Formation mechanism of single-stranded DNA binding protein fibers fractal structure at liquid-solid interface studied by atomic force microscopy
XING Chunyan , ZHANG Miaomiao , QIAO Haiyan ,TANG Jilin , ZHANG Bailin
(1.State Key Laboratory of Electroanalytical Chemistry, Changchun Institute of Applied Chemistry, Chinese Academy of Sciences, Changchun 130022, China; 2.Shaanxi Anke Safety Production Technology Research Institute Company Limited,Xi’an 710065, China; 3.College of Petrochemical Engineering,Liaoning Shihua University, Fushun 113001, China)
Abstract: Single-stranded DNA binding protein(SSBP)could self-assemble into fibrils by 4-aminothiophene(4-ATP)on a gold substrate modified with 1-hexadecyl mercaptan monolayer membrane(HDT/Au).The nanoscale characterization and imaging of the branched patterns formed by the self-assembly of proteins were observed via atomic force microscopy(AFM)for structural analysis.The results showed that amides could induce the formation of protein fibers on HDT/Au substrates, while alkanethiols or carboxythiols could not.It suggested that electrostatic forces between positively charged amides and proteins provided the driving forces to promote aggregation for the self-assembly of protein fibrils.The formation of branched-like protein fibrils rather than the more compact close-packing of single filaments induced by amide substances might provide a basis for the development of a new and spatially controllable protein interface self-assembly method.
Keywords: self-assembly; atomic force microscopy; 4-aminothiophenol; single-stranded DNA binding protein; fibril
0 Introduction
Molecular self-assembly processes at various length scale to form functional complex are ubiquitous in diverse nature systems[1].Owing to the well-ordered organization, high homogeneity and molecular dimensions, the molecular-assembly structures provide insight into the harnessing of unique properties for engineering of new materials.Molecular self-assembly takes place via a subtle balance between noncovalent interactions including hydrogen bonding, van der Waals force, electrostatic interaction, hydrophobic interaction and aromatic π-π stacking to form functional supramolecular structures which are of outstanding interest from fundamental as well as applied perspectives.In addition, understanding the mechanisms of molecular self-assembly can be used to guide to create novel biomaterials which have been the subject of considerable research due to the outstanding properties.
Peptide self-assembly is a wide-ranging phenomenon and is important in several areas of science.The peptide fibrillation is influenced by a variety of environmental triggers, such as pH, ionic strength, metal ions, temperature and oxidative stress, which allows the fabrication of peptide-based nanostructures with designated functions.Among them a variety of self-assembled monolayers(SAMs)play important roles during peptide self-assembly, and it has been found that some organics may induce peptides self-assembly at the surface, which provides a powerful method for fabrication of ordered nanostructures.However, to date, the formation of protein fibrils on a surface has been seldom reported.Here, branched nanostructures from a self-assembling single-stranded DNA binding protein(SSBP)were observed with tapping mode atomic force microscopy(AFM)when the SAM of 1-hexadecanethiol(HDT)on a gold substrate incubated in 4-aminothiophenol(4-ATP).SSBP can readily undergo self-assembly to form well-defined nanostructures such as nanofibers, nanowires and ribbons.Similar phenomena were also observed for platelet-derived growth factor-BB(PDGF-BB)and immunoglobulin E(IgE).However, these fibers could not be formed on the HDT/Au substrate or induced by mercaptan with carboxyl group.Like 4-ATP, cysteamine and butylamine can also induce SSBP into fibrils.The results suggest that electrostatic forces between positively charged amides and proteins provide the driving forces to promote aggregation for the self-assembly of the protein fibrils.
The self-assembling structures of SSBP induced by 4-ATP were similar to the fractal patterns.The diffusion-limited cluster aggregation(DLA)theory can be used to explain the mechanism of the fibril formation.The aggregation progress occurs as the solvent evaporates and the component deposits on the substrate, eventually resulting in a fractal structure.Over a limited range of length scales, many structures have fractal geometry with self-similarity at long and/or short lengths.The fractal structures formed from biomolecules lie down on the surface at designated positions and thereby preventing mutual superimposition in a disordered arrangement.Because of the well-ordered structures, inherent bioactivity, biocompatibility, and chemical and biological modifiability, protein assemblies may be potentially harnessed in the formation of novel, composite bio-or nanomaterials with self-similarity at different length scales.
