丙型肝炎病毒诱发肝细胞肝癌的分子机制
高荣,徐力,徐杰,任浩
第二军医大学微生物学教研室,上海市医学生物防护重点实验室,上海 200433
·综述·
丙型肝炎病毒诱发肝细胞肝癌的分子机制
高荣,徐力,徐杰,任浩
第二军医大学微生物学教研室,上海市医学生物防护重点实验室,上海 200433
肝细胞肝癌(hepatocellular carcinoma,HCC)是影响人类健康的恶性肿瘤之一,丙型肝炎病毒(hepatitis C virus,HCV)是其主要致病因素之一。HCV诱发的HCC是由病毒和宿主免疫介导的多步骤复杂过程,从慢性炎症发展到肝硬化和肝癌,病毒和宿主因子共同参与此过程。其中,宿主基因突变是导致HCC发生的危险因素之一,全面了解HCV诱发HCC的分子机制将有助于解决此问题。
丙型肝炎病毒;肝细胞肝癌;分子机制
肝细胞肝癌(hepatocellular carcinoma,HCC)是人类最常见的恶性肿瘤之一,在男性疾病中居第5位,在女性疾病中居第9位,已成为全球癌症相关死亡的第二大病因。在多数西方国家,丙型肝炎病毒(hepatitis C virus,HCV)是慢性肝病和HCC的主要危险因素[1]。全球约3%的人口感染HCV,大部分感染者不能清除病毒感染并发展为慢性丙型肝炎(chronic hepatitis C,CHC)。CHC可引起进行性肝纤维化相关肝损伤,20~40年后可能演变为肝硬化(10%~20%)和HCC(1%~5%)[2-3]。据估计,全球27%的肝硬化和25%的HCC可归因于HCV感染[4]。慢性HCV感染患者发生HCC的危险与肝纤维化阶段紧密相关,肝硬化患者的HCC发病率远高于轻度纤维化患者[5]。此外,其他危险因素,如乙型肝炎病毒(hepatitis B virus,HBV)/人类免疫缺陷病毒(human immunodeficiency virus,HIV)共感染、肥胖、胰岛素抵抗、非酒精性脂肪性肝炎等,会加快HCV诱发的HCC (HCV-induced HCC,HCV-HCC)进程[6]。基于干扰素治疗的持续病毒学应答(sustained virological response,SVR)可降低大部分HCV患者的HCC发生率,表明清除病毒在阻止肿瘤发生中的重要性[7]。将直接抗病毒药物(direct-acting antiviral agent,DAA)引入抗HCV治疗,可明显提高SVR。然而,DAA在降低HCV相关肝硬化患者HCC发生率及促进HCC复发方面仍有争议。有报道表明,DAA治疗可促进HCV-HCC患者病情恶化,且更难以治疗[8]。因此,阐明HCV-HCC分子机制有助于探讨HCC预防和治疗新策略,本文主要就此进行简要综述。
1 HCV的基因组结构与功能
HCV为黄病毒属单股正链RNA病毒,基因组全长约9.6 kb,具有单一的开放读码框,编码长约 3 000 个氨基酸残基的多蛋白前体。5′和3′端均具有高度保守的非编码区(non-coding region,NCR),这些序列对病毒蛋白的合成及复制是必需的。多蛋白前体在宿主和病毒蛋白酶作用下形成的结构蛋白包括核心(Core)蛋白、包膜蛋白E1和E2,其序列位于近5′区;非结构蛋白包括NS2~NS5,其序列位于近3′区[9]。Core蛋白序列保守,除保护HCV核心结构外,还具有促进HCV复制、调节宿主免疫细胞等功能[10]。包膜蛋白E1和E2均定位于内质网并形成异二聚体,与病毒入侵和病毒颗粒组装有关。E2蛋白还具有高度变异性,参与病毒的免疫逃逸[11]。NS2是一类跨膜蛋白,作用类似金属蛋白酶,除剪切 NS3外,还参与NS5A磷酸化[12]。NS3分为2个结构域,N端的181个氨基酸具有蛋白酶活性,C端的465个氨基酸具有螺旋酶及NTP酶活性,后两者参与病毒复制[13]。NS4A是NS3辅助因子,参与其催化反应,而NS4B是NS5A磷酸化反应的辅助因子[14]。NS5A可能通过与双链RNA依赖的蛋白激酶(double-stranded RNA-dependent protein kinase,PKR)结合,使病毒对干扰素产生耐药,并参与HCV多种蛋白的成熟和RNA复制,调控宿主多种基因表达,刺激细胞增殖,抑制细胞凋亡及影响干扰素疗效[15]。NS5B编码RNA依赖的RNA聚合酶(RNA-dependent RNA polymerase,RdRp),作为核心酶在HCV复制中起重要作用[16]。
2 HCV感染与HCC发生
HCV感染所致肝癌是一个由病毒、宿主和环境等多方因素共同参与的复杂过程。除通过免疫介导的慢性炎症反应间接诱发肝癌外,还可能通过改变宿主的信号通路(如增殖、能量代谢、血管生成、上皮-间质转化(epithelial-mesenchymal transition,EMT)、DNA修复、细胞凋亡和氧化/内质网应激等)直接诱发HCC[17]。慢性炎症可在代谢和基因水平损伤肝细胞并使导致细胞死亡的活性氧(reactive oxygen species,ROS)水平增加。HCV感染后的肝细胞再生可使染色体不稳定性增加,并促进肝细胞新陈代谢,以及恶性克隆进展的遗传/表观遗传发生不可逆的变异[18]。
2.1 HCV直接干预致癌和抗癌通路
2.1.1HCV与细胞周期HCV蛋白可直接作用于某些宿主肿瘤抑制基因和原癌基因[19]。例如,HCV NS5B可结合视网膜母细胞瘤(retinoblastoma,Rb)蛋白并促使其在胞质重新定位和蛋白酶体降解,最终激活促进细胞周期进展的E2F(细胞分裂G1期进入S期的重要转录因子)基因应答[19]。此外,HCV蛋白还可激活Cyclin/Cdk复合物来促进G1/S转换,进而干扰细胞周期进程。如NS2蛋白能激活Cyclin D/Cdk4并上调Cyclin E表达,而Core蛋白上调Cyclin E和Cdk2表达[20-22]。
2.1.2HCV与生长因子信号通路生长因子信号通路激活对HCC的发生和维持起重要作用。HCV Core、E2、NS3和NS5A蛋白通过干扰Raf/丝裂原活化蛋白激酶(mitogen-activated protein kinase,MAPK)/细胞外调节蛋白激酶(extracellular regulated protein kinase,ERK)通路刺激细胞增殖[23-26]。HCV也可激活Wnt/β-catenin通路,通过Wnt配体与Frizzled受体结合活化,阻止β-catenin多聚蛋白复合体形成,从而激活c-myc和Cyclin D等一系列靶基因的转录[27]。体外实验表明,HCV NS5A和Core过表达可激活β-catenin,其间接机制为抑制GSK-3β激活及使β-catenin稳定化。最近研究表明,β-catenin可与NS5A蛋白相互作用而被直接激活[28]。目前为止,HCV感染细胞内β-catenin活化的功能意义尚不清楚。虽然Wnt信号通路失调不足以引起肝细胞恶性转化,但β-catenin表达水平在多数HCC患者中升高,提示其可能参与肿瘤生长的维持[29]。
