawiczet al., 1987), acupuncture method (Wanget al., 2014) and high-pressure spraycoated culture medium investigation method (Wensinget al., 2010). Ashfieldet al. (1995) used injection vaccination to identify high-resistance soybean germplasms, such as Flbmbeau, PI96983 and Jack. Sunet al. (1989) identified high-resistance materials with field spray method among which are Jilin2, Heinong9 and Jilin7. Chenget al. (2016) used outdoor highpressure spray method withPseudomonas syringaephysiological race S1 to identify the 309 soybean breeds of Jianghuai region in China. Zhanget al. (2006)used acupuncture to screen Hefeng15, Dongnong42 and 108 soybean materials in northeast China for identification ofPseudomonas syringaerace 4.
Many resistance genes are still in plants, having many defense systems and response factors, but pathogens are inextricable from plants (Llorenteet al.,2005). Therefore, the development of soybean bacterial spot resistant cultivars can reduce the losses caused by the disease without the expense and negative environmental impact of disease management or control which will be of great significance to identify excellent germplasm resources and mining excellent resistance genes in soybean breeding (Liet al.,2011). In 1999, a RILs population whose parents are BelnebRR-1 and A55 response toPsp(HB16 and 83-Sc2A) associated with QTLs (Ariyarathneet al., 1999). Results showed that leaf response of isolate HB16 was related to the three QTLs (LGPv03,LGPv05 and LGPv10) in common bean (Phaseolus vulgarisL.) and reactions to strain 83-Sc2A were significantly associated with the four regions (LGs Pv02, Pv04, Pv05 and Pv09) (Trabancoet al.,2014). Thapaet al. (2015) worked on four QTLs of introgression lines (ILs) in common bean using composite interval mapping (CIM) method; the results showed that bsRr1-1, bsRr1-2, bsRr1-12a and bsRr1-12b were located on chromosome one, two and 12,respectively. In common bean, two independentRgenes (Rpsar-1 andRpsar-2) are responsible for resistance to AvrRpm1 and are located at the end of LGs 11 and 08 (Gonzálezet al., 2017). In tomato, the inbred backcross population derived from PI114490 and H7998 with quantitative trait loci (QTL) of resistant bacterial spot race T4is obtained on chromosome 11 and the variation is 29.4 (Huttonet al., 2010).Four QTLs show association with partial resistance inArabidopsis thaliana, located on chromosomes one,two, three and five, respectively, and fine mapping narrowed to 62 genes (Rantet al., 2013). Ahmadet al.(2011) investigated the main effect of QTLs on chromosome III, and QTLs with weak chromosome I were identified as RILs population, whose parents were Bayreuth and Shahdara. Kover and Cheverud (2007)found a few QTLs of resistance gene inA. thaliana's response toP. syringae. According to the heterologous inbred lines (HIFs) strategy, two major and two minor QTLs in the Near Isogenic Lines (NILs) population are detected by two different environmental conditions,major QTL (PRP-Ps2) is negatively correlated with salicylic acid (SA) dependent activation, and these results indicate that Pst (DC3000) partial resistance is controlled by relatively few QTLs inArabidopsis(Dobónet al., 2011; Perchepiedet al., 2006).
In this study, 10 germplasms were inoculated withP. sgneau001 to identify disease resistance, and it could be found that of Charleston gave the highest resistance levels to bacterial spot disease, in contrast,Dongnong594 was highly susceptible to this disease.Thus, QTL analysis and qRT-PCR were used to screen resistance genes of RILs population in soybean.
Materials and Methods
Experiment materials
High-density genetic mapping had been obtained by SLAF sequencing of 147 RILs population derived by crossing semi-dwarf soybeans Charleston (♀) from the United States and Dongnong594 (♂) (Qiet al.,2014). There were 20 linkage groups of 5 308 specific length amplified fragments with a total length of 2 655.68 cM and an average of 0.5 cM adjacent marker.P. sgneau001 was used as experimental strain, which was collected from the diseased leaves of soybean in Jiamusi City, and was isolated, purified and identified by Koch's postulates.