1 Experiments
1.1 Chemicals and reagents
4-Aminothiophenol(4-ATP), 1-Hexadecanethiol(HDT), butylamine and platelet-derived growth factor-BB(PDGF-BB)were purchased from Aldrich(USA)and used without further purification.Cysteamine hydrochloride was obtained from Sangon(China).Escherichia coli single-stranded DNA binding protein(SSBP)was obtained from Promega Corp(USA).Immunoglobulin E(IgE)was purchased from Meridian(USA).The relevant reagents of buffer solution(10 mmol·L-1Tris-HCl, 50 mmol·L-1NaCl, pH 7.40)were purchased from Sinopharm Chemical Reagent Co.Ltd(China).
1.2 Preparation of SAMs on gold substrate
The gold slides were immersed in a 1.0 mmol·L-1ethanol solution of HDT at room temperature for at least 24 h and then incubated in a 1.0 mmol·L-1ethanol solution of 4-ATP at room temperature for 3 h.The samples were rinsed copiously with ethanol and Milli-Q water successively.Then the HDT/Au substrates with 4-ATP were incubated in 30 μL solution containing 5 μg·mL-1proteins at 0 ℃ for 22 h.After incubation, the samples were rinsed with Milli-Q water and then dried in air for AFM imaging.
1.3 AFM measurement
All AFM experiments were performed on AFM(SPA-400, SII, Japan)in tapping mode.A tapping mode AFM tip(PPP-SEIHR probe with a spring constant of 8.8 N·m-1, resonant frequency 117 kHz)was used to observe the topography.The AFM images were acquired in air under ambient conditions.The Image J(NIH, Bethesda, MD, http://rsbweb.nih.gov/ij/)was used to estimate the fractal dimensions of the protein nanostructures from the AFM images.The fractal dimensions were calculated using the box-counting algorithm.The topography images were converted to an 8-bit binary format and box values of 2, 4, 8, 16, 32, 64 and 128 were overlaid on the image as described earlier.
2 Results and discussion
2.1 AFM images of anisotropic SSBP fibrils induced by 4-ATP
The HDT SAMs were formed on gold slide by incubation in the solution of HDT for at least 24 h.Then the samples were incubated in the solution of 4-ATP for 3 h.At last, 30 μL solution containing 5 μg·mL-1SSBP were added onto the sample and incubated at 0 ℃ for 22 h.After incubation, the samples were rinsed with Milli-Q water then dried in air for AFM imaging.A series of tapping mode AFM images of SSBP fibrils induced by 4-ATP on HDT/Au SAMs at different magnifications were shown in Fig.1.The images showed significant and intriguing SSBP molecular self-organization on the HDT/Au SAMs induced by 4-ATP.Fig.1(a)featured ordered, branched structures which represented self-assembly proteins on the substrate.The morphology and organization of each fibril were not highly uniform, they varied in the length of the branch.Some of the branches stopped and the other branches grew and fork endlessly.Compared with previous reports, the fibrils in this structure were predominantly orientated to a specific direction.The higher-magnification images in Fig.1(b)and Fig.1(c)showed that each of the fibrils consisted of several continuous SSBP molecules.The branches had similar width((60±5)nm)and height((7.5±1)nm), which was consistent with the size of a single SSBP protein molecule[35], and they forked into several self-similar fibrils.A phase image corresponding to the topography image(Fig.1(b))was shown in Fig.1(d).The diameter of an individual fibril was determined from this phase image to be ca.70 nm.The higher-magnification phase images on the left and right sides of the main image of Fig.1(d)featured anisotropic conformation of the proteins and each bundle consisted of one or two fibrils.SSBP molecules closely packed into the fibrils, and each molecule could be clearly determined.However, fibrils could not form without 4-ATP incubation on HDT SAMs.In addition, fibrils could not form on 4-ATP SAMs, either.

