Chikungunya virus, epidemiology, clinics and phylogenesis: A review
Alessandra Lo Presti, Alessia Lai, Eleonora Cella, Gianguglielmo Zehender, Massimo Ciccozzi,3*
1Department of Infectious Parasitic and Immunomediated Diseases, Epidemiology Unit, Reference Centre on Phylogeny, Molecular Epidemiology and Microbial Evolution (FEMEM), Istituto Superiore di Sanita`, Rome, Italy
2Department of Biomedical and Clinical Sciences, L. Sacco Hospital, University of Milan, Milan, Italy
3University Campus-Biomedico, Rome, Italy
Chikungunya virus, epidemiology, clinics and phylogenesis: A review
Alessandra Lo Presti1, Alessia Lai2, Eleonora Cella1, Gianguglielmo Zehender2, Massimo Ciccozzi1,3*
1Department of Infectious Parasitic and Immunomediated Diseases, Epidemiology Unit, Reference Centre on Phylogeny, Molecular Epidemiology and Microbial Evolution (FEMEM), Istituto Superiore di Sanita`, Rome, Italy
2Department of Biomedical and Clinical Sciences, L. Sacco Hospital, University of Milan, Milan, Italy
3University Campus-Biomedico, Rome, Italy
Chikungunya virus is a mosquito-transmitted alphavirus that causes chikungunya fever, a febrile illness associated with severe arthralgia and rash. Chikungunya virus is transmitted by culicine mosquitoes; Chikungunya virus replicates in the skin, disseminates to liver, muscle, joints, lymphoid tissue and brain, presumably through the blood. Phylogenetic studies showed that the Indian Ocean and the Indian subcontinent epidemics were caused by two different introductions of distinct strains of East/Central/South African genotype of CHIKV. The paraphyletic grouping of African CHIK viruses supports the historical evidence that the virus was introduced into Asia from Africa.Phylogenetic analysis divided Chikungunya virus isolates into three distinct genotypes based on geographical origins: the first, the West Africa genotype, consisted of isolates from Senegal and Nigeria; the second contained strains from East/Central/South African genotype, while the third contained solely Asian.The most recent common ancestor for the recent epidemic, which ravaged Indian Ocean islands and Indian subcontinent in 2004-2007, was found to date in 2002. Asian lineage dated about 1952 and exhibits similar spread patterns of the recent Indian Ocean outbreak lineage, with successive epidemics detected along an eastward path. Asian group splitted into two clades: an Indian lineage and a south east lineage. Outbreaks of Chikungunya virus fever in Asia have not been associated necessarily with outbreaks in Africa. Phylogenetic tools can reconstruct geographic spread of Chikungunya virus during the epidemics wave. The good management of patients with acute Chikungunya virus infection is essential for public health in susceptible areas with current Aedes spp activity.
ARTICLE INFO
Article history:
Received 14 April 2014
Received in revised form 15 July 2014
Accepted 15 October 2014
Available online 20 December 2014
CHIKV
1. Introduction
Chikungunya virus (CHIKV) is a mosquito-transmitted alphavirus that belongs to the Togaviridae family[1]. It causes chikungunya fever (CHIK fever), a febrile illness associated with severe arthralgia and rash[2-5]. Chikungunya is a Makonde word (Bantu language) meaning ‘The one which bends up’ referring to the posture of the affected patient acquired due to excruciating pain in the joints[6].
The CHIKV is a small (about 60-70 nm-diameter), spherical, enveloped, positive-strand RNA virus[7- 9].
Its genome is about 12 kb long and is capped in 5′ and has a polyA tail in the 3′ end. The genome structure includes two open reading frames (ORFs) that encodes for two polyproteins (non-structural polyprotein and structural polyprotein), which can be cleaved respectively into four non-structural proteins (nsP1, nsP2, nsP3, nsP4) and five structural proteins (C, E3, E2, 6K, E1) by viral and cellular proteases[10].
Chikungunya virus is transmitted by culicine mosquitoes and can alternatively affects vertebrates and arthropods[11,12]. The arthropods remain infected throughout all its life. Its transmission to humans is mainly through Aedes species mosquitoes[13]. Aedes aegypti, Aedes albopictus and Aedes polynesiensis are commonly involved in the transmission although Culex has also been reported for the transmission in some cases[1,13,14].
African CHIKV circulates primarily in a sylvatic/ enzootic cycle, transmitted by arboreal primatophilic Aedes mosquitoes (eg., Aedes furcifer and Aedes africanus)and probably relies on nonhuman primates as reservoir hosts[13,15].
A recent Indian study reported transmission of chikungunya virus by Anopheles stephensi too[16]. The Indian Ocean outbreak is caused by transmission by Aedes only[1]. The common reservoirs for chikungunya virus are monkeys and other vertebrates. The role of cattles and rodents has also been reported in the transmission of the virus[13].
The CHIKV usually shows a periodicity with occurrence of disease in the community with latency intervals of 3-4 years, probably due to its cycle in monkeys[11,13]. Following transmission, CHIKV replicates in the skin, and disseminates to the liver, muscle, joints, lymphoid tissue (lymph nodes and spleen) and brain, presumably through the blood (Figure 1)[17-20].
