Generation of monoclonalantibodies against n-3 fatty acid desaturase
Generation of monoclonalantibodies against n-3 fatty acid desaturase
Dear Editor:
Omega-3 polyunsaturated fatty acids(n-3 PUFAs) are essentialfatty acids for normal cellular functions and have been used for prevention and treatment of many diseases,including coronary heart disease,diabetes,and cancers[1-3].n-3 PUFAs and n-6 PUFAs have been shown to decrease and increase the severity of severalhuman diseases,respectively[4].Unfortunately, mammals have no enzymes to synthesize n-6 and n-3 fatty acids.Therefore,they mustrely on a dietary supply.Today,the ratio of n-6 to n-3 PUFAs has reached to an unhealthy 20-30:1 due to people taking n-6 rich diet and eating less sea fish.Caenorhabditis(C.)elegans fat-1 gene encodes an n-3 desaturase thatintroduces a double bond atn-3 position of n-6 fatty acids to form n-3 fatty acids[5].Several fat-1 transgenic mammals(including mouse,pig and cow)have been produced in which n-3 PUFAs were increased and n-6 PUFAs were reduced,resulting in a significantreduction of n-6/n-3 ratio in those transgenic animals[6-8]. However,due to lack of commercially available FAT-1 antibody,FAT-1 expression is indirectly determined by measurement of FAT-1 activity including increase of n-3 PUFAs and decrease of n-6/n-3 ratio in animals.In this study,we used recombinant FAT-1 to immunize mice and to generate FAT-1 specific monoclonalantibody.Then,FAT-1 monoclonalantibodies were used to detect FAT-1 protein in tissue samples of the liver,lung,kidney,heart,brain,and spleen of fat-1 transgenic mice by Western blotting and immunohistochemicalstaining assays.
Escherichia(E.)coli expression system allows rapid and economical production of recombinant FAT-1 protein.To express C.elegans fat-1 gene in E.coli efficiently,we optimized the codons of the full-length C.elegans fat-1 gene sequence using GeneArtⓇGene Synthesis(GeneArt,Regensburg, Germany)followed by subcloning into the expression vector pCold II.The recombinant FAT-1 protein was purified by HisTrap FF affinity chromatography col-umn(GE Healthcare Life Sciences,Piscataway,NJ, USA)and analyzed by SDS-PAGE and Coomassie brilliant blue staining(Fig.1A),which showed a protein band of approximately 46 kDa in size,corresponding to the known molecular weight of FAT-1[5].The identity of the FAT-1 recombinant protein was further confirmed by searching sequence databases using mass spectrometry data.Recombinant FAT-1 was used to generate FAT-1 monoclonal antibodies and after three rounds of subcloning,the subclone 3A11 showed the best titer.Coomassie brilliant blue staining revealed the presence of two protein bands 55 kDa and 25 kDa in size,respectively (Fig.1B).Enzyme-linked immunosorbent assay (ELISA)furthershowed thatthe FAT-1 monoclonalantibodies(3A11)between 0.05 and 100μg/mL exhibited a linear increase in OD450(Fig.1C).Further analysis revealed thatthe antibodies were IgG2a.