Fig.1 AFM topographic(a, b, c)and phase(d, corresponding to image b)images of SSBP fibrils on the HDT/Au SAMs induced by 4-ATP in different scanning ranges.Areas in white squares and rectangles are magnified respectively on the sides of the main image
2.2 Different protein fibrils induced by 4-ATP
In contrast to our observations on other proteins, then the similar operations were performed on another two proteins.Similar to SSBP, PDGF-BB and IgE could also form fibrils on the HDT/Au substrate induced by 4-ATP.Fig.2 showed AFM topographic and phase images of the HDT/Au substrate after being incubated in different proteins, respectively.In terms of shape, distribution and density of the proteins, the structures in Fig.2(a)was very similar to Fig.1(b).SSBP(Fig.1(b)and Fig.1(d))and PDGF-BB(Fig.2(a)and Fig.2(c))formed regular, dense branched aggregates, which were different in structure and dimension to IgE(Fig.2(b)and Fig.2(d)), which also possessed an outward-branching architecture.The corresponding molecular masses of SSBP, PDGF-BB and IgE were 47, 25 and 188 kDa, respectively, which implied that IgE molecule was much bigger than SSBP and PDGF-BB molecules.Thus, the IgE fibrils were wider than SSBP fibrils and PDGF-BB fibrils.The fibrils had uniform widths in each fibril, and they were branching and forking in several areas.In addition, they were predominantly orientated in a direction, which was assumed the growing direction.AFM results of protein fibrils not only demonstrated analogies of different protein fibrils, but also clearly revealed similarities in their self-assembly structures and mechanisms, suggesting a possible general model for protein fibril assembling induced by 4-ATP.

Fig.2 AFM topographic(a, b)and phase(c, d)images of PDGF-BB(a, c)and IgE(b, d)on HDT/Au induced by 4-ATP
2.3 Possible formation mechanism of protein fibrils
To further reinforce the self-assembly mechanisms of the aggregation of proteins into different classes of fibrils, other amides were used to induce the proteins into fibrils.SSBP fibrils induced by cysteamine and butylamine also provided an evidence of branched “tree-like” structures.Like the modification of 4-ATP, the HDT/Au substrate first incubated in cysteamine and butylamine for 3 h respectively, then incubated in SSBP for 22 h.Fig.3 showed the SSBP fibrils on HDT/Au substrate induced by 4-ATP(Fig.3(a)), cysteamine(Fig.3(b))and butylamine(Fig.3(c)), respectively.The structures were fully connected patterns that were denser than the fibrils in Fig.1(12 h incubation)and assembled into superstructures on the order of several micrometres in length.In general, while the incubation time was longer than 20 h, SSBP tended to favor this configuration, whereas the incubation time was shorter than 20 h, SSBP predominantly formed structures of configuration in Fig.1.Different effects might cooperate to produce the final structures that were observed, including variations in the incubation time and the functional groups of the amides that could direct the assembly of the protein into specific structures.These parameters also affected the dimension of the structures.The resulting highly branched structures were similar to the fractal patterns that had been observed in the DLA of colloids[29].The nonspecifically adsorbed particles performed Brownian diffusion and encountered an existing structure(seed)or escaped to infinity.These particles could either escape to infinity or stick to the seed irreversibly, as such process repeated, the assembly grew longer leading to the fractal structures.The resulting clusters were highly branched and fractal in nature, quantitatively described by the fractal dimension.The fractal dimension(D)measured the rate of accumulation of structural detail with increasing magnification, and could be used as a quantitative measure of the self-similarity of the fractal across length scales.For the assembly of SSBP, the fractal dimension of the protein nanostructures from the AFM images was estimated using the Image J(NIH, Bethesda, MD, http://rsbweb.nih.gov/ij/)[6].The fractal dimensions were calculated using the box-counting algorithm.The topography images were converted to an 8-bit binary format and box values of 2, 4, 8, 16, 32, 64 and 128 were overlaid on the image as described earlier.The slope of the plot of log(size)vs.log(count)corresponded to the negative ofD.Fig.3 showed the dimensions which calcula-ted using the box-counting algorithm were corresponding to the AFM images on the left side.From these plots, the dimensions of the SSBP fibrils induced by 4-ATP, cysteamine and butylamine were calculated to be 1.898 6, 1.819 2 and 1.737 8, respectively.The results indicated that the rates of accumulation of structural detail were different with increasing magnification.Because 4-ATP and cysteamine contained sulfhydryl group, it might be beneficial to the increase of rates.
