PARP3生物学功能研究进展
王丽,季鸣,陈晓光
(中国医学科学院/北京协和医学院 药物研究所,北京 100730)
PARP3生物学功能研究进展
(中国医学科学院/北京协和医学院 药物研究所,北京 100730)
PARP3属于聚(ADP-核糖)聚合酶[Poly(ADP-ribose) polymerase,PARP]超家族,与PARP1、2有极高的同源性,均具有DNA依赖的ADP-核糖基团转移活性,但其在组织分布、生物学功能方面却又表现出极大不同。目前PARP1、2研究已较为深入,而PARP3的研究尚处于起步阶段,已有研究证明其可被双链断裂DNA及单链断裂哑铃型DNA所激活并对靶蛋白及自身进行MAR修饰,在DNA单链断裂、双链断裂、PARP1活化、神经系统和体液免疫等方面发挥作用,且与胶质瘤、乳腺癌存在相关性,但具体机制尚不明确。本文旨在从结构、活化、功能及与疾病的关系4个方面对PARP3的研究现状进行总结。
PARP3;结构;激活;功能;肿瘤
聚(ADP-核糖)聚合酶[Poly(ADP-ribose) polymerase,PARP]蛋白家族是一类ADP-核糖基转移酶,以NAD+为底物,将带负电荷的ADP-核糖基团转移至靶蛋白,对蛋白质进行翻译后修饰,从而调控一系列细胞功能,如DNA修复、转录调节、RNA干扰、影响线粒体功能等[1]。在PARP蛋白家族的17个成员中,只有PARP1、PARP2、PARP3具有DNA依赖的ADP-核糖基团转移活性,提示其在DNA损伤应答中发挥着一定的作用[2]。这3种酶通过结合损伤的DNA对自身进行ADP-核糖基化修饰或对其他靶蛋白进行ADP-核糖基化修饰[3]。目前PARP1、PARP2的研究已经较为深入,研究表明,它们可催化靶标蛋白的PAR修饰,主要进行DNA损伤修复的调节、细胞死亡及转录的调节,进而在多种急慢性疾病尤其是肿瘤中发挥着极其重要的作用,其抑制剂Olaparib已经上市用于卵巢癌治疗[4-5]。而蛋白质组学研究表明,PARP3存在于包含DNA-蛋白激酶、PARP-1、DNA连接酶III、DNA连接酶IV、Ku70蛋白和Ku80蛋白的免疫复合物中[6]。由于PARP3与PARP1、2蛋白同源性高,预示PARP3在DNA损伤修复、转录调节等过程中也可能起到重要作用。然而当PARP1表达被抑制后,PARP2的表达会代偿性增加,但PARP3却不会在PARP1、2表达受到抑制后代偿性增加[7-10]。此外,PARP1与PARP2在哺乳动物的组织分布非常相似,但PARP3的组织分布却与2者区别很大[7],这些都提示PARP3具有独特的生物学作用。本文旨在从结构、活化、功能及与疾病的联系等4个方面对PARP3的研究现状进行总结。
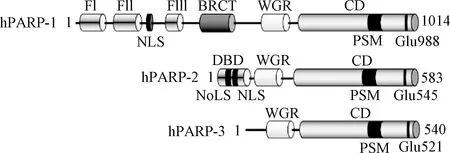
图1 PARP1、PARP2和PARP3结构Fig.1 Structures of PARP1, PARP2 and PARP3
1 PARP3的结构
PARP3由3个结构域组成:一个特殊的N端结构域(NTR)、一个中间的WGR结构域和一个含有螺旋区域的C端催化结构域(CD)[11]。PARP3 WGR结构域和CD结构域与PARP1有很高的同源性,而 NTR区域与PARP1相差较远。PARP1的 NTR区域中含有3个锌指结构和一个BRCT区域,在结合DNA和传递信息至催化区域方面发挥着重要功能,而PARP3 NTR区域只有40个氨基酸[12-14]。研究证明,若PARP1丧失了 NTR区域,则其DNA结合能力以及DNA依赖的催化活性基本完全丧失,而PARP3失去 NTR区域后,其DNA结合能力以及DNA依赖的ADP-核糖基化活性会有降低,但不会完全丧失,表明NTR区域对于PARP3的催化活性不是必需的,而WGR区域对于PARP3的催化活性则是必需的,PARP3的WGR区域可结合DNA并将信息传递至CD区域,具有PARP1 NTR区域所具有的部分功能[15]。同样, Grundy等[16]实验结果表明,人源PARP3的WGR区域若发生突变,就会丧失在DNA损伤部位聚集及加速DNA断裂修复的功能。此外,PARP3自身也存在其他翻译后修饰,包括其N端α氨基酸的甲基化和461位丝氨酸(S461)的磷酸化,其中S461的磷酸化在PARP3介导的DNA双链断裂修复发挥重要作用[17]。
