极地微生物酶资源开发研究进展
王伟 姚从禹3 孙晶晶 郝建华
研究进展
极地微生物酶资源开发研究进展
王伟1,2姚从禹1,2,3孙晶晶1,2郝建华1,2
(1中国水产科学研究院黄海水产研究所, 农业农村部极地渔业开发重点实验室, 山东 青岛 266071;2青岛海洋科学与技术试点国家实验室海洋药物与生物制品功能实验室, 山东 青岛 266235;3上海海洋大学食品学院, 上海 201306)
两极地区的微生物在极端环境中演化出能够耐受低温、高盐等特殊性质的酶。极地微生物的酶有潜在的应用价值, 是重要的生物资源。极地微生物在各种复杂的极地环境中形成了高度多样性, 蕴含着可供开发利用的大量新酶资源。近年来国内外持续发现大量的极地微生物新酶, 说明极地微生物已成为新酶的重要来源。本文简述了近5年来极地微生物新酶开发的研究进展, 分类列举了有较好商用前景的蛋白酶、脂肪酶/酯酶、糖类降解酶等重要工业酶的筛选和性质等研究简况。
极地微生物 酶资源开发 蛋白酶 脂肪酶 酯酶 糖类降解酶
0 引言
极地具有独特的地理、气候及环境特征, 如低温、寡营养、高pH、高盐、极端光照条件等。极地微生物通过演化出一系列特定结构和功能, 适应了上述极端环境。具有在低温下高效催化的低温酶是极地微生物的重要适应特征。
极地是一个微生物资源库, 其中的极端微生物能够产生新型生物活性物质(如酶、多糖、多肽等)[1]。极地微生物的多样性与活性物质研究已成为现代微生物学的研究热点。极地微生物也是酶资源的重要来源, 极地微生物酶的研究从20世纪90年代开始兴起, 以蛋白酶、脂肪酶、淀粉酶等产业价值高的酶类作为主要研究对象。极地微生物的低温酶与耐盐酶等结构和功能新颖的极端酶凭借其独特的催化作用大大拓宽了微生物酶的应用范围, 也给酶工程的研究带来了新的思路和方向。本文对极地微生物来源的蛋白酶、脂肪酶/酯酶、糖类降解酶等近5年来的研究进展进行综述, 以期对极地微生物资源的综合利用提供参考。
1 产酶的极地微生物筛选
虽然宏基因组文库或数据挖掘等新方法已经用于极地微生物新酶筛选, 但以经典的分离培养法对菌株开展酶活性筛选仍是近5年的主流方法(表1)。这些研究多选择工业应用潜力较大的水解酶(蛋白酶、脂肪酶、糖类降解酶等)进行筛选, 主要以平板透明圈法初步确定各种酶的活性。这些研究都能从来源多样的极地环境样品中筛选出多种产酶菌株, 说明极地微生物酶资源丰富, 有待继续广泛深入地研究。

表1 近年从极地环境样品中分离筛选产酶微生物简况
2 蛋白酶
蛋白酶是一类催化蛋白质或肽类的肽键水解的酶类, 其种类多样, 在众多行业及科研中广泛应用, 成为具有重要商业价值的工业酶。21世纪初报道了大量产适冷蛋白酶的极地微生物, 近年来极地微生物蛋白酶研究热度不减。从3个南极淡水湖中分离出细菌18种63株, 真菌1种8株, 它们在4℃均有蛋白酶活力[17]; Kim和Choi[18]研究了温度对四株有生产低温蛋白酶潜力的南极细菌的影响。
从表2可见近年从极地微生物中获得的蛋白酶几乎都是低温酶; 从南极筛选的蛋白酶较多, 且主要为工业应用最多的丝氨酸蛋白酶。来自真菌的酶少于细菌的酶, 说明细菌是极地蛋白酶的主要来源。这些研究的目标是获得有潜在工业应用价值(如洗涤剂和乳制品加工)的低温酶, 工作主要集中在酶纯化、性质及应用潜力, 对酶的空间结构、催化机理等理论研究相对较少。以功能宏基因组筛选北极王湾海底沉积蛋白酶[31], 发现的新型中性金属蛋白酶潜在应用价值不高, 但证明了宏基因组法筛选蛋白酶的潜力。
3 脂肪酶/酯酶
脂肪酶/酯酶是能够催化脂肪酸甘油酯水解和合成的酶, 在食品、制药、洗涤、能源等工业中应用广泛。近年在南北极地区的微生物中获得了大量脂肪酶/酯酶(表3)。