Spray inoculation under indoors condition
The plants grew to V 1 (first trifoliate) inside greenhouse under 16 h photoperiod, with day and night temperature of 22℃±2℃ and 65% relative humidity.The evaluation was performed in the three independent replicates. Single bacterial colony was picked from media and grew over night in NYG liquid media supplemented with Carbenicillin 50 mg • mL-1at 28℃ (Ross and Somssich, 2016).P. sgsuspension OD600nm=1.0 (1OD=5.0×108CFU • mL-1) was diluted 1 000 times to a concentration of approximately 105CFU • mL-1, mixed with 10 mmol • L-1MgCl2and 0.05% Silwet-L77.
The bacterial suspension was applied by spraying inoculum on the back of the leaves, when they were fully expanded (Yanget al., 2013). Plants were kept in 100% relative humidity in a plant growth cabinet for 96 h after inoculation (Fondevillaet al., 2012) and after which an equal volume of the three leaf disks was removed with a punch, ground thoroughly in 300 μL of 10 mmol • L-1MgCl2, diluted to a suitable multiple and applied to NYG solid medium with a 50 mg • mL-1carbenicillin. After 2 days of culture, the number of bacteria on the medium was counted. The rate of disease was determined by number of lesions that were counted on the 12th day afterP. sginoculation.
Spray inoculation under field condition
From 2017 to 2018, RILs population were planted on the Xiangyang Farm of Northeast Agricultural University with a length of 2 m, a row spacing of 60 cm and a plant spacing of 10 cm. Inoculation identification was carried out in the stage of soybean V six (about 55 days of soybean growth). The inoculation method was a high-pressure spray method, and the specific steps were the same as above. The rate of disease was determined from 20-30 days afterP. sginoculation.
Disease scale evaluation
Disease score was on a scale of 0-5, where 0=leaves almost had no change; 1=number of spots was 5-20;2=the lesion area expanded with chlorotic halos formed around, with 20-50 spots; 3=the lesion area spreads, the number of spots was 50-80; 4=the area of diseased lesions exceeded half of the leaf area and the leaves turned yellow; 5=the leaves turned yellow,withered and dropped (Donget al., 2010). Bacterial spot disease phenotype was visually rated by disease index after inoculation ofP. sg. 0 and 1 were resistant,grade 2 was for moderate levels of resistance toP. sgwhile grade 3-5 represented susceptibility. Statistical analysis of data was conducted using SPSS 22.0 software.
Genetic effect analyses of RILs population
SEA (segregation analysis) software package developed in C++ language, which was based on the Windows, was used to observe the quantitative traits of the parental segregation population and identify the quantitative trait main gene and multi-gene mixed genetic model. At the same time, all the genetic models were analyzed, and AIC (akaike information criterion) value was calculated by the likelihood function value of the model. The optimal genetic model was selected, according to the minimum value of AIC. The genetic parameters of the main gene and the polygene were estimated by the distribution parameters of each component, the gene effect values and various genetic variances were estimated by the least square method, and Smirnov test and the Kolmogorov test were performed simultaneously.
QTL analysis
QTL mapping and analysis was conducted using composite interval mapping (CIM) method of Win QTL Cartographer V.2.5. The likelihood ratio (LR)was greater than 11.5 (LOD>2.5) as the threshold for QTL existence, LOD value greater than 2.5 was used to determine the confidence interval, 1 000 iteration test was performed, and the existence of QTL was judged atp<0.05 (Churchill and Doerge, 1994).
Gene model prediction and annotation
Soybase genome browser (http://soybase.org/gbrowse/cgi-bin/gbrowse/gmax1.01/) was used to find all the predicted genes and their functional annotation information in the positioning interval. QuickGO website (http://www.ebi.ac.uk/QuickGO/) and KEGG website (http://www.kegg.jp/) were used to observe the structure of the gene and its associated pathway map. At the same time, NCBI website was used to find and analyze the protein domains against the predicted candidate genes. Phytozome website (www.phytozome.net/soybean) was used to predict the coding DNA sequences (CDS) of resistance genes and screening for a typical domain of a disease-resistant gene or a gene that had been confirmed of plant disease resistance.