2 PARP3的激活及修饰
PARP3近几年来被证实可被双链断裂DNA及含缺口的寡聚核苷酸所激活[18-21],而Langelier等[15]证明PARP-3可选择性的被含有5’磷酸基团的断裂DNA所激活,其中平末端双链DNA中间含单一缺口或由单链DNA所形成的中间部位含有单一缺口哑铃型结构可使PARP3的活性达到最高水平。粘末端即使含有5’磷酸基团,其激活能力也明显弱于平末端或哑铃型结构。
关于PARP3究竟具有单腺苷二磷酸核糖基化(MonoADP-ribose,MAR)活性还是聚腺苷二磷酸核糖基化(PolyADP-ribose,PAR)活性这一问题目前颇有争议。Sejal等[22]用多种水解酶及化学方法水解所形成的DP-核酸基团,通过TLC和高分辨率丙烯酰胺测序凝胶的方法证实了PARP3对于自身修饰表现出MAR活性,而Stuart将PARP3修饰的组蛋白进行了ADP-核酸基团的分离后通过丙烯酰胺测序凝胶的方法证实了PARP3对组蛋白的修饰主要表现为MAR活性,但也可形成少量的低分子量PAR(最多15个ADP-核糖基团)[21]。PARP1与PARP2对蛋白的翻译后修饰是形成PAR,而PARP3主要形成MAR,由此推测,PARP的功能与PARP1、PARP2可能有较大差距。PAR的功能涉及细胞分裂[23-25]、转录调节[26]、蛋白降解[27]和细胞应激反应[28-31]等多个方面,而MAR的功能研究则较为匮乏。有研究表明,外源性白喉毒素可通过在EEF2的白喉酰胺上添加一个ADP-核糖基化从而抑制RNA的转录[32]。提示MAR也可影响蛋白-蛋白相互作用以及蛋白-核酸的相互作用。
3 PARP3的功能
3.1 双链断裂修复 DNA双链断裂修复对于维持细胞的基因稳定性至关重要[33],真核细胞主要通过同源重组、经典非同源末端重组和替代非同源末端重组这几种方式进行DNA双链断裂的修复[34],几种方式之间相互竞争且各有特点,DNA末端剪切可使细胞倾向于同源重组及易于出错的替代非同源末端重组这2个途径[35]。PARP3可使Ku80ADP-核糖基化,限制DNA末端剪切使细胞进行经典非同源末端重组[18]。敲除PARP3后,Ku80在激光导致的DNA损伤处聚积减少,使BRCA1 和53BP1之间的平衡更倾向于BRCA1[18],导致DNA末端剪切增多[36-41],经典非同源末端重组减少。有趣的是,敲除PARP3虽增加了DNA末端剪切,却同时抑制了同源重组,只促进易于导致删除突变的替代非同源末端重组,其中机制尚待进一步探索与阐明。除了在修复方式的选择环节起作用外,PARP3在经典非同源末端重组的修复过程中也发挥着重要的作用。研究表明,当激光导致细胞DNA损伤后,PARP3可被招募至损伤处,而敲除PARP3后导致DNA双链损伤修复的缺陷及DNA损伤相关γ-H2AX灶点消失的延迟[21]。敲除PARP3后加入野生型的PARP3会使DNA双链损伤修复功能恢复,但加入无催化活性的PARP3则无此效果[21]。此外,PARP3也可促进γ射线或基因损伤药物、化疗药物所导致的细胞DNA双链断裂的修复[21]。PARP3被DNA双链断裂损伤激活后,引起其对自身和APLF蛋白进行ADP-核糖基化,并招募APLF至DNA损伤处,过量表达APLF可使PARP3缺失细胞中γ-H2AX灶点的消失速度恢复正常,进而表明PARP3可通过增加细胞内的APLF来促进细胞中的DNA双链断裂损伤修复[21]。而APLF作为DNA双链断裂修复的一个核心蛋白,其与非同源末端重组的核心因子XRCC4和Ku80可产生直接的相互作用[42]。APLF和染色质结合后一方面可使染色质的结构改变易于进行DNA损伤修复[21],另一方面可促进XRCC4-LigIV复合体在DNA损伤处的滞留,过量表达XRCC4-LigIV复合体可促进PARP3和APLF双敲除细胞中的DNA双链损伤修复,提示PARP3和APLF可通过增加XRCC4-LigIV复合体在染色质损伤处的聚集来促进DNA双链损伤的非同源末端重组[21]。虽然目前研究已证明PARP3可促进DNA双链损伤的非同源末端重组,但PARP3与XRCC4-LigIV复合体及APLF与XRCC4-LigIV复合体之间的具体作用关系还有待探讨。