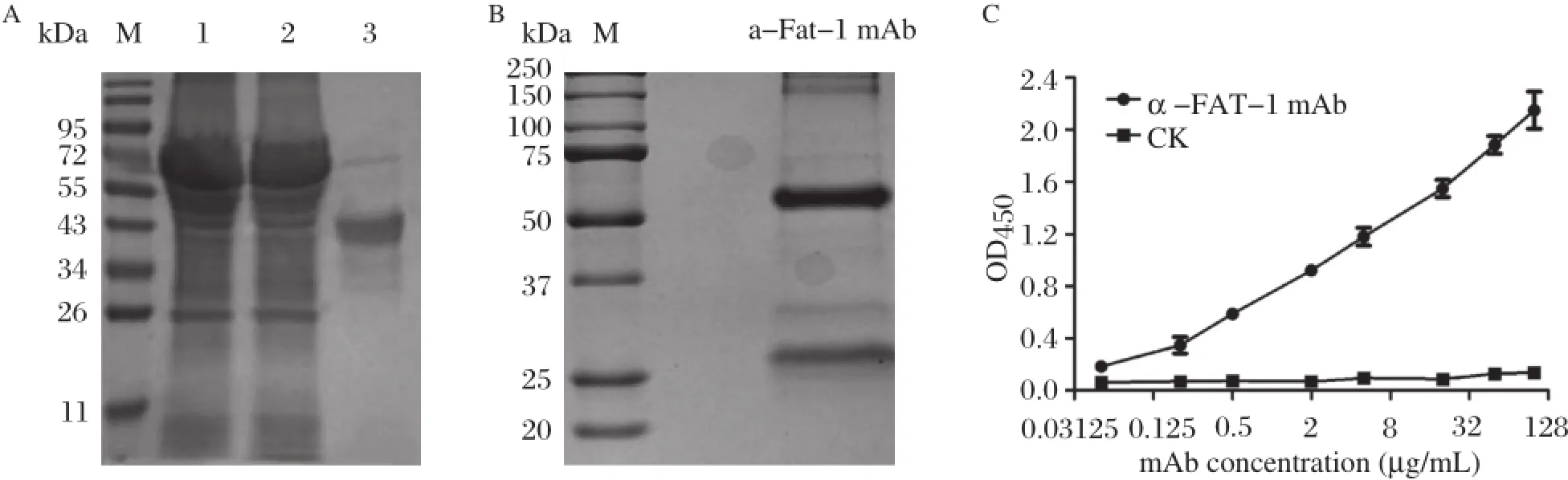
Fig.1 Preparation of FAT-1 monoclonal antibodies.A:The rFAT-1 expression in transfected bacteria.SDS-PAGE pattern after Coomassie Brilliant Blue G-250 staining ofthe transfected bacterialcellpellets(lane 1),celllysate supernatant(lane 2)and the purified rFAT-1 protein(lane 3).The purification of FAT-1 mAb.SDS-PAGE pattern after Coomassie Brilliant Blue G-250 staining of the purified 3A11 mAb concentrations displayed two protein species,the upperband was heavy chain and the lower band was lightchain.The titers of the FAT-1 mAb.The titers of the FAT-1 mAb 3A11 was tested using ELISA reactivity and pepck mAb was used as a negative control.Data representthe average absorbance at450 nm detected by rFAT-1 protein at 0.05,0.20,0.5,5.0,20.0,50.0,100.0μg/mL,the sensitivity of the mAb was 0.05μg/mL.
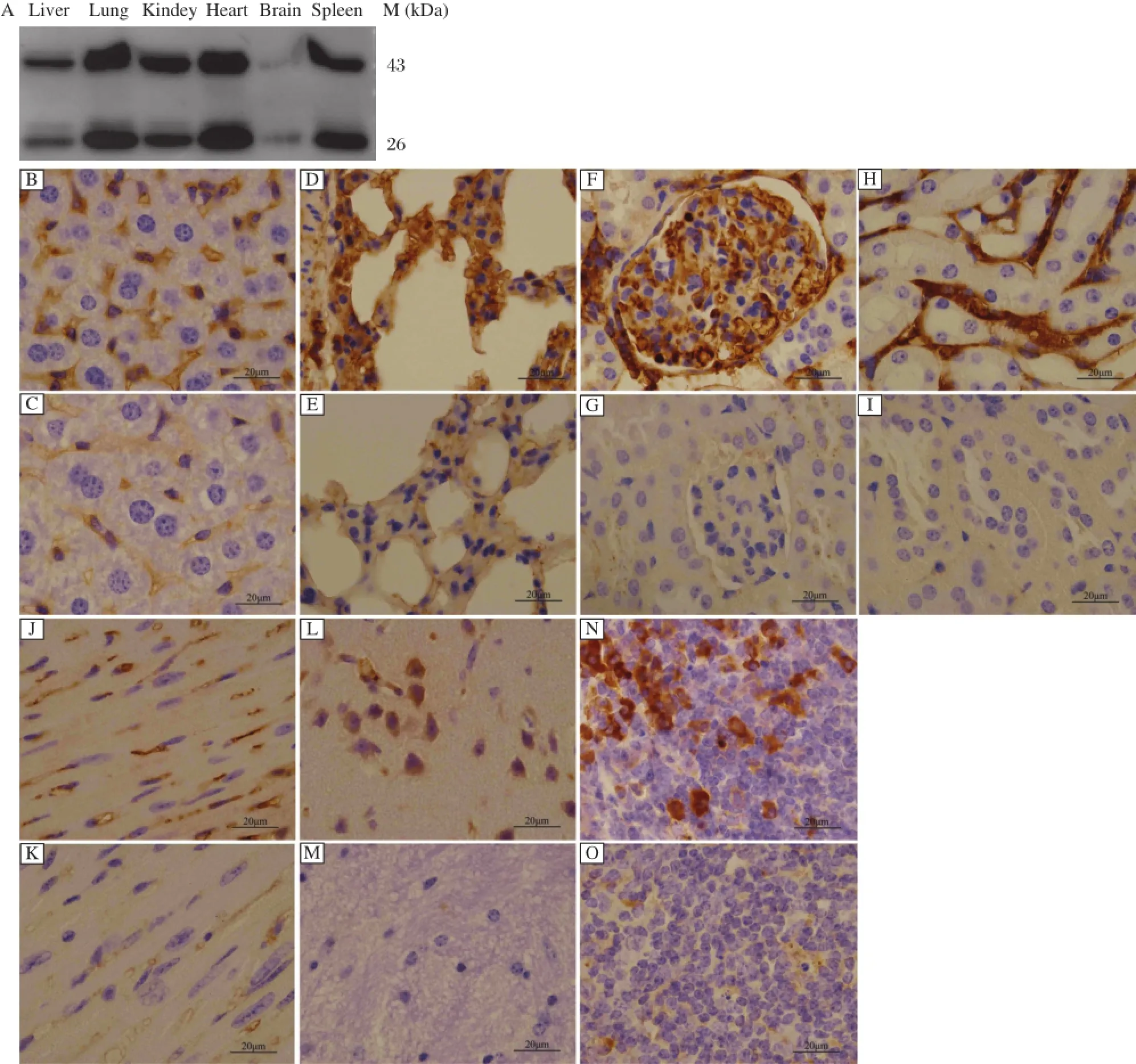
Fig.2Function of FAT-1 monoclonal antibodies.A:Detection ofthe specificity of FAT-1 mAb by Western blotting.Western blotting wasused to analyze binding of FAT-1 mAb 3A11 to denatured FAT-1 in tissues at reducing conditions.Note that the mAb clone binds with two protein species with molecular masses of about46 kD and 26 kD,the full-length FAT-1 protein and its degradation products,respectively.B-O:Determination of FAT-1 mAb’seffectivenessby Immunohistochemicalstains(Magnification×100).Immunohistochemicalstainsshowsthe protein ofFAT-1 detected in livertissue (B),alveolus