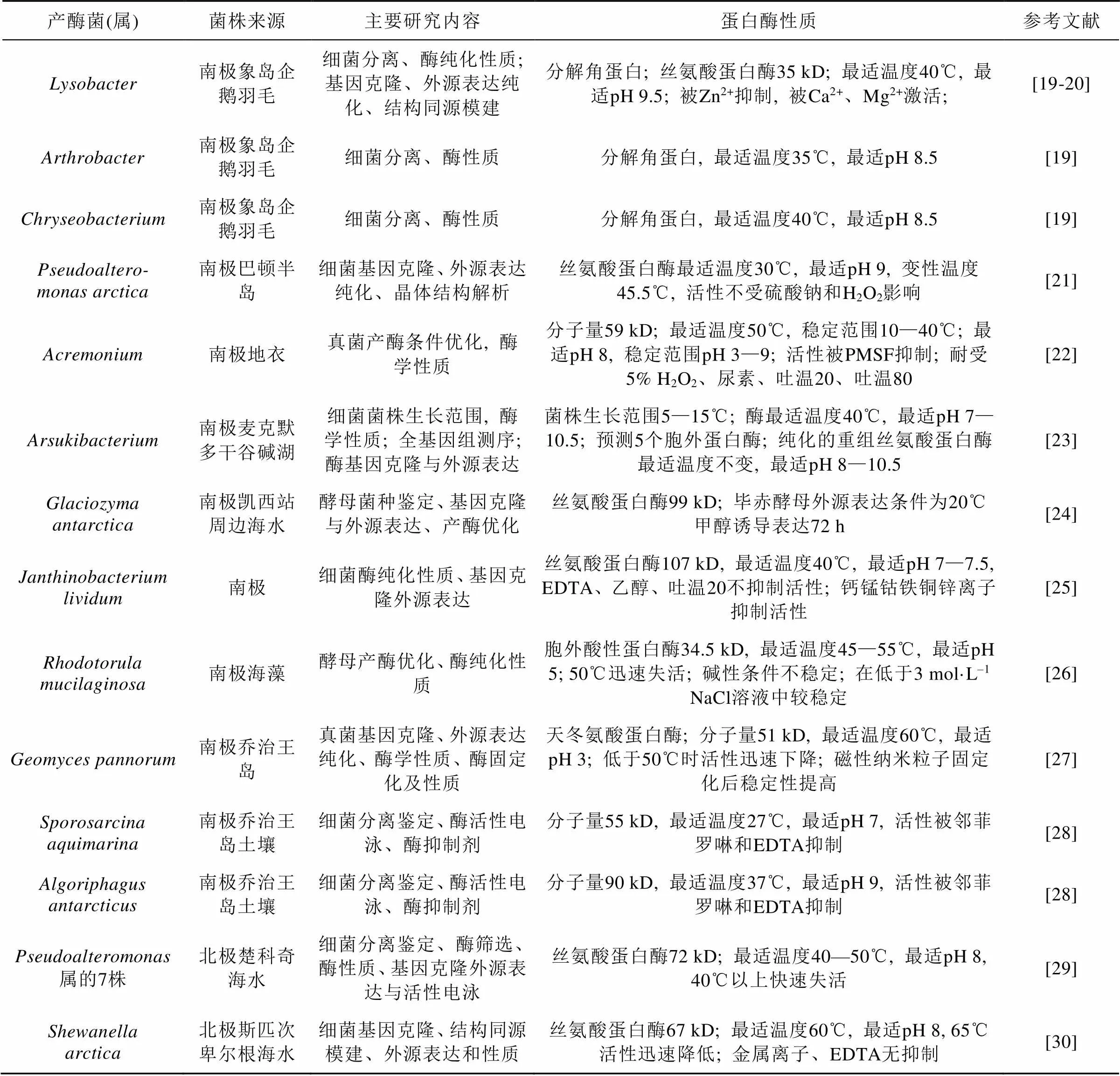
表2 近年新发现的极地微生物蛋白酶简况
近年发现的极地微生物脂肪酶/酯酶种类多样(表3), 主要是来自细菌的低温酶。这些酶多数具有有机溶剂耐受能力, 展现出在不同领域的应用价值。从表3可见宏基因组筛选已经成为极地微生物新型酯酶筛选的主流方法, 用该方法获得的酯酶数量已经超过了传统菌株分离的方法。一般情况下在极地微生物中筛选获得低温酶, 但也有可能获得热稳定性理想的高温酶。表3筛选到的脂肪酶/酯酶都是碱性或中性酶, 无酸性酶, 这可能和研究目标多选择细菌有关, 也和对酸性脂肪酶/酯酶的应用需求没有碱性酶高有关。今后如果更关注极地真菌脂肪酶/酯酶, 可能会发现新的酸性酶。
4 糖类降解酶
近年来从极地微生物中分离的糖类降解酶种类众多, 性质独特。从北极斯匹次卑尔根岛王湾分离的海洋细菌多具有褐藻胶、果胶、淀粉、木聚糖或羧甲基纤维素的低温降解酶[49]。这也说明了极地微生物糖类降解酶资源开发的良好前景。
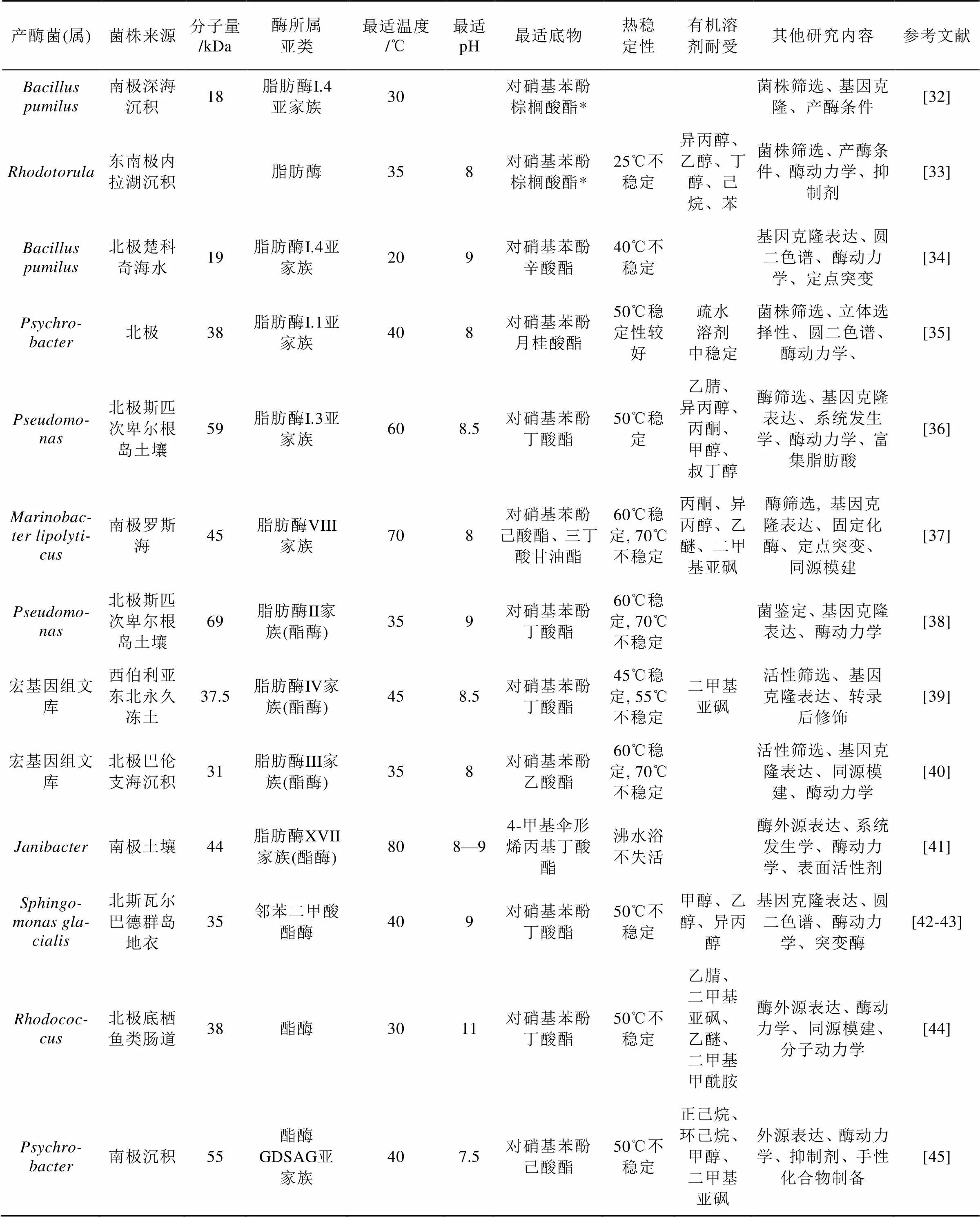
表3 近年新发现的极地微生物脂肪酶/酯酶简况

续表3
*仅使用一种底物, 未作比较
4.1 水解葡萄糖单元的糖苷键的酶
淀粉酶能将淀粉转化为低分子量的糖如葡萄糖、麦芽糖或寡糖。该酶在淀粉工业中使用广泛, 在酶制剂市场占有很大份额。α-淀粉酶是内切酶, 用于纺织品、纸张、食品、生物燃料、洗涤剂和制药工业的多种生物加工工艺[50]。南极乔治王岛海水细菌的α-淀粉酶(属于糖苷水解酶GH13家族)最适条件20℃、pH 8.0, 是已知的α-淀粉酶的最低最适反应温度[50]。南极深海沉积细菌α-淀粉酶Amy172的最适条件50℃、pH 10[51]。南极真菌的耐热α-淀粉酶, 最适条件70℃、pH 6。这是首次在适冷真菌中发现耐热淀粉酶[52]。南极乔治王岛土壤真菌新型适冷葡萄糖淀粉酶最适条件30℃、pH 6, 活性不依赖Ca2+[53]。北极王湾沉积物细菌α-淀粉酶Amy3的最适条件25℃、pH 8.5, 具有良好的低温催化及嗜盐性[54]。该菌株还具有两种α葡萄糖苷酶: GH13家族的Pagl最适条件30℃、pH 8, 最适底物麦芽糖, 耐受葡萄糖[55]; GH97家族的PspAG 97A最适条件30℃、pH 7.5, 可水解α-1,2/1,4/1,6糖苷键[56]。北极斯匹次卑尔根岛海水细菌普鲁兰酶(GH13家族)最适条件35℃、pH 6—7, 只水解α-1,6糖苷键[57]。
南极乔治王岛土壤细菌β-葡糖苷酶EaBgl1A属于GH1家族, 最适条件30℃、pH 7, 对葡萄糖的耐受性好于几种商用酶[58], 晶体结构解析表明其适应低温的主要机制是形成四聚体[59]。北极王湾表层海水细菌的新型内切β-1,4-葡糖苷酶属于GH10家族, 低温耐盐, 纤维素酶活性明显而木聚糖酶活性很低[60]。从北极洋中脊热液口宏基因组序列中得到了嗜热纤维素酶, 最适条件100℃、pH 5.7, 其在85℃活性稳定, 在GH9家族纤维素酶中热稳定性最好[61]。
4.2 卡拉胶酶
卡拉胶是一种硫酸化的红藻多糖, 由半乳糖和3,6-脱水半乳糖通过α-1,3和β-1,4糖苷键交联形成。卡拉胶酶能降解卡拉胶产生水溶性好、生物活性高的卡拉胶寡糖, 在食品工业及医药等领域具有良好的应用前景。潘爱红等[62]优化了南极普里兹湾沉积物细菌卡拉胶酶的产量。胡秋实等[63]从北极海水中筛选出四株高产卡拉胶酶的假交替单胞菌菌株, 酶的最适反应温度均为20℃。林欢等[64]从南极海藻分离一株高产卡拉胶酶的细菌, 优化产酶条件, 确定酶最适条件为37℃、pH 7。