3.2 单链断裂修复 染色体单链断裂(SSBs)是细胞最常见的DNA损伤,每个细胞平均每天都会出现成千上万次染色体单链断裂[43-44]。很大一部分SSBs被ADP-核糖基转移酶所监测并催化形成PAR或MAR,进而进行修复[45-46]。已有研究表明,PARP1、PARP2在单链断裂修复发挥重要作用[47-49],研究表明,在鸡DT40细胞中,PARP3可促进染色体单链断裂修复,其与PARP1均可作为SSB的检测器,但其识别的DNA断裂种类不同,作用方式和靶标也不相同。PARP1可识别多种DNA断裂,而PARP3只对含有5’-磷酸末端和3’-羟基末端的DNA断裂较为敏感;PARP1活化后通过使自身及组蛋白H1形成PAR发挥作用[50],而PARP3则是通过使自身及组蛋白H2BE2形成MAR发挥作用[16]。PARP1和PARP3均可在单链断裂修复中发挥重要作用,识别不同单链断裂,激活不同靶标,2者是否会有空间与时间上的关联与区分呢?比如,2者是否分别识别不同发展阶段的单链断裂,是否进行不同细胞区域的单链断裂修复等。此外,PARP3在鸡DT40细胞中可促进染色体单链断裂修复,但鸡DT40不含PARP2,与人源细胞有一定区别,这些都有待于进一步研究和探讨。
3.3 激活PARP1 研究表明,PARP3和PARP1存在相互作用[51-52],PARP3可在无DNA存在的情况下激活PARP1[53]。在PAR 形成过程中,第一个ADP-核糖基化,即MAR的形成是限速步骤,故PARP3可作为PARP1的活性启动器来促进PARP1的作用[53-54]。虽然其他PARP是否也具有这一相互作用还有待研究,但具有ADP催化活性的PARP共定位的广泛存在[31],提示这种相互作用可能较为普遍,在PARP活性调节中发挥关键作用。
3.4 调控神经发生发展 Michele等[55]在人源SK-N-SH细胞中利用染色质免疫共沉淀的方法分析了PARP3与基因组的相互作用,结果表明,PARP3主要定位于发育基因,尤其是神经形成相关基因的周围。人源PARP3与起始复合体2(PRC2)中的一些PcG蛋白存在相互作用,包括EZH2、 SUZ12、RBAP46/48、YY1和 HDAC1/2[6],而这些蛋白是胚胎发生发展过程中的重要调控因子[56]。SoxE家族转录因子SOX8、SOX9、SOX1与DLX3和DLX4在神经嵴特化和前基板外胚层的分化过程中发挥着关键的作用[57-58],而染色质免疫共沉淀分析结果表明这些基因均为PARP3的靶标,提示PARP3可通过调节这些基因的表达来调控神经基板边界的特化和神经嵴的形成和多样化。脊椎动物模型斑马鱼的体内研究进一步证实了在斑马鱼胚胎发育过程中,这些基因的表达确实依赖于PARP3的表达[55]。斑马鱼早期胚胎中PARP3基因被抑制后,SOX9a、DLX3b、DLX4b和神经源性分化蛋白等的表达降低及胚胎内耳(起源于前基板外胚层)的缺失,色素沉淀(黑色素细胞来源于神经嵴细胞)的延迟进一步表明了PARP3在神经嵴和神经板边界形成过程中发挥着关键的转录调控功能。此外,成年猴的多种组织末梢神经节的神经元中均可检测到大量的PARP3表达,提示PARP3在外周神经系统的神经元中可能发挥一定的作用[59]。此外,在多发性硬化症小鼠的脊髓中可检测到PARP3表达显著上调,提示PARP3在中枢神经系统也可能发挥一定的作用[60]。
3.5 其他 PARP3也与细胞周期有关,过表达PARP3不会影响中心体的复制,但会造成细胞G1/S期阻滞[61]。PARP3还可负性调控免疫球蛋白转换重组(class switch recombination,CSR)[62],PARP3的缺失可增强B细胞的转换重组,从而加强B细胞应对感染等所产生抗体的多样性。PARP3也可抑制端粒酶的活性,在一些肿瘤细胞中抑制PARP3后会导致端粒酶活性的增强从而促进端粒的维持,减少基因的不稳定性[63]。PARP3的功能及机制尚待进一步探讨及挖掘。
4 PARP3与肿瘤的关系