cells(D),glomeruli(F),renal tubulointerstitial cells(H),myocardialcell(J),neurogliocyte,neuron(L)and,splenocyte(N).The normal C57BL/6 tissues yielded some weakly positive staining,especially in the liver tissue(C),alveolus cells(E)and myocardial cell(K).
Immunoblotting assays using FAT-1 monoclonal antibodies(3A11)demonstrated a protein band ofapproximately 46 kDa in size from the homogenates of tissue specimens ofthe liver,lung,kidney,heart,brain, and spleen of mfat-1 transgenic mice,but not from those of normal C57BL/6 mice(Fig.2A).A protein band approximately 26 kDa in size was also detected, which may be FAT-1 degradation products.FAT-1 is rich in cysteine and has three successive cysteines at positions 241-243.As the two disulfide bonds can be broken under denaturing conditions,the stability of pe ptide bo n d ma y be affec te d subseque ntly. Immunohistochemical staining of tissue specimens of transgenic and normal C57BL/6 mice further demonstrated that FAT-1 protein was present in many tissues of the transgenic mice,but was notfound in the wildtype control C57BL/6 tissues(Fig.2B-O).
The expression of fat-1 gene or FAT-1 in transgenic tissues or cells can be examined previously only by reverse transcription-polymerase chain reaction or by gas chromatography to measure the increase of n-3 PUFAs and the decrease of n-6/n-3 ratios.The generation of FAT-1 monoclonalantibodies provides a new toolto detect FAT-1 protein directly.