4.3 琼胶酶
琼脂(琼胶)也是红藻多糖, 由不同形式的半乳糖作为单糖单元构成。琼胶酶能降解琼胶产生有活性的琼寡糖。南极普里兹湾沉积物细菌NJ21有三种外切型β琼胶酶, 产物均为新琼二糖。GH42家族的琼胶酶Aga1161最适条件40℃、pH 8, 40℃活性不稳定[65]; Aga3463属于GH86家族, 最适条件50℃、pH 7, 50℃活性不稳定[66]; 琼胶酶Aga3311也属于GH42家族, 最适条件35℃、pH 7[67]。分离自东南极近岸海冰硅藻的细菌β琼胶酶属于GH16家族, 最适条件40℃、pH 7, 50℃不稳定[68]。
4.4 β-半乳糖苷酶
β-半乳糖苷酶有两种活性: 水解乳糖/低聚半乳糖; 连接半乳糖生成低聚半乳糖。该酶在乳制品工业中应用广泛。东南极湖泊嗜盐古菌的半乳糖苷酶是GH42家族的单体酶, 比较其野生酶和六个突变酶的低温催化参数及空间结构模型, 说明一个氨基酸突变就能使低温催化能力明显改变[69]。南极乔治王岛土壤细菌β-半乳糖苷酶属于GH2家族, 最适条件28℃、pH7, 低温下具有水解和聚合两种酶活[70]。该酶晶体结构解析发现其适应低温的主要因素是二聚体酶分子表面增加了溶剂的可及性[71]。来自北极加拿大海盆海冰冰芯的海单胞菌有两个半乳糖苷酶: BGAL584-1为不耐热的低温酶, 最适条件30℃、pH 7[72]; 而MaBGA最适条件60℃、pH 6[73]。两个酶分别属于GH2和GH42家族。北极王湾沉积物细菌低温半乳糖苷酶为同源四聚体, 最适条件45℃、pH 7—8, 活性在45℃不稳定[74]。
4.5 木聚糖酶
木聚糖是木糖以β-1,4糖苷键形成的多糖, 带有阿拉伯糖和葡萄糖醛酸的侧链。木聚糖酶可以将木聚糖降解为木糖及木寡糖, 广泛应用于造纸、纺织、食品、饲料等行业。从南极乔治王岛海绵分离的真菌低温木聚糖酶属于GH10家族, 最适条件50℃、pH 6, 最适底物为阿拉伯木聚糖, 活性在35℃不稳定, 是已知的最不耐热的真菌木聚糖酶[75-76]。南极乔治王岛海水细菌的GH10家族木聚糖酶最适条件35℃、pH 7—9, 以定向进化和随机突变结合的策略优化了该酶的热稳定性[77]。从北极洋中脊热液口宏基因组序列中筛选到GH10家族的高温木聚糖酶, 最适条件80℃、pH 5.6, 其降解纤维素的的活性高于木聚糖[78]。
4.6 褐藻酸裂解酶
褐藻酸是甘露糖醛酸和古洛糖醛酸以多种排列方式组成的线性共聚物。褐藻酸裂解酶能将褐藻酸降解成具有多种生物活性的寡糖。
东升[79]从北极海带中分离了21株产褐藻酸裂解酶的细菌, 酶的最适温度在20—50℃。高杨[80]从南大洋沉积物分离了10株能降解褐藻酸的真菌, 并表达了曲霉22-5的褐藻酸裂解酶Aly-i7, 其最适条件50℃、pH 7, 最适底物聚古洛糖醛酸, 裂解褐藻酸的产物为二糖。前述产琼胶酶的南极细菌NJ21的褐藻酸裂解酶Al163属于多糖裂解酶6家族(PL6)的内切酶, 最适条件40℃、pH 7, 最适底物聚古洛糖醛酸[81]。从北极洋中脊热液口宏基因组序列中获得的褐藻酸裂解酶属于PL7家族, 最适条件65℃、pH 6, 最适底物聚甘露糖醛酸, 该酶是已知最耐热的褐藻酸裂解酶[82]。
4.7 果胶酶
果胶是植物细胞壁的组分, 是部分甲酯化的α-1,4-D-聚半乳糖醛酸。果胶酶在食品、纺织和造纸行业应用广泛, 可分为聚半乳糖醛酸酶、裂解酶和果胶甲酯酶三类[83]。南极阿斯曼山细菌低温碱性果胶裂解酶属于PL6家族, 最适条件30℃、pH 10, 40℃活性不稳定, 最适底物为聚半乳糖醛酸[84]。南极乔治王岛土壤真菌聚半乳糖醛酸酶(GH28家族)在15℃、pH 3时活性明显高于商用酶[83]。