2.1.3HCV与转化因子信号通路转化生长因子β(transforming growth factor β,TGF-β)是一种多向性和多效性的细胞因子,可通过细胞表面受体信号转导途径发挥抗增殖、促凋亡作用,在EMT过程中具有促肿瘤活性[30]。HCV NS5A蛋白与TGF-β受体1(transforming growth factor β receptor 1,TGFBR1)结合抑制TGF-β介导的Smad2磷酸化和核易位及Smad3/Smad4异源二聚体形成,从而阻断TGF-β信号转导[31]。此外,TGF-β通路也是HCV Core蛋白的作用靶点[32]。值得注意的是,与经典的TGFBR1通路不同,由c-Jun氨基末端激酶(c-Jun N-terminal kinase,JNK)介导的Smad3蛋白磷酸化可通过上调纤溶酶原激活物抑制剂1(plasminogen activator inhibitor 1,PAI-1)促进细胞外基质沉积,并在慢性肝病-肝硬化-HCC这一疾病演变过程中表达[33]。此外,HCV可间接通过激活磷脂酰肌醇-3激酶(phosphoinositide-3 kinase,PI3K)/蛋白激酶B(protein kinase B,Akt)通路抑制细胞凋亡,这种作用可能是通过NS5A下调PTEN并减弱PI3K/Akt通路的抑制作用实现的[34]。
2.1.4HCV与细胞凋亡细胞凋亡是为维持内环境稳定,由基因控制的细胞自主有序的死亡,是一个主动过程。凋亡抑制是肿瘤发生多步骤过程中的重要事件,主要表现为恶性转化细胞在细胞周期中逃避关键检查点。HCV感染与凋亡之间的相互作用尚未完全阐明。细胞实验中,细胞凋亡随时间变化,凋亡活性在感染早期较高,并随时间减少[35]。过度凋亡在急性肝炎、急性重型肝炎及慢性肝炎患者中均可观察到。持续凋亡与持续性炎症和纤维化有关,而缺乏细胞凋亡则有助于HCC的发生[36]。
P53是一个关键的肿瘤抑制蛋白,是细胞生长周期的负调控因子,可阻滞细胞周期,促进细胞凋亡。HCV Core蛋白可直接激活P53信号通路,但这种相互作用仍有争议[37]。此外,HCV NS2、NS3/4A和NS5A可诱导P53蛋白从细胞核到细胞质/核周区域重新定位从而干扰P53通路[22]。HCV Core蛋白通过诱导FLICE阻断肿瘤坏死因子α(tumor necrosis factor α,TNF-α)介导的凋亡信号,HCV NS5A通过阻断天冬氨酸特异性半胱氨酸蛋白酶3(cysteinyl aspartate specific proteinase 3,caspase-3)和抑制多聚 ADP核糖聚合酶1〔poly (ADP-ribose) polymerase 1,PARP1〕,防止TNF-α介导细胞凋亡[38]。
2.1.5HCV与自噬自噬又称Ⅱ型细胞死亡,是细胞在自噬相关基因调控下,利用溶酶体降解自身受损的细胞器和大分子物质的过程。HCV可抑制宿主细胞通过自噬对病毒的清除,从而起促进肿瘤形成的作用[39]。自噬缺陷性小鼠可引起HCC自发形成,这主要是由于P62蛋白堆积和随后转录因子Nrf2(可调节多种细胞保护基因)的持续激活。P62水平在HCV-HCC患者体内增高,提示自噬损伤在肿瘤发展中可能具有重要作用[40]。此外,自噬缺乏的肝细胞也可促进EMT[41]。另一方面,在抗肝细胞大鼠模型中,自噬可促进癌前病变的肝脏结节生长[42]。总之,HCV感染引发的自噬在病毒复制/分泌、炎症和脂质代谢平衡等方面发挥重要作用。
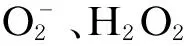
2.2 HCV诱导的免疫反应与HCC
HCV通过多种机制(抑制Ⅰ型IFN产生,改变CD4+T细胞向Th2细胞、Th17细胞、调节性T细胞分化等)逃避宿主先天性免疫和获得性免疫而建立慢性感染[46-47]。这些改变导致肝脏轻度慢性炎症,并诱导ROS和一氧化氮释放,增强脂质过氧化作用及毒性细胞因子的异常表达,同时破坏宿主免疫监视而促进肝癌发展[48]。
TNF-α、白细胞介素1(interleukin 1,IL-1)、IL-23和IL-6、淋巴毒素α(lymphotoxin α,LTα)和LT β等多种炎性细胞因子参与慢性肝炎和HCC的发生和发展。其中,LTα和LTβ在HCC发展中起重要作用,LT高表达诱导的严重肝损伤与肝浸润淋巴细胞(liver infiltrating lymphocyte,LIL)数量增加相关[49]。HCV感染期间,丧失病毒清除作用的LIL在肝脏中积累并建立慢性炎症。值得注意的是,Ramzan等发现,与无HCC的HCV患者相比,HCV-HCC患者的肝硬化组织中存在较高水平的肝浸润性T和B细胞。特别是大量存在于肝硬化区域而不是HCV-HCC患者肿瘤组织中的CD8+T细胞与HCC发生相关,且是手术切除后复发的预后因素[50]。此外,在HCV-HCC患者中,炎症性CD8+T细胞的增加伴随着参与肿瘤免疫监视的自然杀伤(natural killer,NK)细胞和自然杀伤T(natural killer T,NKT)细胞的减少[50]。相反,具有抗肿瘤免疫关键作用的调节性T细胞在HCC和HCV-肝硬化组织中增加[51]。这些数据表明,在HCV到肝硬化再到HCC的发展过程中,肿瘤促进炎症反应和抗肿瘤免疫受损的协同效应可能至关重要。
2.3 HCV诱导的脂质代谢异常与HCC
HCV-HCC常伴有脂肪性肝炎,表明与炎症相关的脂质代谢改变可参与癌症发生[52]。HCV蛋白主要通过活化脂肪生成途径和降低脂质分解代谢干扰宿主脂质代谢,从而触发感染肝细胞中的脂毒性[53]。慢性HCV患者应用他汀类药物可使伴随的肝硬化和HCC呈剂量依赖性减少,证实脂肪变性与HCC关系密切[54]。近期有研究提出HCV和肥胖可协同诱导HCC的新机制。NS5A通过增加Toll样受体4(Toll-like receptor 4,TLR4)水平,增强干细胞转录因子NANOG的诱导作用,从而产生肿瘤起始干细胞样细胞(tumor-initiating stem-like cell,TIC)[55]。此外,干细胞转录因子NANOG还可诱导EMT主调节器Twist1协同瘦素(leptin)-STAT3通路促进肿瘤发生,活化脂肪酸氧化,支持TIC自我更新和耐药性产生[56]。
HCV诱导脂肪变性促进HCC发展的一个重要方面是改变肝脏T细胞功能。最近,在脂肪性肝炎和肥胖复合小鼠模型中观察到大量CD8+T和NKT细胞肝浸润,它们的激活可诱导肝细胞脂肪变性,进而发挥炎症和致癌作用[57]。在肥胖诱导小鼠模型中,脂质生成增加(特别是亚油酸)与选择性CD4+T细胞丢失直接相关,而选择性CD4+T细胞可削弱抗肿瘤免疫而促进HCC进展[58]。