RNA extraction and qRT-PCR analysis
Functional genes associated with pathogen infestation and plant disease resistance were selected through functional annotation in gene annotation, and qRT-PCR analysis was performed on these genes. The inoculated plants were cultured in a greenhouse at 22℃±2℃,and kept in 100% relative humidity. Primary and trifoliate leaves were taken at 0, 2, 4, 8, 12 and 24 h after inoculation; however, MgCl2was inoculated as a control experiment. qRT-PCR experiment was repeated with three organisms for each sample. Liquid nitrogen was used to froze the sample and placed the sample in a refrigerator at –80℃. The total RNA from soybean leaves was extracted using Trizol Reagent and c-DNA was synthesized using the Prime ScriptTMRT reverse transcription reagent kit. Using cDNA as a template, the total reaction system was 20 μL. Based on the predicted candidate gene sequences, fluorescent primers were designed, andGmUNK1 (Glyma.12g020500) was used as an internal reference gene. Primer Premier 5.0 was used to design the primer sequences based on phytozome website (http:/www. Phyzome.Org/). PCR amplification standard procedure was used as the followings: first step, pre-denaturation 1 cycle at 50℃ for 30 min; second step, PCR reaction for 1 cycle 85℃ 30 s. The amplification and melting curves after the completion of qRT-PCR reaction were analyzed, and relative quantitative analysis of gene expression levels was done by comparing 2-ΔΔCTmethods (Liuet al., 2014) using the following equation:ΔΔCT=(CtTarget–CtUNK1)X–(CtTarget–CtUNK1)Y,XandYrepresented test and control treatments,respectively.
Results
Phenotype analysis of RILs population
After the 14th day of inoculation, the infected leaves of RILs population parents showed a different reaction; compared with Charleston, Dongnong594 had visibly chlorotic and necrotic patches (Figs. 1 and 2).Results indicated that 10 germplasm resources had four germplasms which were highly resistant cultivars,four susceptible varieties and two moderately resistant cultivars (Fig. 1). Meanwhile, the number of bacterium colonies of Charleston medium was 104, while that of Dongnong594 had 528 indicated that the number of bacteria in Charleston was significantly lower than that of Dongnong594 and this was closely related to the lesions of leaves. RILs population showed significant segregation of resistance with average number of bacterial colonies that ranged from 9 to 2 861 in the 147 RILs lines, thus the maximum value exceeded the highest valued parent and the minimum value was lower than the lowest valued parent (Fig. 3). The average value of phenotypic data in RILs had 414 bacterial colonies. Standard deviation and kurtosis of the phenotypic trait were 371.29 and 14.88, respectively. Skewness of the phenotypic trait was 2.94.Charleston and Dongnong594 had disease scale of 1 and 4, respectively. Thus, indicating that the character had both positive and negative heterosis, which was in accordance with a normal distribution of disease scores and it was suitable for QTL mapping analysis (Fig. 4).