5 其他酶
5.1 磷酸酶
磷酸酶能水解蛋白、核苷酸、生物碱等底物中的磷酸酯键, 在分子生物学、免疫学等领域应用广泛[85]。南极乔治王岛土壤酵母磷酸酶最适条件47℃、pH 9.5, 其低温活性较高而在47℃活性不稳定[86]。乔治王岛近岸表层海水细菌磷酸酶最适条件20—22℃、pH 7, 在48℃活性不稳定, Mg2+能提高其活性和热稳定性[87]。
5.2 超氧化物歧化酶
超氧化物歧化酶(superoxide dismutase, SOD)能降解强氧化剂超氧阴离子自由基, 在制药、化妆品等行业有应用前景。南极海冰细菌SOD最适条件30℃、pH 8.0, 50℃活性不稳定, 属于Fe-SOD亚类[88]。南极海冰酵母的Fe-SOD在pH 1.0—9.0和50℃的稳定性都较好[89]。南极利文斯顿岛曲霉有两种Cu/Zn-SOD, 低温诱导酶产量增加[90]。
6 展望
上文列举了近年来极地微生物酶资源开发简况, 主要关注了文献报道较多、应用价值较高的几类酶。报道的新酶以热稳定性较低的低温酶为主, 个别酶热稳定性较好, 这些低温酶大多有耐盐能力。值得注意的是高温酶的报道, 挪威生命科学大学Eijsink等从北极扬马延岛北部的扬马延热液口发现了三个高温酶: 木聚糖酶[78]、褐藻酸裂解酶[82]和纤维素酶[61], 都具有理想的应用前景。说明极地的高温环境虽然罕见, 但也蕴藏着独特的酶资源。该研究将酶的底物(预处理的欧洲云杉木屑)在70℃的海底沉积中放置1年, 再回收测宏基因组获取新酶基因。这种原位诱导筛选的方式很少用于极地微生物的新酶筛选, 有望成为将来常见的高效筛选策略。
目前传统的平板培养法筛选菌株的新酶仍是国内外最常用的方法。由于其简便易操作, 预计将来也不会被宏基因组等非培养方法替代。宏基因组作为重要的非培养筛选方法, 能克服众多极地菌株无法培养的困难, 直接在基因水平发现新酶, 其基于功能[31,39-40,46]和基于序列[47,61,78,82]的筛选策略已经在极地微生物资源开发中获得了众多新酶, 今后必将成为常规的新酶筛选方法。此外, 从极地菌株的全基因组序列中发掘新酶也成为了常见的方法[23, 42, 44-45, 51, 53, 58, 60, 68-69, 72, 81, 83-84, 87]。随着微生物全基因组测序费用的下降和公开的极地微生物全基因组数据的迅速增长, 可以推测从全基因组数据中获取新酶基因的方法将逐渐替代传统的酶基因克隆方法, 如基因组文库筛选或简并引物PCR扩增法。
多数极地微生物新酶筛选工作的应用目标明确, 获得了大量有潜在工业应用价值的酶, 不过未见真正转化为商用酶制剂的报道, 可能受限于酶性质、酶制剂开发周期以及厂商对产品来源保密。
在极地微生物酶的研究内容上, 筛选、基因克隆表达和酶的纯化与性质研究是主流, 酶催化机理和晶体结构解析等更深入的研究较少, 蛋白质工程改造工作也不多。酶分子改造能进一步提高酶的工作效能, 而理论研究是蛋白质工程的重要指导, 可以预期今后会有更多极地微生物酶理论研究及分子改造工作。
1 DE Pascale D, De Santi C, Fu J, et al. The microbial diversity of Polar environments is a fertile ground for bioprospecting[J]. Marine Genomics, 2012, 8: 15-22.