HCV患者的HCC发病风险也与肝纤维化程度紧密联系。促纤维形成细胞因子已被鉴定为HCC进展的关键调节剂。TGF-β是参与纤维形成的关键细胞因子,直接由HCV Core蛋白或氧化、内质网应激和核因子κB(nuclear factor κB,NF-κB)通路活化诱导产生[59]。最近,Jee等证实HCV患者肝细胞表达高水平TGF-β,且体外TGF-β的分泌量足以活化肝星状细胞[60]。刺激后的肝星状细胞在肌成纤维细胞中转分化并释放促纤维形成介质、炎性细胞因子和趋化因子,加重肝脏炎症和纤维化[61]。TGF-β除具有促纤维形成作用,还是免疫抑制剂,有利于肿瘤转化细胞的免疫逃逸[62]。
2.4 HCV诱导的促血管生成与HCC
新血管生成是癌细胞生长和存活的必要阶段。许多研究表明,所有HCC患者中,HCV阳性者微血管密度较高[63]。HCV结构和非结构蛋白在此过程中发挥直接作用。HCV Core蛋白通过上调低氧诱导因子1α(hypoxia inducible factor 1α,HIF-1α)促进血管生成,HIF-1α可上调血管内皮生长因子(vascular endothelial growth factor,VEGF)和环加氧酶2,也可激活基质金属蛋白酶(matrix metalloproteinase,MMP)如MMP-2和MMP-9[64]。VEGF是HCC中重要的内皮特异性生长因子,可作为HCC的预后因子[65]。HCV感染上调促血管生成素2(angiopoietin 2,Ang-2)水平,Ang-2可与Tie-1受体结合抑制血管生成。在VEGF刺激下,Ang-2可从内皮细胞释放并促进VEGF的促血管生成作用[66]。
EMT在癌细胞转移过程中发挥核心作用,此过程中上皮细胞获得间充质细胞特征,失去细胞极性,失去与基底膜的连接,从而使细胞运动性和侵袭性增加[67]。值得注意的是,NS5A通过激活Twist2触发EMT,而Twist2是转移过程中的关键转录调节子[68]。另一种EMT诱导途径由E1/E2蛋白触发,并通过TGF-β和VEGF信号通路转导。TGF-β和VEGF也受HIF-1α调节,HIF-1α被HCV Core蛋白上调,并增强肝癌细胞的迁移性和渗透性[69]。
2.5 宿主因子在HCV-HCC中的作用
2.5.1癌基因突变与HCC目前已确定几个与肝癌发生相关的基因突变[70]。肿瘤抑制基因p53突变导致基因失活或产生显性阴性型[28]。全基因组测序发现,编码Wnt信号通路的β-catenin蛋白的基因具有较高突变率(>30%)[71]。端粒酶反转录酶(telomerase reverse transcriptase,TERT)基因是HCC中另一频繁发生遗传性改变的基因,通常涉及其启动子区,导致其表达增加[61]。
2.5.2宿主基因单核苷酸多态性(singlenucleotidepolymorphism,SNP)与HCCSNP与疾病的关系近年来备受关注。对与HCV治疗和自然清除相关的λ干扰素成员IFNL3/4的研究发现,IFNL3 mRNA的3′-非翻译区(untranslated region,UTR)多态性通过影响与HCV感染诱导产生的微小RNA(microRNA,miRNA)结合而控制其转录稳定性[72-73]。据报道,IFNL3的多态性与HCC发生风险升高相关,特别是在未达到SVR的患者,但有待进一步证实[74]。其他细胞因子和(或)相关受体的多态性也与HCC发生相关。如携带低单倍型IL-10和TNF-α GG基因型的患者发生HCC的风险更高,IL-23R的罕见突变体与埃及患者中HCV-HCC风险降低相关,与VEGF基因高表达相关的多态性也与HCC相关[65,75-76]。最近,日本一项大型全基因组关联研究发现了与HCV-HCC相关的遗传变异基因:DEPDC5和人类主要组织相容性复合体Ⅰ类链相关基因A(major histocompatibility complex class I-related chain A,MICA)[77]。DEPDC5功能未知,MICA可结合NK细胞活化性受体凝集素样同型二聚体(natural killer group 2, member D,NKG2D),在肿瘤免疫监视中介导靶细胞的毒性。此外,已发现位于MICA上游的HCP5基因的新型突变体,其对HCC的预测性更好[78]。在不同种族中进一步验证分析将有助于确认这些标记的预测价值。
2.5.3HCV基因序列变异与HCCHCV基因变异也影响HCC发展。有研究发现,Core、NS3和NS5A蛋白在HCV 1b及Core在HCV 1a中的多态性与HCC发生相关[79-80]。Harouaka等比较肿瘤与非肿瘤组织HCV序列后,发现存在不同HCV准种,提示HCV特定变异株更易诱发HCC[81]。
2.5.4表观遗传学改变与HCC表观遗传调节基因的失调也与HCC发生相关,如具有DNA甲基化作用的组蛋白赖氨酸N-甲基转移酶EZH2和重组人蛋白质精氨酸甲基转移酶1(protein arginine methyltransferase 1,PRMT1)[82-83]。其中,EZH2通过靶向多种基因在HCC中异常表达,PRMT1抑制则与HCV介导的蛋白磷酸酶2A催化性C亚基(protein phosphatase 2A catalytic subunit C,PP2Ac)相关。此外,多种肿瘤抑制基因(包括来自HCV的CDKN2A、GSTP1、RUNX3、APC、SOCS-1和RASSF1A)的高度甲基化也与HCC发生相关[84-85]。
2.6 非编码RNA与HCV-HCC
非编码RNA泛指一类不编码蛋白质的RNA,根据长度可划分为miRNA和长链非编码RNA(long non-coding RNA,lncRNA)等。miRNA和lncRNA的变化均可介导HCC相关的表观遗传改变。
2.6.1miRNA与HCV-HCC已报道多种miRNA可调节HCV复制。其中最具代表性的是miR-122,可与HCV基因组RNA 5′-UTR结合而增强肝细胞内的病毒复制[86]。miRNA在HCV-HCC中扮演不同角色,有些抑制增殖、脂质代谢和肝细胞生长的miRNA(如miR-122、miR-27a和miR-181c)被下调,而其他可调节代谢和免疫反应的miRNA(如miR-21、miR-221、miR-130a、Let7b、miR-155和miR-200c)被上调[87]。最近发现,miR-200c和miR-21的上调与HCV患者的肝纤维化相关[87]。