4.1 胶质瘤 胶质瘤患者5年生存率为20%,而恶性胶质瘤,作为一种高侵袭、高转移且易复发的脑部肿瘤,其患者5年生存率只有不到3%[64]。目前临床治疗包括手术、化疗、放疗等多种方法,然而患者生存期依然非常短,中位生存期仅为12~15个月[64]。放疗是一种传统有效的治疗方法,但随之产生的放疗抵抗却非常棘手[65-66],近期研究表明,PARP3有可能是克服化疗抵抗的一个潜在靶点[67]。恶性胶质瘤患者肿瘤组织中PARP3表达显著高于正常组织,如若敲除恶性胶质瘤细胞中PARP3基因则会抑制其在体内、体外的生长速度,且对放疗更加敏感,这可能与PARP3参与细胞DNA损伤修复有关。机制研究表明,敲除PARP3可抑制FOXM1的活性从而增强恶性胶质瘤细胞的放疗敏感性,但其下游通路及具体机制尚待阐明,PARP3是否还通过其他途径抑制胶质瘤细胞生长和克服放疗抵抗也值得探索。
4.2 乳腺癌 Ivan等[68]检测了443例单侧侵袭性乳腺癌患者肿瘤组织中PARP3的mRNA水平,并将其与正常乳腺组织中PARP3 mRNA含量相比较,结果发现,PARP3在肿瘤组织中的mRNA含量较正常组织减少,有10.4%的乳腺癌患者肿瘤组织中PARP3表达低;并且PARP3表达低多出现在雌激素受体及孕激素受体阴性的患者中,在这类患者中,低表达PARP3比例为25.7%,而在雌激素受体或孕激素受体阳性的患者中,该比例为5.4%。此外,低表达PARP3更常见于SBR(Scarff-Bloom -Richardson)分级标准中的III期乳腺癌患者(20.1%),而I期乳腺癌患者中的比例为1.9%[68]。以上结果提示,低表达PARP3与乳腺癌恶性程度相关,但其因果关系是否存在还需进一步研究。此外,低表达PARP3常见于正常表达PARP1的乳腺癌患者(13.4%),少见于高表达PARP1的乳腺癌患者(4.6%)。PARP3可促进DNA双链修复的非同源末端重组途径,而PARP1可通过抑制Ku蛋白与损伤DNA的结合从而抑制非同源末端重组途径[69],PARP3与PARP1之间表达的互斥现象提示,乳腺癌的DNA双链断裂修复主要依赖于同源重组。
5 结语
PARP3与PARP1、PARP 2具有极高的同源性,均可被断裂DNA所激活,在DNA损伤修复、转录调节等过程中起到重要作用,但其具体发挥作用又有极大不同。PARP3在神经系统发生发展、体液免疫抗体多样化、细胞周期及端粒酶调控方面也扮演着重要的角色,且与胶质瘤、乳腺癌存在一定相关性,提示PARP3可作为潜在的抗肿瘤靶点。然而PARP3的研究仍处于起步阶段,其分子作用机制、生物学功能以及在疾病发生发展中的作用亟待深入探索。本实验室目前正在建立PARP3的酶学筛选方法,用于选择性的PARP3抑制剂活性筛选,期待利用抑制剂对PARP3的作用机制进行深入研究。
[1] Robert I,Karicheva O,Reina San MB,et al.Functional aspects of PARylation in induced and programmed DNA repair processes:preserving genome integrity and modulating physiological events[J].Mol Aspects Med,2013,34(6):1138-1152.
[2] De Vos M,Schreiber V,Dantzer F.The diverse roles and clinical relevance of PARPs in DNA damage repair:current state of the art[J].Biochem Pharmacol,2012,84(2):137-146.
[3] Ali SO,Khan FA,Galindo-Campos MA,et al.Understanding specific functions of PARP-2:new lessons for cancer therapy[J].Am J Cancer Res,2016,6(9):1842-1863.