Fig. 1 Number of Pseudomonas colonies reflected in culture medium
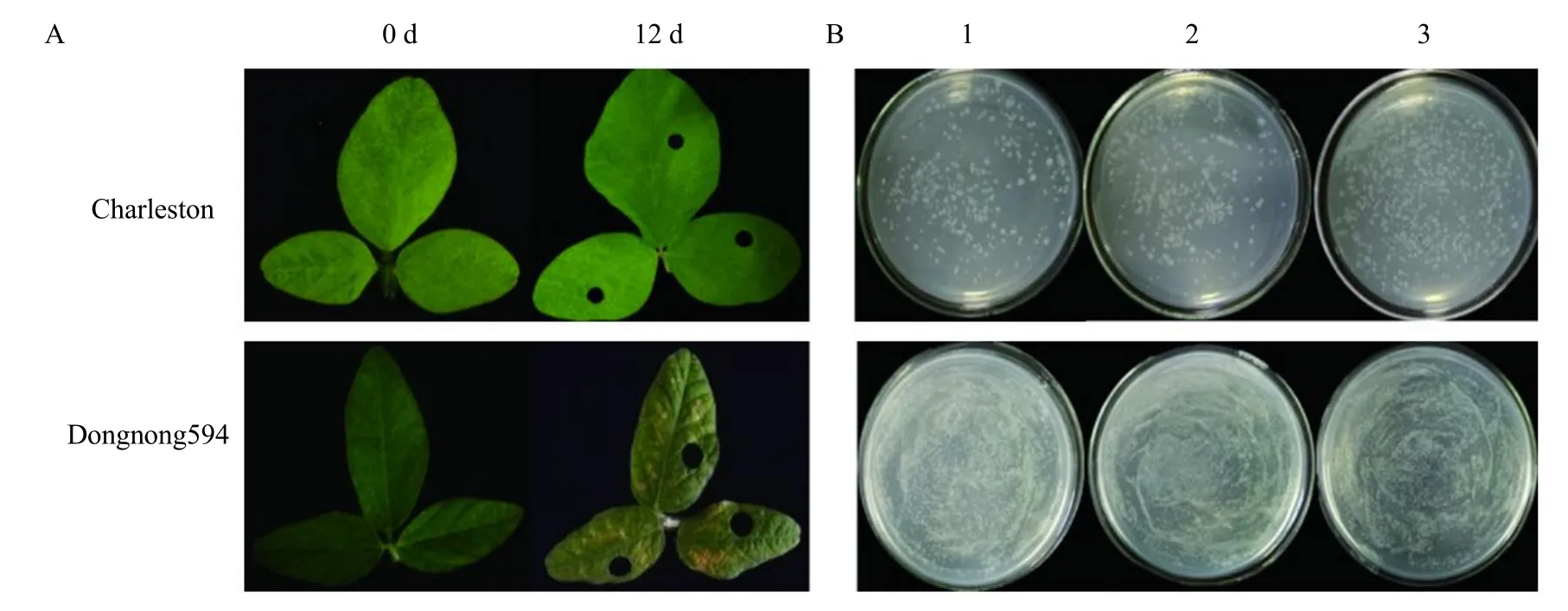
Fig. 2 Phenotype of soybean inoculated with P. sg
QTL mapping analysis for bacterial spot disease in RILs population
The average number of bacteria of RILs population was used to identify QTL associate with pathogen resistance. Five QTLs with CIM were identified with LOD>2.5, located on LG D1b (chromosome 02), C2(chromosome 06) and H (chromosome 12), respectively (Fig. 5).
LOD scores ranged from 2.68 to 4.95 with significance threshold, which was determined by 1 000 times permutation test (p<0.05). LG D1b and H contained two QTLs, which were named according to bacterial spot resistanceP. sg(P. sg-d1b-1,P. sg-d1b-2,P.sg-h-1 andP. sg-h-2) (Table 1). Two QTLs,P. sg-d1b-1 andP. sg-d1b-2, were distributed on LG D1b, having two LOD scores of 2.68 and 3.13, accounted for 6%and 7% of the phenotypic variance, and additive effect of –62.56, –63.43, respectively.P. sg-d1b-1 andP. sgd1b-2 were located on 66.90 cM and 72.10 cM, Marks 1032085 and 1022191, Marks 997567 and 1052231,respectively. The main-effect QTLs were detected in LG H (P. sg-h-1 andP. sg-h-2). This could explain 11%of the phenotypic contribution for the resistance ofP.sg, and LOD scores of 4.72 and 4.95, respectively. The main QTLP. sg-h-2 on LG H was located on many genes that involved in disease resistance, mapped between 2 076 180 bp and 6 229 601 bp. However,QTL on LG C2 namedP. sg-C2-1 was located at 35.80 cM below Marks 444279 and 458983, respectively. It explained about 6% of the variation and an additive effect of –95.36.
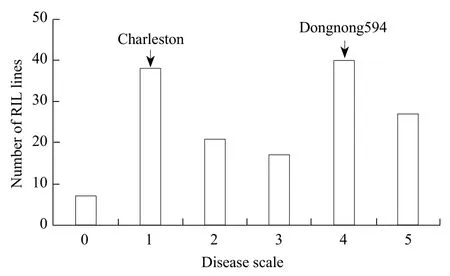
Fig. 3 Frequency distribution of disease scale in 'Dongnong594×Charleston' RILs population
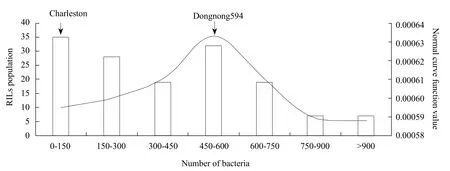
Fig. 4 Frequency distribution of number of bacteria after 4 days of inoculation in RILs
qRT-PCR expression analysis of candidate genes
With the results of gene annotation information, candidate genes were predicted through the phytozome website (http://www.phytozome.net/cgi-bin/gbrowse/soybean) within the five QTL regions, six genes out of them showed a relationship with disease resistance protein (TIR-NBS-LRR class) family (Fig. 6). According to it, all the genes contained in GO:0005515 and they were found in a large pathway which refered to the innate immune response caused by external infestation based on GO annotation results. GO:0007165 was involved in intracellular signal transduction and with genes shown, responded to biological stress, such as tolerance to environmental stress.