2.6.2lncRNA与HCV-HCClncRNA可调节肿瘤抑制基因或癌基因的表达[88]。HCC相关lncRNA通过表观遗传沉默、剪接调节、lncRNA-miRNA相互作用、lncRNA-蛋白相互作用和遗传变异等多种机制,参与HCC进展的多样化生物过程,如细胞增殖、凋亡、侵袭、转移和血管发生等[89-90]。研究发现,在HCV-HCC的不同发展阶段,lncRNA LINC01419的表达在早期HCC组织中较发育不良组织明显增加,而lncRNA AF070632在晚期HCC组织中比早期HCC组织减少。此外,与非肿瘤性肝组织相比,LINC01419和AK021443的表达在HCC组织中上调。基因共表达网络分析结果提示,LINC01419和AK021443主要参与细胞周期进展,而AF070632与辅因子结合、氧化还原和羧酸分解代谢过程相关[91]。Kamel等报道了两种lncRNA:lncRNA-尿路上皮癌胚抗原1(urothelial carcinoma associated-1,UCA1)和p53反义转录本WRAP53(WD repeat containing,antisense to TP53)。他们发现HCC患者的UCA1和WRAP53血清水平高于慢性HCV感染患者或健康志愿者,肿瘤组织中UCA1和WRAP53的表达高于相邻的非肿瘤组织,血清和组织中UCA1与WRAP53的表达存在显著相关性。此外,UCA1和WRAP53的上调与Child-Pugh评分相关。Kaplan-Meier分析显示,UCA1和WRAP53阳性HCC患者具有显著降低的无复发生存期(relapse-free survival,RFS)。Cox多变量分析显示,WRAP53是一个独立的RFS预后因子[92]。这些发现有助于更好地了解lncRNA在HCV-HCC发展中的作用。
2.6.3环状RNA分子(circularRNA,circRNA)与HCV-HCCcircRNA是一类不具有5′端帽子和3′端poly(A)尾巴并以共价键形成环形结构的非编码RNA分子。相较于miRNA和lncRNA,circRNA具有更高的稳定性和序列保守性[93]。多项研究已报道circRNA与HCC有关。Shang等发现hsa_circ_0005075在HCC组织中明显升高,并与肿瘤大小相关,在HCC发展中促进细胞黏附[94]。Xu等发现ciRS-7在HCC组织中明显升高,与肝癌微血管侵犯、甲胎蛋白(α fetoprotein,AFP)水平及低龄明显相关,且部分影响HCC恶化[95]。Qin等发现hsa_circ_0001649在HCC组织中明显降低,其可能在HCC形成及转移过程中起重要作用[96]。circRNA在HCV-HCC发展中的作用仍需进一步阐明。
3 结语
HCV慢性感染是HCC发生的重要危险因素,对HCV-HCC发病机制的研究在世界范围内已引起极大关注。阐明HCV如何通过直接和间接机制触发HCC,鉴别与HCC相关的基因和表观遗传因子,将有助于监测慢性肝炎-肝硬化-肝癌的发展,确认高风险HCV患者并探索相应干预措施。随着DAA的不断上市,HCV被称为可“治愈”的肝炎病毒。一项前瞻性研究表明,尽管DAA不会增加HCV患者患肝硬化和肝癌的风险,但如果患者在DAA治疗前已存在未发现的肝硬化或肝癌,DAA的使用将会加重肝癌恶化且更加难以治疗[8]。近期一项HCV-HCC患者接受单纯DAA治疗的研究显示,早期HCC的高复发率与HCV的清除率一致,提示DAA对肿瘤免疫监视可能存在抑制作用[97]。尽管目前HCV蛋白对宿主信号通路调节作用的研究已取得可喜成果,但这些结果多基于体外细胞模型获得,未来仍需进一步的体内研究证实。总之,研究HCV-HCC的分子机制,阐明肝脏免疫学及分子微环境在HCV-HCC进展中的作用,识别肝癌发生和发展的预测指标,将为人类战胜HCV感染奠定基础。
[1] El-Serag HB. Hepatocellular carcinoma [J]. N Engl J Med, 2011, 365(12): 1118-1127.
[2] Buchanan R, Hydes T, Khakoo SI. Innate and adaptive genetic pathways in HCV infection [J]. Tissue Antigens, 2015, 85(4): 231-240.
[3] Arzumanyan A, Reis HM, Feitelson MA. Pathogenic mechanisms in HBV and HCV-associated hepatocellular carcinoma [J]. Nat Rev Cancer, 2013, 13(2): 123-135.
[4] Westbrook RH, Dusheiko G. Natural history of hepatitis C [J]. J Hepatol, 2014, 61 (1 Suppl): S58-S68.
[5] Mohamed AA, Elbedewy TA, El-Serafy M, El-Toukhy N, Ahmed W, Ali El Din Z. Hepatitis C virus:a global view [J]. World J Hepatol, 2015, 7(26): 2676-2680.
[6] Webster DP, Klenerman P, Dusheiko GM. Hepatitis C [J]. Lancet, 2015, 385(9973): 1124-1135.
[7] Yoshida H, Tateishi R, Arakawa Y, Sata M, Fujiyama S, Nishiguchi S, Ishibashi H, Yamada G, Yokosuka O, Shiratori Y, Omata M. Benefit of interferon therapy in hepatocellular carcinoma prevention for individual patients with chronic hepatitis C [J]. Gut, 2004, 53(3): 425-430.
[8] Cheung MC, Walker AJ, Hudson BE, Verma S, McLauchlan J, Mutimer DJ, Brown A, Gelson WT, MacDonald DC, Agarwal K, Foster GR, Irving WL; HCV Research UK. Outcomes after successful direct-acting antiviral therapy for patients with chronic hepatitis C and decompensated cirrhosis [J]. J Hepatol, 2016, 65(4): 741-747.
[9] Barth H, Liang TJ, Baumert TF. Hepatitis C virus entry: molecular biology and clinical implications [J]. Hepatology, 2006, 44(3): 527-535.