[4] Baldwin P,Van De Ven AL,Seitzer N,et al.Nanoformulation of the PARP Inhibitor Olaparib Enables Radiosensitization of a Radiation-Resistant Prostate Cancer Model[J].Int J Radiat Oncol Biol Phys,2016,96(2S):E595.
[5] McCormick A,Swaisland H.In vitro assessment of the roles of drug transporters in the disposition and drug-drug interaction potential of olaparib[J].Xenobiotica,2016:1-47.
[6] Rouleau M,McDonald D,Gagne P,et al.PARP-3 associates with polycomb group bodies and with components of the DNA damage repair machinery[J].J Cell Biochem,2007,100(2):385-401.
[7] Beck C,Robert I,Reina-San-Martin B,et al.Poly(ADP-ribose) polymerases in double-strand break repair:focus on PARP1,PARP2 and PARP3[J].Exp Cell Res,2014,329(1):18-25.
[8] Ghosh R,Roy S,Kamyab J,et al.Common and unique genetic interactions of the poly(ADP-ribose) polymerases PARP1 and PARP2 with DNA double-strand break repair pathways[J].DNA Repair (Amst),2016,45:56-62.
[9] Sukhanova MV,Abrakhi S,Joshi V,et al.Single molecule detection of PARP1 and PARP2 interaction with DNA strand breaks and their poly(ADP-ribosyl)ation using high-resolution AFM imaging[J].Nucleic Acids Res,2016,44(6):e60.
[10] Tulin AV.Poly (ADP-ribose) polymerase :methods and protocols[M].New York,Humana Press,2011:521.
[11] Lehtio L,Jemth AS,Collins R,et al.Structural basis for inhibitor specificity in human poly(ADP-ribose) polymerase-3[J].J Med Chem,2009,52(52):3108-3111.
[12] Eustermann S,Videler H,Yang JC,et al.The DNA-binding domain of human PARP-1 interacts with DNA single-strand breaks as a monomer through its second zinc finger[J].J Mol Biol,2011,407(1-4):149-170.
[13] Langelier MF,Planck JL,Roy S,et al.Crystal structures of poly(ADP-ribose) polymerase-1 (PARP-1) zinc fingers bound to DNA:structural and functional insights into DNA-dependent PARP-1 activity[J].J Biol Chem,2011,286(12):10690-10701.