Interestingly,Glyma.02g193400 andGlyma.12g027100 occurred in this pathway, which was the main class of disease resistance genes; both encoding tobacco mosaic virus (TMV) resistance protein N(Djebbiet al., 2015).Glyma.02g193400 was associate with transmembrane receptor activities and took part in intracellular defense response and innate immune response. Qiet al.(2014) revealed thatGmCHX1 contained TMV resistance protein N inGlycine soja,thus making it a potential salt tolerance determinant in soybean through qRT-PCR and the rapid gainof-function tests.Glyma.12g027100 encoded TMV resistance protein N-like and the homologous gene wasAT5G17680, they participated in the process that modulated the expression ofDEK3 target genes(Lobanenkovet al., 1985).Glyma.12g040000 andGlyma.06g175100 were associate with the role of protein kinase activity and protein phosphorylation,also had the same GO in the upstream of this pathway(GO:0006468, GO:0004672).

Fig. 5 Composite interval mapping (CIM) with QTL Cartographer identified five QTLs

Table 1 QTLs for resistance to P. sg in Dongnong594 (susceptible) and Charleston (resistant) recombinant inbred population
The homologous gene ofGlyma.12g040000 wasCLAVATA1, a kind of trans-membrane receptor kinase, which interacted withCLV2, these two receptors were involved inCLV3 during plant development, and contributed to bacterial wilt through a signaling pathway (Hanemianet al., 2016;Nimchuk, 2017).SOBIR1 was the homologous gene ofGlyma.06g175100, this appeared to be a crucial component of RLP-containing complexes, which could encode a putative leucine rich repeat (LRR)transmembrane protein that was expressed in response toPseudomonas syringae(Biet al., 2014; Liebrandet al., 2013; Takahashiet al., 2016). LRR motifs played an important role in recognizing and binding various pathogen associated molecular patterns(PAMPs) and promoted innate immune response (Bejet al., 2014).Glyma.12g055600 andGlyma.12g062500 occurred in GO pathway (GO:0005515) and participated in the process of DNA-damage-repair and encoding of Rab5-interacting family protein.AT3G20820 was homologous gene ofGlyma.12g055600, associate with ubiquitination. Ubiquitination had implicated in various cellular functions which included endocytosis,signal transduction and DNA damage repaired (Kobe and AV, 2001; Haglund and Dikic, 2014; Igawaet al.,2009; Yamaokaet al., 2013). The homologous gene ofGlyma.12g062500 wasAT5G59410, containing Rabaptin-5, a factor of endocytosis and early endosome fusion, functioned as a molecular linker to regulate endocytic and recycling traffic (Vitaleet al., 2014).

Fig. 6 Soybean bacterial spot disease QTL and distribution of candidate genes
The resistant cultivar Charleston and the susceptible cultivar Dongnong594 were inoculated withP. sgand analyzed by qRT-PCR. Compared with the control and in regards to the genes,Glyma.02g193400 andGlyma.12g027100 had no detect signal, while other four candidate genes (Glyma.06g175100,Glyma.12g040000,Glyma.12g055600 andGlyma.12g062500)were expressed in soybean leaves within 24 h after inoculation (Fig. 7).