2 丁新彪, 丛柏林, 张扬, 等. 南极普里兹湾及邻近海域沉积物微生物多样性与生理生化研究[J]. 海洋科学进展, 2014, 32(2): 209-218.
3 董宁. 东南极格罗夫山土壤微生物多样性及其可培养细菌的产酶和抗菌活性筛选[D]. 青岛: 中国海洋大学, 2014.
4 郝文惠. 大西洋与南海深海沉积物真菌多样性分析及极地产低温酶菌种的筛选[D]. 厦门: 厦门大学, 2014.
5 张丽珉, 赵琳, 丛柏林. 南极罗斯海区域可培养微生物分离鉴定及产低温酶能力初步筛选[J]. 海洋学报, 2018, 40(8): 152-164.
6 LEE Y M, JUNG Y J, HONG S G, et al. Diversity and physiological characteristics of culturable bacteria from marine sediments of Ross Sea, Antarctica[J]. The Korean Journal of Microbiology, 2014, 50(2): 119-127.
7 TOMOVA I, STOILOVA-DISHEVA M, VASILEVA-TONKOVA E. Characterization of heavy metals resistant heterotrophic bacteria from soils in the Windmill Islands region, Wilkes Land, East Antarctica[J]. Polish Polar Research, 2014, 35(4): 593-607.
8 TSUJI M. Genetic diversity of yeasts from East Ongul Island, East Antarctica and their extracellular enzymes secretion[J]. Polar Biology, 2018, 41(2): 249-258.
9 BARAHONA S, YUIVAR Y, SOCIAS G, et al. Identification and characterization of yeasts isolated from sedimentary rocks of Union Glacier at the Antarctica[J]. Extremophiles, 2016, 20(4): 479-491.
10 LEE Y M, KIM E H, LEE H K, et al. Biodiversity and physiological characteristics of Antarctic and Arctic lichens-associated bacteria[J]. World Journal of Microbiology and Biotechnology, 2014, 30(10): 2711-2721.
11 SINGH P, SINGH S M, ROY U. Taxonomic characterization and the bio-potential of bacteria isolated from glacier ice cores in the High Arctic[J]. Journal of Basic Microbiology, 2016, 56(3): 275-285.
12 SINGH P, SINGH S M, DHAKEPHALKAR P. Diversity, cold active enzymes and adaptation strategies of bacteria inhabiting glacier cryoconite holes of High Arctic[J]. Extremophiles, 2014, 18(2): 229-242.
13 SINGH P, ROY U, TSUJI M. Characterisation of yeast and filamentous fungi from Brøggerbreen glaciers, Svalbard[J]. Polar Record, 2016, 52(4): 442-449.
14 Salam S, Lekshmi S, Silvester R, et al. Effect of environmental factors on growth and enzyme production of cold adapted bacteria from water and sediment of Kongsjord, Ny-Alesund, Arctic[J]. Journal of Environmental Biology, 2017, 38(4): 579-585.
15 DE SANTI C, ALTERMARK B, DE PASCALE D, et al. Bioprospecting around Arctic Islands: Marine bacteria as rich source of biocatalysts[J]. Journal of Basic Microbiology, 2016, 56(3): 238-253.
16 张良. 北极海洋沉积物细菌群落结构及其胞外水解酶研究[D]. 青岛: 国家海洋局第一海洋研究所, 2018.
17 MATSUI M, KAWAMATA A, KOSUGI M, et al. Diversity of proteolytic microbes isolated from Antarctic freshwater lakes and characteristics of their cold-active proteases[J]. Polar Science, 2017, 13: 82-90.
18 KIM H D, CHOI J I. Effect of temperature on growth rate and protease activity of Antarctic microorganisms[J]. Korean Journal of Microbiology and Biotechnology, 2014, 42(3): 293-296.
19 Pereira J Q, Lopes F C, Petry M V, et al. Isolation of three novel Antarctic psychrotolerant feather-degrading bacteria and partial purification of keratinolytic enzyme fromsp. A03[J]. International Biodeterioration and Biodegradation, 2014, 88: 1-7.
20 Pereira J Q, Ambrosini A, Passaglia L M P, et al. A new cold-adapted serine peptidase from Antarcticsp. A03: Insights about enzyme activity at low temperatures[J]. International Journal of Biological Macromolecules, 2017, 103: 854-862.
21 PARK H J, LEE C W, KIM D, et al. Crystal structure of a cold-active protease (Pro21717) from the psychrophilic bacterium,PAMC 21717, at 1.4 Å resolution: Structural adaptations to cold and functional analysis of a laundry detergent enzyme[J]. PLoS One, 2018, 13(2): e0191740.
22 da Silva Nascimento T C E, de Sena A R, Gomes J E G, et al. Extracellular serine proteases bysp. L1-4B isolated from Antarctica: Overproduction using cactus pear extract with response surface methodology[J]. Biocatalysis and Agricultural Biotechnology, 2015, 4(4): 737-744.
23 LYLLOFF J E, HANSEN L B S, JEPSEN M, et al. Genomic and exoproteomic analyses of cold- and alkaline-adapted bacteria reveal an abundance of secreted subtilisin-like proteases[J]. Microbial Biotechnology, 2016, 9(2): 245-256.
24 Alias N, AHMAD MAZIAN M, Salleh A B, et al. Molecular cloning and optimization for high level expression of cold-adapted serine protease from Antarctic yeastPI12[J]. Enzyme Research, 2014, 2014: 1-20.