[14] Lonskaya I,Potaman VN,Shlyakhtenko LS,et al.Regulation of poly(ADP-ribose) polymerase-1 by DNA structure-specific binding[J].J Biol Chem,2005,280(17):17076-17083.
[15] Langelier MF,Riccio AA,Pascal JM.PARP-2 and PARP-3 are selectively activated by 5’ phosphorylated DNA breaks through an allosteric regulatory mechanism shared with PARP-1[J].Nucleic Acids Res,2014,42(12):7762-7775.
[16] Grundy GJ,Polo LM,Zeng Z,et al.PARP3 is a sensor of nicked nucleosomes and monoribosylates histone H2B(Glu2)[J].Nat Commun,2016,7:12404.
[17] Dai X,Rulten SL,You C,et al.Identification and Functional Characterizations of N-Terminal alpha-N-Methylation and Phosphorylation of Serine 461 in Human Poly(ADP-ribose) Polymerase 3[J].J Proteome Res,2015,14(6):2575-2582.
[18] Beck C,Boehler C,Guirouilh Barbat J,et al.PARP3 affects the relative contribution of homologous recombination and nonhomologous end-joining pathways[J].Nucleic Acids Res,2014,42(9):5616-5632.
[19] Boehler C,Gauthier LR,Mortusewicz O,et al.Poly(ADP-ribose) polymerase 3 (PARP3),a newcomer in cellular response to DNA damage and mitotic progression[J].Proc Natl Acad Sci U S A,2011,108(7):2783-2788.
[20] Lindgren AE,Karlberg T,Thorsell AG,et al.PARP inhibitor with selectivity toward ADP-ribosyltransferase ARTD3/PARP3[J].ACS Chem Biol,2013,8(8):1698-1703.
[21] Rulten SL,Fisher AE,Robert I,et al.PARP-3 and APLF function together to accelerate nonhomologous end-joining[J].Mol Cell,2011,41(1):33-45.
[22] Vyas S,Matic I,Uchima L,et al.Family-wide analysis of poly(ADP-ribose) polymerase activity[J].Nat Commun,2014,5(65):121-132.
[23] Chang P,Coughlin M,Mitchison TJ.Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function[J].Nat Cell Biol,2005,7(11):1133-1139.
[24] Ha GH,Kim HS,Go H,et al.Tankyrase-1 function at telomeres and during mitosis is regulated by Polo-like kinase-1-mediated phosphorylation[J].Cell Death Differ,2012,19(2):321-332.
[25] Ozaki Y,Matsui H,Asou H,et al.Poly-ADP ribosylation of Miki by tankyrase-1 promotes centrosome maturation[J].Mol Cell,2012,47(5):694-706.
[26] Ji Y,Tulin AV.The roles of PARP1 in gene control and cell differentiation[J].Curr Opin Genet Dev,2010,20(5):512-518.
[27] Huang SM,Mishina YM,Liu S,et al.Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling[J].Nature,2009,461(7264):614-620.
[28] Malanga M,Althaus FR.The role of poly(ADP-ribose) in the DNA damage signaling network[J].Biochem Cell Biol,2005,83(3):354-364.
[29] Petesch SJ,Lis JT.Activator-induced spread of poly(ADP-ribose) polymerase promotes nucleosome loss at Hsp70[J].Mol Cell,2012,45(1):64-74.
[30] Di Giammartino DC,Shi Y,Manley JL.PARP1 represses PAP and inhibits polyadenylation during heat shock[J].Mol Cell,2013,49(1):7-17.
[31] Leung AK,Vyas S,Rood JE,et al.Poly(ADP-ribose) regulates stress responses and microRNA activity in the cytoplasm[J].Mol Cell,2011,42(4):489-499.
[32] Collier RJ.Understanding the mode of action of diphtheria toxin:a perspective on progress during the 20th century[J].Toxicon,2001,39(11):1793-1803.
[33] Jackson SP,Bartek J.The DNA-damage response in human biology and disease[J].Nature,2009,461(7267):1071-1078.