The gene expressions ofGlyma.06g175100,Glyma.12g040000,Glyma.12g055600 andGlyma.12g062500 reached their peaks at 4 h after inoculation withP. sgin Charleston, which had the most prominent phenotypic trait of disease resistance. In contrast, after inoculation 8 h, the four candidate genes had the highest expression level in Dongnong594, cultivar associated with susceptibility diseases. The expression level ofGlyma.06g175100 reached two times in Charleston at 4 h after inoculation and reached the highest value at 8 h, which was 2.5 times higher than at 0 h in Dongnong594, indicating that the gene played a key role in the plants, but mainly in susceptible varieties(Fig. 7A).Glyma.12g040000 andGlyma.12g055600 exhibited the highest expression level in Charleston at 4 h after inoculation, which was seven and five times of 0 h, respectively, and it was considered as stress response process, when the expression level increased rapidly (Figs. 7B and 7C).

Fig. 7 Expression of resistance related genes at different times by qRT-PCR in Dongnong594, Charleston
The expression levels ofGlyma.12g040000 andGlyma.12g055600 at 4 h after inoculation in diseaseresistant cultivar were significantly higher than those at 8 h after inoculation in disease susceptible cultivar,indicating that they play a greater role in diseaseresistant cultivar and performed their functions very quickly after-inoculation. The expression level ofGlyma.12g062500 changed in both resistant and susceptible cultivars, and in resistant varieties the highest value of the expression level was slightly higher than that of the susceptible varieties, suggesting that the gene played a major role in resistant varieties (Fig. 7D). The changing expressions ofGlyma.12g062500 andGlyma.06g175100 were generally consistent at the early time after inoculation of the plants, especially, when the expression decreased in 2 h and then rose to the highest in 4 h.
Discussion
Resistance to bacterial spot disease in RILs population controlled by QTLs
One way to increase our knowledge of resistantP. sgwas to study its control by using QTLs in developing efficient breeding strategies to produce resistant soybean cultivars. In this study, the severity of bacterial spot disease in RILs populations of soybean was evaluated by using both disease scale and culture medium coating test underP. sginoculation.Different from other quantitative trait loci results,this work suggested that five QTLs of bacterial spot disease were detected in RILs populations, located on LG D1b, C2 and H, respectively. In addition, two QTL intervals were in LG D1b and H respectively.Some of these QTLs had been reported in the past,showing association with other soybean pathogens.Approximately 18 cM upstream of region spanned from 66.61 cM to 68.65 cM on LG D1b, QTL for soybean mosaic virus (SMV) strain SC7 resistance was identified and mapped in 184 RILs, derived a cross between Kefeng1 (resistant) and Nannong1138-2(susceptible) (Yanet al., 2015). One QTL on LG C2 for soybean plant sudden death syndrome (SDS)resistance was identified for the foliar disease severity(FDS) three traits (qFDS003-03).
In the study reported herein, the geneGlyma.06g175 500 was obtained by QTL in 34.42-36.12 cM of LG C2. It was a homologous gene with CNL(coiled-coil (CC) domain-encoding) disease resistance geneGlyma.06g175600 that was a key gene (or gene cluster) influencedP. sg(Nepal and Benson, 2015).Duet al. (2018) reported that genehrpZmwith over expression ofP. sgcould enhance the resistance of PRR in transgenic soybean and the expression level substantially increased during multiple defense pathways. Therefore, it could be said that some resistance genes controlled multiple disease pathogens sometimes (Xianet al., 2002). QTL on linkage group'H' regioned in 54.92-59.81 cM was also associated with resistant to soybean pest, such as antibiosis that was resistance to corn earworm and soybean pod borer(SPB) (Rectoret al., 2000; Zhaoet al., 2008).
Candidate genes analysis of soybean bacterial spot disease
Four candidate genes (Glyma.06g175100,Glyma.12g040000,Glyma.12g055600 andGlyma.12g062500)were expressed in soybean leaves within 24 h after inoculation, also had similar expression levels. The resistant cultivar reached its peak expression at 4 h after inoculation, while the susceptible cultivar reached its peak expression at 8 h after inoculation suggested that resistant plant showed a rapid effect factor in triggering innate immune response. Nemchinovet al.(2017) reported that both Dongnong594 and Charleston showed signs of necrosis at 24 h. To further confirmed howP. syringaeresistance genes respond to SA and JA (jasmonic acid), the expression patterns response toP. syringaeresistance genes were analyzed by using qRT-PCR after exposing tobacco samples to 100 μmol • L-1SA or JA for 0-24 h periods of time (Liuet al., 2013). Stefanowiczet al.(2016)reported thatAt2g02360 was a stress-responsive gene,and the expression levels had significantly changed inA. thalianaseedlings after SA treatment for 24 h by using qRT-PCR.