[10] Domínguez-Villar M, Muoz-Suano A, Anaya-Baz B, Aguilar S, Novalbos JP, Giron JA, Rodríguez-Iglesias M, Garcia-Cozar F. Hepatitis C virus core protein up-regulates anergy-related genes and a new set of genes, which affects T cell homeostasis [J]. J Leukoc Biol, 2007, 82(5): 1301-1310.
[11] Lyu J, Imachi H, Fukunaga K, Yoshimoto T, Zhang H, Murao K. Roles of lipoprotein receptors in the entry of hepatitis C virus [J]. World J Hepatol, 2015, 7(24): 2535-2542.
[12] Pavio N, Romano PR, Graczyk TM, Feinstone SM, Taylor DR. Protein synthesis and endoplasmic reticulum stress can be modulated by the hepatitis C virus envelope protein E2 through the eukaryotic initiation factor 2alpha kinase PERK [J]. J Virol, 2003, 77(6): 3578-3585.
[13] Lenz O, Verbinnen T, Lin TI, Vijgen L, Cummings MD, Lindberg J, Berke JM, Dehertogh P, Fransen E, Scholliers A, Vermeiren K, Ivens T, Raboisson P, Edlund M, Storm S, Vrang L, de Kock H, Fanning GC, Simmen KA. In vitro resistance profile of the hepatitis C virus NS3/4A protease inhibitor TMC435 [J]. Antimicrob Agents Chemother, 2010, 54(5): 1878-1887.
[14] Gretton SN, Taylor AI, McLauchlan J. Mobility of the hepatitis C virus NS4B protein on the endoplasmic reticulum membrane and membrane-associated foci [J]. J Gen Virol, 2005, 86(Pt 5): 1415-1421.
[15] Krishnan P, Beyer J, Mistry N, Koev G, Reisch T, DeGoey D, Kati W, Campbell A, Williams L, Xie W, Setze C, Molla A, Collins C, Pilot-Matias T. In vitro and in vivo antiviral activity and resistance profile of ombitasvir, an inhibitor of hepatitis C virus NS5A [J]. Antimicrob Agents Chemother, 2015, 59(2): 979-987.
[16] Samimi-Rad K, Nategh R, Malekzadeh R, Norder H, Magnius L. Molecular epidemiology of hepatitis C virus in Iran as reflected by phylogenetic analysis of the NS5B region [J]. J Med Virol, 2004, 74(2): 246-252.
[17] Bartosch B, Thimme R, Blum HE, Zoulim F. Hepatitis C virus-induced hepatocarcinogenesis [J]. J Hepatol, 2009, 51(4): 810-820.
[18] Hoshida Y, Fuchs BC, Bardeesy N, Baumert TF, Chung RT. Pathogenesis and prevention of hepatitis C virus-induced hepatocellular carcinoma [J]. J Hepatol, 2014, 61(1 Suppl): S79-S90.
[19] Mesri EA, Feitelson MA, Munger K. Human viral oncogenesis: a cancer hallmarks analysis [J]. Cell Host Microbe, 2014, 15(3): 266-282.
[20] Mitchell JK, Lemon SM, McGivern DR. How do persistent infections with hepatitis C virus cause liver cancer? [J]. Curr Opin Virol, 2015, 14: 101-108.
[21] McGivern DR, Villanueva RA, Chinnaswamy S, Kao CC, Lemon SM. Impaired replication of hepatitis C virus containing mutations in a conserved NS5B retinoblastoma protein-binding motif [J]. J Virol, 2009, 83(15): 7422-7433.
[22] Bittar C, Shrivastava S, Bhanja Chowdhury J, Rahal P, Ray RB. Hepatitis C virus NS2 protein inhibits DNA damage pathway by sequestering p53 to the cytoplasm [J]. PLoS One, 2013, 8(4): e62581.
[23] Hayashi J, Aoki H, Kajino K, Moriyama M, Arakawa Y, Hino O. Hepatitis C virus core protein activates the MAPK/ERK cascade synergistically with tumor promoter TPA,but not with epidermal growth factor or transforming growth factor alpha [J]. Hepatology, 2000, 32(5): 958-961.
[24] Zhao LJ, Wang L, Ren H, Cao J, Li L, Ke JS, Qi ZT. Hepatitis C virus E2 protein promotes human hepatoma cell proliferation through the MAPK/ERK signaling pathway via cellular receptors [J]. Exp Cell Res, 2005, 305(1): 23-32.
[25] Bürckstümmer T, Kriegs M, Lupberger J, Pauli EK, Schmittel S, Hildt E. Raf-1 kinase associates with hepatitis C virus NS5A and regulates viral replication [J]. FEBS Lett, 2006, 580(2): 575-580.
[26] Feng DY, Sun Y, Cheng RX, Ouyang XM, Zheng H. Effect of hepatitis C virus nonstructural protein NS3 on proliferation and MAPK phosphorylation of normal hepatocyte line [J]. World J Gastroenterol, 2005, 11(14): 2157-2161.
[27] Higgs MR, Lerat H, Pawlotsky JM. Hepatitis C virus-induced activation of beta-catenin promotes c-Myc expression and a cascade of pro-carcinogenetic events [J]. Oncogene, 2013, 32(39): 4683-4693.
[28] Tornesello ML, Buonaguro L, Izzo F, Buonaguro FM. Molecular alterations in hepatocellular carcinoma associated with hepatitis B and hepatitis C infections [J]. Oncotarget, 2016, 7(18): 25087-25102.
[29] Whittaker S, Marais R, Zhu AX. The role of signaling pathways in the development and treatment of hepatocellular carcinoma [J]. Oncogene, 2010, 29(36): 4989-5005.
[30] Syed V. TGF-beta signaling in cancer [J]. J Cell Biochem, 2016, 117(6): 1279-1287.
[31] Choi SH, Hwang SB. Modulation of the transforming growth factor-beta signal transduction pathway by hepatitis C virus nonstructural 5A protein [J]. J Biol Chem, 2006, 281(11): 7468-7478.
[32] Pavio N, Battaglia S, Boucreux D, Arnulf B, Sobesky R, Hermine O, Brechot C. Hepatitis C virus core variants isolated from liver tumor but not from adjacent non-tumor tissue interact with Smad3 and inhibit the TGF-beta pathway [J]. Oncogene, 2005, 24(40): 6119-6132.
[33] Matsuzaki K, Murata M, Yoshida K, Sekimoto G, Uemura Y, Sakaida N, Kaibori M, Kamiyama Y, Nishizawa M, Fujisawa J, Okazaki K, Seki T. Chronic inflammation associated with hepatitis C virus infection perturbs hepatic transforming growth factor beta signaling, promoting cirrhosis and hepatocellular carcinoma [J]. Hepatology, 2007, 46(1): 48-57.
[34] Cheng D, Zhang L, Yang G, Zhao L, Peng F, Tian Y, Xiao X, Chung RT, Gong G. Hepatitis C virus NS5A drives a PTEN-PI3K/Akt feedback loop to support cell survival [J]. Liver Int, 2015, 35(6): 1682-1691.