25 KIM H D, KIM S M, CHOI J I. Purification, characterization, and cloning of a cold-adapted protease from Antarctic[J]. Journal of Microbiology and Biotechnology, 2018, 28(3): 448-453.
26 LARIO L D, CHAUD L, ALMEIDA M G, et al. Production, purification, and characterization of an extracellular acid protease from the marine Antarctic yeastL7[J]. Fungal Biology, 2015, 119(11): 1129-1136.
27 GAO B, HE L, WEI D Z, et al. Identification and magnetic immobilization of a pyrophilous aspartic protease from Antarctic psychrophilic fungus[J]. Journal of Food Biochemistry, 2018, 42(6): e12691.
28 SANTOS A F, PIRES F, JESUS H E, et al. Detection of proteases fromandisolated from Antarctic soil[J]. Annals of the Brazilian Academy of Sciences, 2015, 87(1): 109-119.
29 PARK H J, LEE Y M, KIM S, et al. Identification of proteolytic bacteria from the Arctic Chukchi Sea expedition cruise and characterization of cold-active proteases[J]. Journal of Microbiology, 2014, 52(10): 825-833.
30 Qoura F, Kassab E, Reiße S, et al. Characterization of a new, recombinant thermo-active subtilisin-like serine protease derived from[J]. Journal of Molecular Catalysis B: Enzymatic, 2015, 116: 16-23.
31 王光龙. 利用功能宏基因组技术对北极、大西洋海底沉积物中的新型蛋白酶、酯酶进行筛选、鉴定和性质研究[D]. 济南: 山东大学, 2014.
32 郝文惠, 王凡羽, 郭玉, 等. 南极深海沉积物中产低温脂肪酶菌株的筛选与基因克隆[J]. 应用海洋学学报, 2014, 33(3): 306-311.
33 Maharana A K, SINGH S M. A cold and organic solvent tolerant lipase produced by Antarctic strainsp. Y-23[J]. Journal of Basic Microbiology, 2018, 58(4):331-342.
34 WI A R, JEON S J, KIM S, et al. Characterization and a point mutational approach of a psychrophilic lipase from an arctic bacterium,[J]. Biotechnology Letters, 2014, 36(6): 1295-1302.
35 ZHANG Y, JI F L, WANG J Y, et al. Purification and characterization of a novel organic solvent-tolerant and cold-adapted lipase fromsp. ZY124[J]. Extremophiles, 2018, 22(2): 287-300.
36 杨文娟. 极端环境细菌脂肪酶资源挖掘及其在生物制药领域的应用探索[D]. 武汉: 华中科技大学, 2017.
37 PARK S H, KIM S J, PARK S, et al. Characterization of organic solvent-tolerant lipolytic enzyme fromisolated from the Antarctic Ocean[J]. Applied Biochemistry and Biotechnology, 2019, 187(3): 1046-1060.
38 Wicka M, Wanarska M, Krajewska E, et al. Cloning, expression, and biochemical characterization of a cold-active GDSL-esterase of asp. S9 isolated from Spitsbergen island soil[J]. Acta Biochimica Polonica, 2016, 63(1): 117-125.
39 PETROVSKAYA L E, NOVOTOTSKAYA-VLASOVA K A, SPIRINA E V, et al. Expression and characterization of a new esterase with GCSAG motif from a permafrost metagenomic library[J]. FEMS Microbiology Ecology, 2016, 92(5): fiw046.
40 DE SANTI C, ALTERMARK B, PIERECHOD M M, et al. Characterization of a cold-active and salt tolerant esterase identified by functional screening of Arctic metagenomic libraries[J]. BMC Biochemistry, 2016, 17: 1.
41 Castilla A, Panizza P, Rodríguez D, et al. A novel thermophilic and halophilic esterase fromsp. R02, the first member of a new lipase family (Family XVII)[J]. Enzyme and Microbial Technology, 2017, 98: 86-95.
42 HONG D K, JANG S H, LEE C W. Gene cloning and characterization of a psychrophilic phthalate esterase with organic solvent tolerance from an Arctic bacteriumPAMC 26605[J]. Journal of Molecular Catalysis B: Enzymatic, 2016, 133(Suppl.1): 337-345.
43 KASHIF A, TRAN L H, JANG S H, et al. Roles of active-site aromatic residues in cold adaption ofesterase EstSP1[J]. ACS Omega, 2017, 2(12): 8760-8769.
44 De Santi C, Tedesco P, Ambrosino L, et al. A new alkaliphilic cold-active esterase from the psychrophilic marine bacteriumsp.: Functional and structural studies and biotechnological potential[J]. Applied Biochemistry and Biotechnology, 2014, 172(6): 3054-3068.
45 邓盾, 张云, 孙爱君, 等. 一个新颖南极微生物酯酶EST112-2的功能鉴定和在手性叔醇(S)-芳樟醇制备中的应用[J]. 有机化学, 2018, 38(5): 1185-1192.
46 TCHIGVINTSEV A, TRAN H, POPOVIC A, et al. The environment shapes microbial enzymes: five cold-active and salt-resistant carboxylesterases from marine metagenomes[J]. Applied Microbiology and Biotechnology, 2015, 99(5): 2165-2178.
47 DE SANTI C, WILLASSEN N P, WILLIAMSON A. Biochemical characterization of a family 15 carbohydrate esterase from a bacterial marine arctic metagenome[J]. PLoS One, 2016, 11(7): e0159345.
48 DE SANTI C, GANI O A, HELLAND R, et al. Structural insight into a CE15 esterase from the marine bacterial metagenome[J]. Scientific Reports, 2017, 7: 17278.
49 JAIN A, KRISHNAN K P. A glimpse of the diversity of complex polysaccharide-degrading culturable bacteria from Kongsfjorden, Arctic Ocean[J]. Annals of Microbiology, 2017, 67(2): 203-214.