The nucleotide binding site leucine rich repeat(NBS-LRR) gene had been confirmed as an important gene family of bacteria spot disease resistant (Muylleet al., 2005). Genes encoding NBS-LRR proteins could recognize the plant pathogens, thus serving as major resistance mechanisms, this had also been confirmed in common beans (Collinset al., 2001;Elliset al., 2000; Molinaet al., 1999).Arabidopsis-Pseudomonasinteraction was a model path system,controlling plant defense responses and signal transduction pathways (Collinset al., 2001; Elliset al.,2000; Molinaet al., 1999). The genome of the model dicotArabidopsisincluded 149NBS-LRRgenes and 11 of that had been regarded as disease resistance genes (Madsenet al., 2003). For instance,Glyma.06g175100, which orthologs withArabidopsis thalianageneSOBIR1, encoding putative leucine rich repeat (LRR) transmembrane protein. Similarly,RPS4 and RRS1 that contained a leucine zipper (LZ)motif and a C-terminal WRKY domain, which was leucine-rich repeat-containing (NLR) proteins inArabidopsis thaliana, confered resistance to multiple pathogen isolates (Ahmadet al., 2017; Narusakaet al., 2016). Protein kinase activity and protein phosphorylation occurred inGlyma.12g040000 andGlyma.06g175100 signal pathway. Therefore, resistance gene in soybean might be mediated also by WRKY family transcription factors, showing association with protein phosphorylation, and defense response which included synthesis of isoflavonoids and signal molecules of salicylic acid (SA)(Pieterseet al., 2012). Liuet al.(2013) thought that it could be presented that SA accumulation could be induced to a higher level than that of JA afterPseudomonas syringaepv. Tomato DC3000 (Pst DC3000) infection. DNA-damage-repair, endocytosis and ubiquitination occurred inGlyma.12g055600 andGlyma.12g062500 signal transduction pathway.Ubiquitination was involved in multiple aspects,during the plant development, Ewanet al.(2011)reported that inArabidopsis, AtUBP12 and NtUBP12 deubiquitinating activity was to suppress plant cell death, when infested withP. st. PARP was automatically activated, when DNA was damaged, to complete DNA repair process. Its family including PARP2 and PARP1, interact with each other, made strong contributions to plant immune responses by restricting the growth ofP. stpathogens (Songet al.,2015). Therefore, it could be considered that the proteins produced by the genesGlyma.12g055600 andGlyma.12g062500 were like that of PARP2 or AtUBP12.
Conclusions
In this study, 10 germplasm and recombinant inbred lines (RILs) population were used to identify the resistant bacterial spot disease trait, and five genomic regions associated withP. sgneau001 were reported.The detection of QTL had great significance, especially in the screening of resistant bacterial spot disease of soybean germplasm resources and molecular marker-assisted breeding. Four of potential candidate genes showed up-regulation at 4 and 8 h in resistant and susceptible cultivar using qRT-PCR of gene expression detection. These candidate genes had been reported for resistance function in diverse crop species.
 Journal of Northeast Agricultural University(English Edition)2021年2期
Journal of Northeast Agricultural University(English Edition)2021年2期
- Journal of Northeast Agricultural University(English Edition)的其它文章
- Development of an Artificial Diet for Effective Oral Delivery of dsRNA to Soybean Pod Borer, Leguminivora glycinivorella (Lepidoptera: Tortricidae)
- Grain Yield and Nitrogen Use Efficiency of Hybrid Rice in Response to High Plant Density and Nitrogen Rate
- Index Design and Comprehensive Evaluation of Germplasm Resources of Fruits Based on Mathematical Model of AHP and FCE: Sterculia nobilis Smith as a Case
- Effects of Interaction of Soil Moisture and Organic Matter on Powdery Mildew Disease and Growth of Heracleum moellendorffii Hance
- Comparative Analysis of Bacillus thuringiensis Vip3Aa57 and Vip3Aa62 Insecticidal Activities
- Comprehensive Evaluation of Processing Quality of Tibetan Native Hulless Barley Variety by Factor Analysis