[35] Zekri AR, Bahnassy AA, Hafez MM, Hassan ZK, Kamel M, Loutfy SA, Sherif GM, El-Zayadi AR, Daoud SS. Characterization of chronic HCV infection-induced apoptosis [J]. Comp Hepatol, 2011. doi: 10.1186/1476-5926-10-4.
[36] Guicciardi ME, Gores GJ. Apoptosis: a mechanism of acute and chronic liver injury [J]. Gut, 2005, 54(7): 1024-1033.
[37] Kao CF, Chen SY, Chen JY, Wu Lee YH. Modulation of p53 transcription regulatory activity and post-translational modification by hepatitis C virus core protein [J]. Oncogene, 2004, 23(14): 2472-2483.
[38] Jiang X, Kanda T, Wu S, Nakamoto S, Wakita T, Shirasawa H, Yokosuka O. Hepatitis C virus nonstructural protein 5A inhibits thapsigargin-induced apoptosis [J]. PLoS One, 2014, 9(11): e113499.
[39] Galluzzi L, Pietrocola F, Bravo-San Pedro JM, Amaravadi RK, Baehrecke EH, Cecconi F, Codogno P, Debnath J, Gewirtz DA, Karantza V, Kimmelman A, Kumar S, Levine B, Maiuri MC, Martin SJ, Penninger J, Piacentini M, Rubinsztein DC, Simon HU, Simonsen A, Thorburn AM, Velasco G, Ryan KM, Kroemer G. Autophagy in malignant transformation and cancer progression [J]. EMBO J, 2015, 34(7): 856-880.
[40] Al-Shenawy HA. Expression of beclin-1, an autophagy-related marker, in chronic hepatitis and hepatocellular carcinoma and its relation with apoptotic markers [J]. APMIS, 2016, 124(3): 229-237.
[41] Grassi G, Di Caprio G, Santangelo L, Fimia GM, Cozzolino AM, Komatsu M, Ippolito G, Tripodi M, Alonzi T. Autophagy regulates hepatocyte identity and epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions promoting Snail degradation [J]. Cell Death Dis, 2015, 6: e1880. doi: 10.1038/cddis.2015.249.
[42] Kowalik MA, Perra A, Ledda-Columbano GM, Ippolito G, Piacentini M, Columbano A, Falasca L. Induction of autophagy promotes the growth of early preneoplastic rat liver nodules [J]. Oncotarget, 2016, 7(5): 5788-5799.
[43] Rebbani K, Tsukiyama-Kohara K. HCV-induced oxidative stress: battlefield-winning strategy [J]. Oxid Med Cell Longev, 2016. doi: 10.1155/2016/7425628.
[44] Lozano-Sepulveda SA, Bryan-Marrugo OL, Cordova-Fletes C, Gutierrez-Ruiz MC, Rivas-Estilla AM. Oxidative stress modulation in hepatitis C virus infected cells [J]. World J Hepatol, 2015, 7(29): 2880-2889.
[45] Tsukiyama-Kohara K. Role of oxidative stress in hepatocarcinogenesis induced by hepatitis C virus [J]. Int J Mol Sci, 2012, 13(11): 15271-15278.
[46] Park SH, Rehermann B. Immune responses to HCV and other hepatitis viruses [J]. Immunity, 2014, 40(1): 13-24.
[47] Lee HC, Sung SS, Krueger PD, Jo YA, Rosen HR, Ziegler SF, Hahn YS. Hepatitis C virus promotes T-helper (Th)17 responses through thymic stromal lymphopoietin production by infected hepatocytes [J]. Hepatology, 2013, 57(4): 1314-1324.
[48] Zampino R, Marrone A, Restivo L, Guerrera B, Sellitto A, Rinaldi L, Romano C, Adinolfi LE. Chronic HCV infection and inflammation: Clinical impact on hepatic and extra-hepatic manifestations [J]. World J Hepatol, 2013, 5(10): 528-540.
[49] Haybaeck J, Zeller N, Wolf MJ, Weber A, Wagner U, Kurrer MO, Bremer J, Iezzi G, Graf R, Clavien PA, Thimme R, Blum H, Nedospasov SA, Zatloukal K, Ramzan M, Ciesek S, Pietschmann T, Marche PN, Karin M, Kopf M, Browning JL, Aguzzi A, Heikenwalder M. A lymphotoxin-driven pathway to hepatocellular carcinoma [J]. Cancer Cell, 2009, 16(4): 295-308.
[50] Ramzan M, Sturm N, Decaens T, Bioulac-Sage P, Bancel B, Merle P, Tran van Nhieu J, Slama R, Letoublon C, Zarski JP, Jouvin-Marche E, Marche PN, Leroy V. Liver-infiltrating CD8+lymphocytes as prognostic factor for tumour recurrence in hepatitis C virus-related hepatocellular carcinoma [J]. Liver Int, 2016, 36(3): 434-444.
[51] Piconese S, Timperi E, Pacella I, Schinzari V, Tripodo C, Rossi M, Guglielmo N, Mennini G, Grazi GL, Di Filippo S, Brozzetti S, Fazzi K, Antonelli G, Lozzi MA, Sanchez M, Barnaba V. Human OX40 tunes the function of regulatory T cells in tumor and nontumor areas of hepatitis C virus-infected liver tissue [J]. Hepatology, 2014, 60(5): 1494-1507.
[52] Hwang SJ, Lee SD. Hepatic steatosis and hepatitis C: Still unhappy bedfellows? [J]. J Gastroenterol Hepatol, 2011, 26(Suppl 1): 96-101.
[53] Syed GH, Amako Y, Siddiqui A. Hepatitis C virus hijacks host lipid metabolism [J]. Trends Endocrinol Metab, 2010, 21(1): 33-40.
[54] Simon TG, Bonilla H, Yan P, Chung RT, Butt AA. Atorvastatin and fluvastatin are associated with dose-dependent reductions in cirrhosis and hepatocellular carcinoma, among patients with hepatitis C virus: Results from ERCHIVES [J]. Hepatology, 2016, 64(1): 47-57.
[55] Chen CL, Uthaya Kumar DB, Punj V, Xu J, Sher L, Tahara SM, Hess S, Machida K. NANOG metabolically reprograms tumor-initiating stem-like cells through tumorigenic changes in oxidative phosphorylation and fatty acid metabolism [J]. Cell Metab, 2016, 23(1): 206-219.
[56] Uthaya Kumar DB, Chen CL, Liu JC, Feldman DE, Sher LS, French S, DiNorcia J, French SW, Naini BV, Junrungsee S, Agopian VG, Zarrinpar A, Machida K. TLR4 signaling via NANOG cooperates with STAT3 to activate twist1 and promote formation of tumor-initiating stem-like cells in livers of mice [J]. Gastroenterology, 2016, 150(3): 707-719.