50 Sanchez A, Ravanal M C, Andrews B A, et al. Heterologous expression and biochemical characterization of a novel cold-active α-amylase from the Antarctic bacteriasp. 2-3[J]. Protein Expression and Purification, 2019, 155: 78-85.
51 潘爱红, 李江, 谷晓倩, 等. 南极菌sp. A211-5产碱性α-淀粉酶Amy172的克隆表达及酶学性质研究[J]. 化学与生物工程, 2019, 36(5): 36-42.
52 GAO B, MAO Y, ZHANG L, et al. A novel saccharifying α-amylase of Antarctic psychrotolerant fungi: Gene cloning, functional expression, and characterization[J]. Starch, 2016, 68(1/2): 20-28.
53 CARRASCO M, ALCAÍNO J, CIFUENTES V, et al. Purification and characterization of a novel cold adapted fungal glucoamylase[J]. Microbial Cell Factories, 2017, 16: 75.
54 薛毅, 王梅, 方泽民, 等. 低温、嗜盐α-淀粉酶Amy3的克隆、表达及重组酶性质[J]. 微生物学报, 2018, 58(2): 336-345.
55 LI W, XUE Y, LI J, et al. A cold-adapted and glucose-stimulated type II α-glucosidase from a deep-sea bacteriumsp. K8[J]. Biotechnology Letters, 2016, 38(2): 345-349.
56 LI W, FAN H, HE C, et al. PspAG97A: a halophilic α-glucoside hydrolase with wide substrate specificity from glycoside hydrolase family 97[J]. Journal of Microbiology and Biotechnology, 2016, 26(11): 1933-1942.
57 Elleuche S, Qoura F M, Lorenz U, et al. Cloning, expression and characterization of the recombinant cold-active type-I pullulanase from[J]. Journal of Molecular Catalysis B: Enzymatic, 2015, 116: 70-77.
58 Crespim E, Zanphorlin L M, de Souza F H M, et al. A novel cold-adapted and glucose-tolerant GH1 β-glucosidase fromB7[J]. International Journal of Biological Macromolecules, 2016, 82: 375-380.
59 Zanphorlin L M, de Giuseppe P O, Honorato R V, et al. Oligomerization as a strategy for cold adaptation: Structure and dynamics of the GH1 β-glucosidase fromB7[J]. Scientific Reports, 2016, 6: 23776.
60 ZHAO F, CAO H, ZHAO L, et al. A novel subfamily endo-β-1,4-glucanases in glycoside hydrolase family 10[J]. Applied and Environmental Microbiology, 2019, 85(18): e01029-19.
61 STEPNOV A A, FREDRIKSEN L, STEEN I H, et al. Identification and characterization of a hyperthermophilic GH9 cellulase from the Arctic Mid-Ocean Ridge vent field[J]. PLoS One, 2019, 14(9): e0222216.
62 潘爱红, 李江, 王蕾, 等. 南极交替单胞菌R11-5产卡拉胶酶的发酵条件优化[J]. 微生物学通报, 2018, 45(9): 2022-2034.
63 胡秋实, 苏忠亮, 李江. 两种极端环境微生物产卡拉胶酶的研究[J]. 化学与生物工程, 2014, 31(5): 17-20.
64 林欢, 李然, 王瑞玉, 等. 南极低温降解卡拉胶菌株的筛选、鉴定、产酶条件及酶学性质的初步研究[J]. 化学与生物工程, 2018, 35(5): 47-52.
65 LI J, SHA Y. Expression and enzymatic characterization of a cold-adapted β-agarase from Antarctic bacteriumsp. NJ21[J]. Chinese Journal of Oceanology and Limnology, 2015, 33(2): 319-327.
66 LI J, XIE M S, GAO Y. Identification and biochemical characterization of a novel exo-type β-agarase Aga3463 from an Antarcticsp. strain[J]. International Journal of Biological Macromolecules, 2019, 129: 162-170.
67 刘秀萌, 李江, 侯旭光, 等. 南极菌产琼胶酶aga3311的表达、性质及其降解特性[J]. 微生物学报, 2016, 56(9): 1468-1476.
68 HAN Z, ZHANG Y, YANG J. Biochemical characterization of a new β-agarase from[J]. International Journal of Molecular Sciences, 2019, 20(9): 2143.
69 LAYE V J, KARAN R, KIM J M, et al. Key amino acid residues conferring enhanced enzyme activity at cold temperatures in an Antarctic polyextremophilic β-galactosidase[J]. Proceedings of the National Academy of Sciences of the United States of America, 2017, 114(47): 12530-12535.
70 Pawlak-Szukalska A, Wanarska M, Popinigis A T, et al. A novel cold-active β-D-galactosidase with transglycosylation activity from the Antarcticsp. 32cB – Gene cloning, purification and characterization[J]. Process Biochemistry, 2014, 49(12): 2122–2133.
71 RUTKIEWICZ M, BUJACZ A, BUJACZ G. Structural features of cold-adapted dimeric GH2β-D-galactosidase fromsp. 32cB[J]. Biochimica et Biophysica Acta-Proteins and Proteomics, 2019, 1867(9): 776-786.
72 孙茜. 北极海单胞菌基因组学分析及其β-半乳糖苷酶基因的功能研究[D]. 上海: 华东理工大学, 2015.
73 DING H T, ZENG Q, ZHOU L L, et al. Biochemical and structural insights into a novel thermostable β-1, 3-galactosidase fromsp. BSi20414[J]. Marine Drugs, 2017, 15(1): 13.
74 Alikunju A P, Joy S, Salam J A, et al. Functional characterization of a new cold-adapted β-galactosidase from an arctic fjord sediment bacteriaMCC 3423[J]. Catalysis Letters, 2018, 148(10): 3223-3235.
75 DEL-CID A, UBILLA P, RAVANAL M C, et al. Cold-active xylanase produced by fungi associated with Antarctic marine sponges[J]. Applied Biochemistry and Biotechnology, 2014, 172(1): 524-532.
76 Gil-Durán C, Ravanal M C, Ubilla P, et al. Heterologous expression, purification and characterization of a highly thermolabile endoxylanase from the Antarctic fungussp.[J]. Fungal Biology, 2018, 122(9): 875-882.