[34] Chapman JR,Taylor MR,Boulton SJ.Playing the end game:DNA double-strand break repair pathway choice[J].Mol Cell,2012,47(4):497-510.
[35] Huertas P.DNA resection in eukaryotes:deciding how to fix the break[J].Nat Struct Mol Biol,2010,17(1):11-16.
[36] Barlow JH,Lisby M,Rothstein R.Differential regulation of the cellular response to DNA double-strand breaks in G1[J].Mol Cell,2008,30(1):73-85.
[37] Clerici M,Mantiero D,Guerini I,et al.The Yku70-Yku80 complex contributes to regulate double-strand break processing and checkpoint activation during the cell cycle[J].EMBO Rep,2008,9(8):810-818.
[38] Bunting SF,Callen E,Kozak ML,et al.BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair[J].Mol Cell,2012,46(2):125-135.
[39] Bunting SF,Callen E,Wong N,et al.53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks[J].Cell,2010,141(2):243-254.
[40] Bouwman P,Aly A,Escandell JM,et al.53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers[J].Nat Struct Mol Biol,2010,17(6):688-695.
[41] Cao L,Xu X,Bunting SF,et al.A selective requirement for 53BP1 in the biological response to genomic instability induced by Brca1 deficiency[J].Mol Cell,2009,35(4):534-541.
[42] Kanno S,Kuzuoka H,Sasao S,et al.A novel human AP endonuclease with conserved zinc-finger-like motifs involved in DNA strand break responses[J].EMBO J,2007,26(8):2094-2103.
[43] Caldecott KW.Single-strand break repair and genetic disease[J].Nat Rev Genet,2008,9(8):619-631.
[44] Caldecott KW.XRCC1 and DNA strand break repair[J].DNA Repair (Amst),2003,2(9):955-969.
[45] Caldecott KW.Protein ADP-ribosylation and the cellular response to DNA strand breaks[J].DNA Repair (Amst),2014,19:108-113.
[46] Hottiger MO,Hassa PO,Luscher B,et al.Toward a unified nomenclature for mammalian ADP-ribosyltransferases[J].Trends Biochem Sci,2010,35(4):208-219.
[47] Boehler C,Gauthier L,Yelamos J,et al.Phenotypic characterization of Parp-1 and Parp-2 deficient mice and cells[J].Methods Mol Biol,2011,780:313-336.
[48] El-Khamisy SF,Masutani M,Suzuki H,et al.A requirement for PARP-1 for the assembly or stability of XRCC1 nuclear foci at sites of oxidative DNA damage[J].Nucleic Acids Res,2003,31(19):5526-5533.
[49] Okano S,Lan L,Caldecott KW,et al.Spatial and temporal cellular responses to single-strand breaks in human cells[J].Mol Cell Biol,2003,23(11):3974-3981.
[50] Poirier GG,Savard P.ADP-ribosylation of pancreatic histone H1 and of other histones[J].Can J Biochem,1980,58(58):509-515.
[51] Schreiber V,Dantzer F,Ame JC,et al.Poly(ADP-ribose):novel functions for an old molecule[J].Nat Rev Mol Cell Biol,2006,7(7):517-528.
[52] Ahel I,Ahel D,Matsusaka T,et al.Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins[J].Nature,2008,451(7174):81-85.
[53] Loseva O,Jemth AS,Bryant HE,et al.PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA[J].J Biol Chem,2010,285(11):8054-8060.
[54] Mao Z,Hine C,Tian X,et al.SIRT6 promotes DNA repair under stress by activating PARP1[J].Science,2011,332(6036):1443-1446.
[55] Rouleau M,Saxena V,Rodrigue A,et al.A key role for poly(ADP-ribose) polymerase 3 in ectodermal specification and neural crest development[J].PLoS One,2011,6(1):1116-1121.
[56] Schuettengruber B,Chourrout D,Vervoort M,et al.Genome regulation by polycomb and trithorax proteins[J].Cell,2007,128(4):735-745.