[57] Wolf MJ, Adili A, Piotrowitz K, Abdullah Z, Boege Y, Stemmer K, Ringelhan M, Simonavicius N, Egger M, Wohlleber D, Lorentzen A, Einer C, Schulz S, Clavel T, Protzer U, Thiele C, Zischka H, Moch H, Tschöp M, Tumanov AV, Haller D, Unger K, Karin M, Kopf M, Knolle P, Weber A, Heikenwalder M. Metabolic activation of intrahepatic CD8+T cells and NKT cells causes nonalcoholic steatohepatitis and liver cancer via cross-talk with hepatocytes [J]. Cancer Cell, 2014, 26(4): 549-564.
[58] Ma C, Kesarwala AH, Eggert T, Medina-Echeverz J, Kleiner DE, Jin P, Stroncek DF, Terabe M, Kapoor V, ElGindi M, Han M, Thornton AM, Zhang H, Egger M, Luo J, Felsher DW, McVicar DW, Weber A, Heikenwalder M, Greten TF. NAFLD causes selective CD4+T lymphocyte loss and promotes hepatocarcinogenesis [J]. Nature, 2016, 531(7593): 253-257.
[59] Chusri P, Kumthip K, Hong J, Zhu C, Duan X, Jilg N, Fusco DN, Brisac C, Schaefer EA, Cai D, Peng LF, Maneekarn N, Lin W, Chung RT. HCV induces transforming growth factor β1 through activation of endoplasmic reticulum stress and the unfolded protein response [J]. Sci Rep, 2016, 6: 22487. doi: 10.1038/srep22487.
[60] Jee MH, Hong KY, Park JH, Lee JS, Kim HS, Lee SH, Jang SK. New mechanism of hepatic fibrogenesis: hepatitis C virus infection induces transforming growth factor beta 1 production through glucose-regulated protein 94 [J]. J Virol, 2016, 90(6): 3044-3055.
[61] Pellicoro A, Ramachandran P, Iredale JP, Fallowfield JA. Liver fibrosis and repair: immune regulation of wound healing in a solid organ [J]. Nat Rev Immunol, 2014, 14(3): 181-194.
[62] Stauffer JK, Scarzello AJ, Jiang Q, Wiltrout RH. Chronic inflammation, immune escape, and oncogenesis in the liver: a unique neighborhood for novel intersections [J]. Hepatology, 2012, 56(4): 1567-1574.
[63] Messerini L, Novelli L, Comin CE. Microvessel density and clinicopathological characteristics in hepatitis C virus and hepatitis B virus related hepatocellular carcinoma [J]. J Clin Pathol, 2004, 57(8): 867-871.
[64] Zhu C, Liu X, Wang S, Yan X, Tang Z, Wu K, Li Y, Liu F. Hepatitis C virus core protein induces hypoxia-inducible factor 1α-mediated vascular endothelial growth factor expression in Huh7.5.1 cells [J]. Mol Med Rep, 2014, 9(5): 2010-2014.
[65] Yvamoto EY, Ferreira RF, Nogueira V, Pinhe MA, Tenani GD, Andrade JG, Baitello ME, Gregório ML, Fucuta PS, Silva RF, Souza DR, Silva RC. Influence of vascular endothelial growth factor and alpha-fetoprotein on hepatocellular carcinoma [J]. Genet Mol Res, 2015, 14(4): 17453-17462.
[66] Li Y, Chen J, Wu C, Wang L, Lu M, Chen X. Hepatitis B virus/hepatitis C virus upregulate angiopoietin-2 expression through mitogen-activated protein kinase pathway [J]. Hepatol Res, 2010, 40(10): 1022-1033.
[67] Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions [J]. Nat Rev Mol Cell Biol, 2006, 7(2): 131-142.
[68] Akkari L, Grégoire D, Floc’h N, Moreau M, Hernandez C, Simonin Y, Rosenberg AR, Lassus P, Hibner U. Hepatitis C viral protein NS5A induces EMT and participates in oncogenic transformation of primary hepatocyte precursors [J]. J Hepatol, 2012, 57(5): 1021-1028.
[69] Wilson GK, Brimacombe CL, Rowe IA, Reynolds GM, Fletcher NF, Stamataki Z, Bhogal RH, Simões ML, Ashcroft M, Afford SC, Mitry RR, Dhawan A, Mee CJ, Hübscher SG, Balfe P, McKeating JA. A dual role for hypoxia inducible factor-1α in the hepatitis C virus lifecycle and hepatoma migration [J]. J Hepatol, 2012, 56(4): 803-809.
[70] Shibata T, Aburatani H. Exploration of liver cancer genomes [J]. Nat Rev Gastroenterol Hepatol, 2014, 11(6): 340-349.
[71] Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, Arai Y, Takahashi H, Shirakihara T, Nagasaki M, Shibuya T, Nakano K, Watanabe-Makino K, Tanaka H, Nakamura H, Kusuda J, Ojima H, Shimada K, Okusaka T, Ueno M, Shigekawa Y, Kawakami Y, Arihiro K, Ohdan H, Gotoh K, Ishikawa O, Ariizumi S, Yamamoto M, Yamada T, Chayama K, Kosuge T, Yamaue H, Kamatani N, Miyano S, Nakagama H, Nakamura Y, Tsunoda T, Shibata T, Nakagawa H. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators [J]. Nat Genet, 2012, 44(7): 760-764.
[72] Matsuura K, Tanaka Y. Host genetic variants influencing the clinical course of hepatitis C virus infection [J]. J Med Virol, 2016, 88(2): 185-195.
[73] McFarland AP, Horner SM, Jarret A, Joslyn RC, Bindewald E, Shapiro BA, Delker DA, Hagedorn CH, Carrington M, Gale M Jr, Savan R. The favorable IFNL3 genotype escapes mRNA decay mediated by AU-rich elements and hepatitis C virus-induced microRNAs [J]. Nat Immunol, 2014, 15(1): 72-79.
[74] Chang KC, Tseng PL, Wu YY, Hung HC, Huang CM, Lu SN, Wang JH, Lee CM, Chen CH, Tsai MC, Yen YH, Lin MT, Wu CK, Huang CC, Chen HH, Hu TH. A polymorphism in interferon L3 is an independent risk factor for development of hepatocellular carcinoma after treatment of hepatitis C virus infection [J]. Clin Gastroenterol Hepatol, 2015, 13(5): 1017-1024.
[75] Aroucha DC, Carmo RF, Vasconcelos LR, Lima RE, Mendonça TF, Arnez LE, Cavalcanti Mdo S, Muniz MT, Aroucha ML, Siqueira ER, Pereira LB, Moura P, Pereira LM, Coêlho MR. TNF-α and IL-10 polymorphisms increase the risk to hepatocellular carcinoma in HCV infected individuals [J]. J Med Virol, 2016, 88(9): 1587-1595.