77 ACEVEDO J P, REETZ M T, ASENJO J A, et al. One-step combined focused epPCR and saturation mutagenesis for thermostability evolution of a new cold-active xylanase[J]. Enzyme and Microbial Technology, 2017, 100: 60-70.
78 FREDRIKSEN L, STOKKE R, JENSEN M S, et al. Discovery of a thermostable GH10 xylanase with broad substrate specificity from the arctic mid-ocean ridge vent system[J]. Applied and Environmental Microbiology, 2019, 85(6): e02970-18.
79 东升. 北极褐藻酸裂解酶分泌菌株的多样性分析和褐藻酸裂解酶的成熟与催化机制研究[D]. 济南: 山东大学, 2014.
80 高杨. 南极真菌产褐藻胶裂解酶Aly-i7的基因克隆、表达和酶学性质研究[D]. 青岛: 青岛科技大学, 2018.
81 XIE M S, LI J, HE P Q, et al. Expression and characterization of a bifunctional alginate lyase named Al163 from the Antarctic bacteriumsp. NJ-21[J]. Journal of Oceanology and Limnology, 2018, 36(4): 1304-1314.
82 VUORISTO K S, FREDRIKSEN L, OFTEBRO M, et al. Production, characterization, and application of an alginate lyase, AMOR_PL7A, from hot vents in the arctic mid-ocean ridge[J]. Journal of Agricultural and Food Chemistry, 2019, 67(10): 2936-2945.
83 Carrasco M, Rozas J M, Alcaíno J, et al. Pectinase secreted by psychrotolerant fungi: identification, molecular characterization and heterologous expression of a cold-active polygalacturonase fromsp.[J]. Microbial Cell Factories, 2019, 18: 45.
84 TANG Y M, WU P, JIANG S J, et al. A new cold-active and alkaline pectate lyase from Antarctic bacterium with high catalytic efficiency[J]. Applied Microbiology and Biotechnology, 2019, 103(13): 5231-5241.
85 Nalini P, Ellaiah P, Prabhakar T, et al. Microbial alkaline phosphatases in bioprocessing[J]. International Journal of Current Microbiology and Applied Sciences, 2015, 4(3): 384-396.
86 Yuivar Y, Barahona S, Alcaíno J, et al. Biochemical and thermodynamical characterization of glucose oxidase, invertase, and alkaline phosphatase secreted by Antarctic yeasts[J]. Frontiers in Molecular Biosciences, 2017, 4: 86.
87 Pellizza L A, Smal C, Ithuralde R E, et al. Structural and functional characterization of a cold-adapted stand-alone TPM domain reveals a relationship between dynamics and phosphatase activity[J]. The FEBS Journal, 2016, 283(23): 4370-4385.
88 WANG Q F, WANG Y F, HOU Y H, et al. Cloning, expression and biochemical characterization of recombinant superoxide dismutase from Antarctic psychrophilic bacteriumsp. ANT506[J]. Journal of Basic Microbiology, 2016, 56(7): 753-761.
89 KAN G F, WEN H, WANG X F, et al. Cloning and characterization of iron-superoxide dismutase in Antarctic yeast strainAN5[J]. Journal of Basic Microbiology, 2017, 57(8): 680-690.
90 ABRASHEV R, FELLER G, KOSTADINOVA N, et al. Production, purification, and characterization of a novel cold-active superoxide dismutase from the Antarctic strain363[J]. Fungal Biology, 2016, 120(5): 679-689.
A REVIEW OF NOVEL POLAR MICROBIAL ENZYMES FOR INDUSTRIAL APPLICATIONS
Wang Wei1,2, Yao Congyu1,2,3, Sun Jingjing1,2, Hao Jianhua1,2
(1Key Laboratory of Sustainable Development of Polar Fishery, Ministry of Agriculture and Rural Affairs, Yellow Sea Fisheries Research Institute, Chinese Academy of Fishery Sciences, Qingdao 266071, China;2Laboratory for Marine Drugs and Bioproducts, Pilot National Laboratory for Marine Science and Technology, Qingdao 266235, China;3College of Food Science and Technology, Shanghai Ocean University, Shanghai 201306, China)
Special challenges for microorganisms in cold ecosystems include reduced enzymatic reaction rates, limited bioavailability of nutrients, and frequent extremes in pH and salinity. To thrive successfully in low temperature environments, psychrophiles have evolved a complex range of structural and functional adaptations. Psychrophiles produce cold-active enzymes, which can be up to ten times more active at low and moderate temperatures compared with their mesophilic homologues. The enzymes of polar microorganisms are shaped by their adaptations to the permanently low temperatures. In addition, strongly differing environments, such as permafrost, glaciers and sea ice, have contributed to additional functional diversity. Microorganisms that thrive in the polar zones are a vast reservoir of cold-adapted enzymes. These enzymes could be beneficial in many industrial applications. Research using polar microorganisms to find new bioproducts has been mainly focused on enzymes that can be used in a range of industrial processes. The biotechnological value of cold-adapted enzymes stems from their high turnover (kcat) at low to moderate temperatures and their high thermolability at elevated temperatures. In recent years, a large number of new polar microbial enzymes have been continuously discovered, indicating that polar microbes have become an important source of novel enzymes. This review describes the research progress of new microbial enzymes over the past five years, and focuses on the discovery of important industrial enzymes, such as protease, lipase/esterase and carbohydrate-degrading enzymes, with good commercial prospects.
polar microorganism, exploitation of enzyme resource, protease, lipase, esterase, carbohydrate- degrading enzymes
2019年7月收到来稿, 2019年11月收到修改稿
中国工程院战略研究(2018-ZD-08)、农业农村部极地渔业开发重点实验室开放课题(2019OPF02)和中国水产科学研究院基本科研业务费(2020TD67)资助
王伟, 男, 1980年生。博士, 副研究员, 主要从事极地海洋微生物资源开发研究。E-mail: weiwang@ysfri.ac.cn
郝建华, E-mail: haojh@ysfri.ac.cn
10. 13679/j.jdyj.20190039