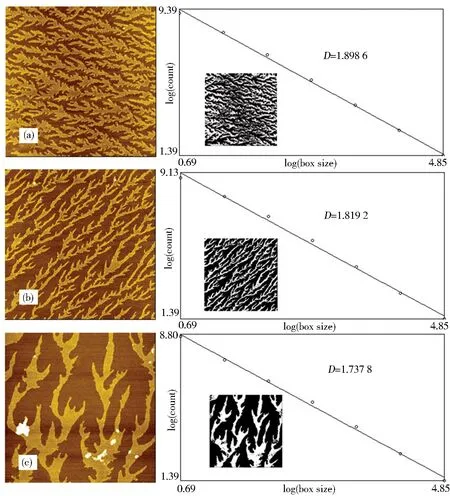
Fig.3 AFM topographic images of SSBP fibrils on HDT/Au induced by 4-ATP(a), cysteamine(b)and butylamine(c), respectively(The scanning size: 2.0 μm × 2.0 μm, calculation of the fractal dimension of the SSBP nanostructure using the box-counting algorithm.Inset:the relative AFM image of SSBP that was used for this calculation(inverted for clarity))
The mechanism of the SSBP fibrils formation induced by amides could be explained by the DLA theory[29].The observed SSBP fibrils were quite similar to the DLA generated fractal shape by experiments and simulations.Fig.4 was the proposed models for SSBP self-assembly on the HDT SAMs induced by amides.The nonspecifically adsorbed amides on the HDT SAMs might act as seeds for the subsequent DLA process.During the incubation in proteins, SSBP molecules would either escape to infinity or irreversibly stick to the seed.When the second SSBP molecule sticked to the first SSBP, it would become a dimer.As such process repeated, the assembly grew longer, leading to a branched shape.

Fig.4 Proposed models for SSBP self-assembly on the HDT SAMs induced by amides
Further studies demonstrated that SSBP could not form fibrils on the HDT/Au substrate without being modified by 4-ATP.The electrostatic interaction between positively charged amides and proteins might provide the driving forces for the self-assembly of the protein fibrils.In order to verify this hypothesis, mercaptans with different terminal groups instead of amides were explored.Fibrils could not be formed on the HDT/Au substrate induced by 16-mercaptohexadecanoic acid(MHA)and 1-dodecanethiol(DDT).For MHA was with negative electric charge and DDT was neutral in the buffer(pH = 7.40), this observation implied that protein fibrils might prefer to forming on the HDT/Au substrate induced by positively charged amides.
3 Conclusion
This study explored the formation mechanism of SSBP fiber fractal structure at liquid-solid interface induced by 4-ATP on HDT/Au.The nanoscale characterization and imaging of the branched patterns formed by the self-assembly of proteins were observed via AFM for structural analysis.The proteins fibrils could induce by amides on HDT/Au substrate, whereas the fibrils could not be formed on the HDT/Au substrate induced by alkanethiols or carboxylthiols.It suggested that electrostatic forces between positively charged amides and proteins provided the driving forces to promote aggregation for the self-assembly of the protein fibrils.This two-dimensional assembly approach generated long proteins fibrils with dimensions in the range of 1.7 and 1.9.The method described here should be useful, not only for the construction of protein patterns, but also for enriching the mechanistic knowledge about the self-assembly of proteins.Importantly, by proper adjustment of the incubation time, different structures could be controlled and immobilized on the surface which might provide an ideal template for potential technological applications.The occurrence of branched-like protein fibrils rather than the more compact close-packing of single filaments induced by amide substances might develop a new and spatially controllable protein interface self-assembly approach.