This work wassupported by grants from the National Natural Science Foundation of China(81202370)and Jiangsu Key Laboratory of Xenotransplantation (BM2012116).Yifan Daiis a Fellow atthe Collaborative Innovation Center For Cardiovascular Disease TranslationalMedicine.
Yours Sincerely,
Yanfei Zhang1,2,△,Yilin Yuan2,3,△,Meilei Sheng2,3, WeiZhang2,3,Ying Wang2,Xiao Zhang4,✉,Yifan Dai2,3,✉
1Departmentof Genetics,2Jiangsu Key Laboratory of Xenotransplantation,3Department of Pathology,4Key Laboratory of Antibody Technique,
Ministry of Public Health,
Nanjing MedicalUniversity,
140 Hanzhong Road,
Nanjing, Jiangsu 210029, China.
✉Corresponding author:Dr.Yifan Dai,
Tel/fax:86-025-86862085/86-025-86862076,
E-mail:daiyifan@njmu.edu.cn;
Dr.Xiao Zhang,
Tel/fax:+86-025-86863100/+86-025-86863100,
E-mail:zhangxiao@njmu.edu.cn.
△These authorscontributed equally to this work.
[1]Calder PC,Yaqoob P.Marine omega-3 fatty acids and coronary heartdisease[J].Curr Opin Cardiol,2012,27(4): 412-419.
[2]Jafari T,Fallah AA,Azadbakht L.Role of dietary n-3 polyunsaturated fatty acids in type 2 diabetes:a review of epidemiological and clinical studies[J].Maturitas, 2013,74(4):303-308.
[3]Chung H,Lee YS,Mayoral R,etal.Omega-3 fatty acids reduce obesity-induced tumor progression independentof GPR120 in a mouse model of postmenopausal breast cancer[J].Oncogene,2014,1-10.
[4]Wood KE,Lau A,Mantzioris E,et al.A low omega-6 polyun saturated fatty acid(n-6 PUFA)d iet increases omega-3(n-3)long chain PUFA status in plasma phospholipids in humans[J].Prostaglandins Leukot Essent Fatty Acids,2014,90(4):133-138.
[5]Kang ZB,Ge Y,Chen Z,et al.Adenoviral gene transfer of Caenorhabditis elegans n--3 fatty acid desaturase o ptimizes fatty acid co mpo sitio n in mammalian cells[J],Proc NatlAcad SciU S A,2001,98(7):4050-4054.
[6]Kang JX.A transgenic mouse model for gene-nutrient interactions[J].J Nutrig enet Nutrigen omics,2008, 1(4):172-177.
[7]Lai L,Kang JX,Li R,et al.Generation o f cloned transgenic pigs rich in omega-3 fatty acids[J].Nat Biotechnol,2006,24(4):435-436.
[8]Wu X,Ouyang H,Duan B,etal.Production ofcloned transg enic cow expressing omega-3 fatty acids[J].Transgenic Res,2012,21(3):537-543.
Received 02 February 2015,Accepted 15 February 2015,Epub 28 May 2015
R335,Code document:B
The authors reported no conflict of interests.
©2015 by the Journal of Biomedical Research.All rights reserved.
10.7555/JBR.29.20150020
 THE JOURNAL OF BIOMEDICAL RESEARCH2015年6期
THE JOURNAL OF BIOMEDICAL RESEARCH2015年6期
- THE JOURNAL OF BIOMEDICAL RESEARCH的其它文章
- Platelets in hemostasis and thrombosis:Novelmechanisms of fibrinogen-independent platelet aggregation and fibronectinmediated protein wave of hemostasis
- Retinolbinding protein 4 correlates with and is an early predictor of carotid atherosclerosis in type 2 diabetes mellitus patients
- CC-chemokine receptor 7 and its ligand CCL19 promote mitralvalve interstitialcellmigration and repair
- Effectofburden and origin sites ofpremature ventricular contractions on left ventricular function by 7-day Holter monitor
- Prevention ofatrialfibrillation with renin-angiotensin system inhibitors on essentialhypertensive patients:a meta-analysis of randomized controlled trials
- Epirubicin-gold nanoparticles suppress hepatocellular carcinoma xenograft growth in nude mice