[76] Labib HA, Ahmed HS, Shalaby SM, Wahab EA, Hamed EF. Genetic polymorphism of IL-23R influences susceptibility to HCV-related hepatocellular carcinoma [J]. Cell Immunol, 2015, 294(1): 21-24.
[77] Miki D, Ochi H, Hayes CN, Abe H, Yoshima T, Aikata H, Ikeda K, Kumada H, Toyota J, Morizono T, Tsunoda T, Kubo M, Nakamura Y, Kamatani N, Chayama K. Variation in the DEPDC5 locus is associated with progression to hepatocellular carcinoma in chronic hepatitis C virus carriers [J]. Nat Genet, 2011, 43(8): 797-800.
[78] Lange CM, Bibert S, Dufour JF, Cellerai C, Cerny A, Heim MH, Kaiser L, Malinverni R, Müllhaupt B, Negro F, Semela D, Moradpour D, Kutalik Z, Bochud PY; Swiss Hepatitis C Cohort Study Group. Comparative genetic analyses point to HCP5 as susceptibility locus for HCV-associated hepatocellular carcinoma [J]. J Hepatol, 2013, 59(3): 504-509.
[79] Sharma SD. Hepatitis C virus 1b viral factors (core, NS3, and NS5A) and increased risk of hepatocellular carcinoma [J]. Hepatology, 2013, 58(2): 491-493.
[80] Korba B, Shetty K, Medvedev A, Viswanathan P, Varghese R, Zhou B, Roy R, Makambi K, Ressom H, Loffredo CA. Hepatitis C virus genotype 1a core gene nucleotide patterns associated with hepatocellular carcinoma risk [J]. J Gen Virol, 2015, 96(9): 2928-2937.
[81] Harouaka D, Engle RE, Wollenberg K, Diaz G, Tice AB, Zamboni F, Govindarajan S, Alter H, Kleiner DE, Farci P. Diminished viral replication and compartmentalization of hepatitis C virus in hepatocellular carcinoma tissue [J]. Proc Natl Acad Sci USA, 2016, 113(5): 1375-1380.
[82] Au SL, Wong CC, Lee JM, Fan DN, Tsang FH, Ng IO, Wong CM. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis [J]. Hepatology, 2012, 56(2): 622-631.
[83] Duong FH, Christen V, Lin S, Heim MH. Hepatitis C virus-induced up-regulation of protein phosphatase 2A inhibits histone modification and DNA damage repair [J]. Hepatology, 2010, 51(3): 741-751.
[84] Kataoka J, Shiraha H, Horiguchi S, Sawahara H, Uchida D, Nagahara T, Iwamuro M, Morimoto H, Takeuchi Y, Kuwaki K, Onishi H, Nakamura S, Takaki A, Nouso K, Yagi T, Yamamoto K, Okada H. Loss of Runt-related transcription factor 3 induces resistance to 5-fluorouracil and cisplatin in hepatocellular carcinoma [J]. Oncol Rep, 2016, 35(5): 2576-2582.
[85] Inokawa Y, Inaoka K, Sonohara F, Hayashi M, Kanda M, Nomoto S. Molecular alterations in the carcinogenesis and progression of hepatocellular carcinoma: Tumor factors and background liver factors [J]. Oncol Lett, 2016, 12(5): 3662-3668.
[86] Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA [J]. Science, 2005, 309(5740): 1577-1581.
[87] Thurnherr T, Mah WC, Lei Z, Jin Y, Rozen SG, Lee CG. Differentially expressed miRNAs in hepatocellular carcinoma target genes in the genetic information processing and metabolism pathways [J]. Sci Rep, 2016, 6:20065. doi: 10.1038/srep20065.
[88] Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease [J]. J Pathol, 2010, 220(2): 126-139.
[89] Huang JL, Zheng L, Hu YW, Wang Q. Characteristics of long non-coding RNA and its relation to hepatocellular carcinoma [J]. Carcinogenesis, 2014, 35(3): 507-514.
[90] Yang X, Xie X, Xiao YF, Xie R, Hu CJ, Tang B, Li BS, Yang SM. The emergence of long non-coding RNAs in the tumorigenesis of hepatocellular carcinoma [J]. Cancer Lett, 2015, 360(2): 119-124.
[91] Zhang H, Zhu C, Zhao Y, Li M, Wu L, Yang X, Wan X, Wang A, Zhang MQ, Sang X, Zhao H. Long non-coding RNA expression profiles of hepatitis C virus-related dysplasia and hepatocellular carcinoma [J]. Oncotarget, 2015, 6(41): 43770-43778.
[92] Kamel MM, Matboli M, Sallam M, Montasser IF, Saad AS, El-Tawdi AH. Investigation of long noncoding RNAs expression profile as potential serum biomarkers in patients with hepatocellular carcinoma [J]. Transl Res, 2016, 168: 134-145.
[93] Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency [J]. Nature, 2013, 495(7441): 333-338.
[94] Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular carcinoma development [J]. Medicine (Baltimore), 2016, 95(22): e3811.
[95] Xu L, Zhang M, Zheng X, Yi P, Lan C, Xu M. The circular RNA ciRS-7 (Cdr1as) acts as a risk factor of hepatic microvascular invasion in hepatocellular carcinoma [J]. J Cancer Res Clin Oncol, 2017, 143(1): 17-27.
[96] Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: A circular RNA and potential novel biomarker for hepatocellular carcinoma [J]. Cancer Biomark, 2016, 16(1): 161-169.
[97] Reig M, Mario Z, Perelló C, Iarrairaegui M, Ribeiro A, Lens S, Díaz A, Vilana R, Darnell A, Varela M, Sangro B, Calleja JL, Forns X, Bruix J. Unexpected high rate of early tumor recurrence in patients with HCV-related HCC undergoing interferon-free therapy [J]. J Hepatol, 2016, 65(4): 719-726.
. REN Hao, E-mail: hren2013@139.com
MolecularmechanismsofhepatitisCvirus-inducedhepatocellularcarcinoma
GAO Rong, XU Li, XU Jie, REN Hao
DepartmentofMicrobiology,ShanghaiKeyLaboratoryofMedicalBiodefense,TheSecondMilitaryMedicalUniversity,Shanghai200433,China
Hepatitis C virus (HCV) is a leading cause of hepatocellular carcinoma (HCC). HCV-induced hepatocarcinogenesis is a sequential process caused by viral factors and (or) chronic host inflammation status. Host genetic variation is now emerging as an additional element that contributes to one’s risk of developing HCC. Therefore, a comprehensive understanding of the molecular mechanisms for HCV-induced HCC will help to solve this problem.
Hepatitis C virus; Hepatocellular carcinoma; Molecular mechanism
国家自然科学基金(31370196)
任浩
2017-04-11)