[57] Yan YL,Willoughby J,Liu D,et al.A pair of Sox:distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development[J].Development,2005,132(5):1069-1083.
[58] Sauka-Spengler T,Bronner-Fraser M.A gene regulatory network orchestrates neural crest formation[J].Nat Rev Mol Cell Biol,2008,9(7):557-568.
[59] Rouleau M,El-Alfy M,Levesque MH,et al.Assessment of PARP-3 distribution in tissues of cynomolgous monkeys[J].J Histochem Cytochem,2009,57(7):675-685.
[60] Selvaraj V,Soundarapandian MM,Chechneva O,et al.PARP-1 deficiency increases the severity of disease in a mouse model of multiple sclerosis[J].J Biol Chem,2009,284(38):26070-26084.
[61] Augustin A,Spenlehauer C,Dumond H,et al.PARP-3 localizes preferentially to the daughter centriole and interferes with the G1/S cell cycle progression[J].J Cell Sci,2003,116(8):1551-1562.
[62] Robert I,Gaudot L,Rogier M,et al.Parp3 negatively regulates immunoglobulin class switch recombination[J].PLoS Genet,2015,11(5):e1005240.
[63] Fernandez-Marcelo T,Frias C,Pascua I,et al.Poly (ADP-ribose) polymerase 3 (PARP3),a potential repressor of telomerase activity[J].J Exp Clin Cancer Res,2014,33(1):1-8.
[64] Van Meir EG,Hadjipanayis CG,Norden AD,et al.Exciting new advances in neuro-oncology:the avenue to a cure for malignant glioma[J].CA Cancer J Clin,2010,60(3):166-193.
[65] Kruh GD.Introduction to resistance to anticancer agents[J].Oncogene,2003,22(47):7262-7264.
[66] Perona R,Sanchez-Perez I.Control of oncogenesis and cancer therapy resistance[J].Br J Cancer,2004,90(3):573-577.
[67] Quan JJ,Song JN,Qu JQ.PARP3 interacts with FoxM1 to confer glioblastoma cell radioresistance[J].Tumour Biol,2015,36(11):8617-8624.
[68] Bieche I,Pennaneach V,Driouch K,et al.Variations in the mRNA expression of poly(ADP-ribose) polymerases,poly(ADP-ribose) glycohydrolase and ADP-ribosylhydrolase 3 in breast tumors and impact on clinical outcome[J].Int J Cancer,2013,133(12):2791-2800.
[69] Paddock MN,Bauman AT,Higdon R,et al.Competition between PARP-1 and Ku70 control the decision between high-fidelity and mutagenic DNA repair[J].DNA Repair (Amst),2011,10(3):338-343.
(编校:吴茜)
Research progress on biological function of PARP3
WANG Li-yuan, JI Ming, CHEN Xiao-guangΔ
(Institute of Materia Medica, Chinese Academy of Medical Sciences, Peking Union Medical College, Beijing 100730, China)
As a member of PARP superfamily, PARP3 shares a high homology with PARP1 and PARP2, which are all DNA-dependent enzymes that are catalytically activated by DNA strand breaks. Compared to PARP1 and PARP2, PARP3 exerted some special properties in tissue expression pattern and biological function. The evidence has shown that PARP3 could be activated by DNA double strand breaks and special DNA single strand breaks and synthesize mono(ADP-ribose) (MAR) covalently attached to target proteins including itself. PARP3 plays an important role in DNA double strand breaks, DNA single strand breaks, activation of PARP1 and development of nervous system. It has been reported that PARP3 is associated with glioma and breast cancers. In this review, PARP3 structure, activation mechanism, biological function and its relationship with diseases will be presented.
PARP3; structure; activation; function; tumor
10.3969/j.issn.1005-1678.2016.12.004
王丽嫄,女,硕士在读,研究方向:抗肿瘤分子药理,E-mail:wangliyuan@imm.ac.cn;陈晓光,通信作者,研究员,研究方向:抗肿瘤分子药理,E-mail:chxg@imm.ac.cn。
R966
A