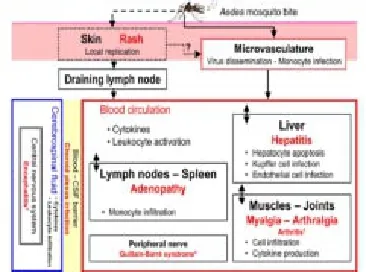
Figure 1. Virus dissemination and target organs.CHIKV spreads rapidly in the body after initial infection. Following inoculation with CHIKV through a mosquito bite, the virus directly enters the subcutaneous capillaries, with some viruses infecting susceptible cells in the skin, such as macrophages or fibroblasts and endothelial cells. Local viral replication seems to be minor and limited in time, with the locally produced virus probably being transported to secondary lymphoid organs close to the site of inoculation. Virus dissemination through the blood and pathological events associated. True arthritis remains a rare event (from 2% to 10%). (Dupuis-Maguiraga et al, 2012).
The pathological events associated with tissue infection are mostly subclinical in the liver (hepatocyte apoptosis) and lymphoid organs (adenopathy), whereas in the muscles and joints are associated with very strong pain, with some of the patients presenting arthritis (Figure 1)[19,20].
Symptoms of CHIKV infection include high fever, rigors, headache, photophobia and a petechial rash or maculopapular rash. In addition, most infected individuals complain of severe joint pain that is often incapacitating and a painful inguinal lymphadenopathy was also reported in a case study of a 28-year-old woman[21-24].
‘Silent’ infections do occur but are rare, being observed in around 15% of infected individuals[25].
The acute phase of CHIKV infection typically lasts from a few days to a couple of weeks. However, arthralgia and/ or myalgia may persist for weeks, months, or even years. Some patients go on to develop a genuine, chronic arthritic syndrome[26,27].
Typically, joint damage fluctuates over time, but always affects the same parts of the body, mostly the extremities (hands, ankles, knuckles)[20,28-30]. The mortality rate is low (0.4%), but is higher in babies less than 1 year old (2.8%) and increases in the elderly with concurrent diseases[29].
There is no specific treatment for CHIK and no vaccine is currently available. The illness is usually self-limiting and resolves with time. Supportive care with rest is indicated during the acute joint symptoms[31]. Infective persons should be protected from further mosquito exposure (staying indoors and/or under mosquito net during the first few days of illness) so that they cannot contribute to the transmission cycle[32].
Laboratory diagnosis relies upon the detection of the virus on early samples and/or specific anti-CHIKV IgM and IgG on blood samples[33]. Commercial kits are available, sometimes with excellent sensitivity and specificity[34] For example the commercial Chikungunya virus real-time reverse transcription-PCR (RT-PCR) kit, by Panning et al, 2009, was 100% sensitive and specific in comparison to a published real-time RT-PCR[34]. This commercial CHIKV kit may assist laboratories in affected regions and serve the needs of outpatient travel medicine clinics worldwide. The capability of quantifying virus RNA concentrations may facilitate the monitoring of disease progression and the assessment of risks of transmission in the nosocomial situation[35]. In addition, this kit may help in regions where CHIKV vectors Aedes aegypti and Aedes albopictus are subject to virus surveillance[34].
Anti-CHIKV antibodies can be detected in patients shortly after symptom onset, usually after 5 days for IgM and only a few days later for IgG. Commercial enzyme immunoassays and immunofluorescence assays are available[36].
Possible problems in the interpretation of the serological results could be a) possible false negativity due to CHIKV induced mixed cryoglobulinemia[37], b) cross-reactivity with viruses of the Semliki Forest serocomplex requiring seroneutralization, and c) long-term persistence of anti-CHIKV IgM months after disease onset[33]. To demonstrate a recent CHIKV infection in most cases is sufficient a synchronous testing of a sample from the acute stage and a sample collected at least 3 weeks later[33].
For diagnosis, monitoring, detection and genotyping of CHIKV, conventional reverse transcription-polymerase chain reaction (RT-PCR) methods have been used. Regarding the detection of CHIKV-RNA, it can be detected in plasma samples within the first week after symptom onset,commonly with extremely high levels of viremia[38].
Real-time quantitative RT-PCR to detect and quantify the CHIKV was also developed[39]. This method is sensitive and specific and detects a wide range of CHIKV concentrations. More recently, a positive- and negative-strand quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) assays for CHIKV nsP3 was designed for diagnosis and studying virus replication[40]. This positive- and negative-strand qRT-PCR assays had limits of quantification of 1 and 3 log10 RNA copies/reaction, respectively. Compared to a published E1 diagnostic assay using 30 laboratory-confirmed clinical samples, the positive-strand nsP3 qRT-PCR assay had higher R2and efficiency and detected more positive samples[40].
A SYBR Green I based quantitative RT-PCR assay was also recently developed[41]. This assay was found to be 10-fold more sensitive than conventional RT-PCR and no cross reactivity was observed with related alphaviruses and flaviviruses.
A novel molecular diagnostic platform that ensures a rapid and cost-effective one-step RT-PCR assay, with high sensitivity and specificity, for the early detection of the Chikungunya virus (CHIKV) was also developed[42]. It uses 2,7-diamino-1,8-naphthyridine derivative (DANP)-labeled cytosine-bulge hairpin primers to amplify the nsP2 region of the CHIKV genome, followed by measurement of the fluorescence emitted from DANP-primer complexes after PCRs.
The detection limit of othis assay was 0.01 plaque-forming units per reaction of CHIKV. An advantage of this assay could be represented by the fact that the HP-nsP2 primers were highly specific in detecting CHIKV, without any crossreactivity with the panel of RNA viruses validated in this study. The feasibility of the DANP-coupled hairpin RTPCR for clinical diagnosis was evaluated[42] using clinical serum samples from CHIKV-infected patients, and the specificity and sensitivity were 100% (95% CI, 80.0% to 100%) and 95.5% (95% CI, 75.1% to 99.8%), respectively. This Novel DANP-Coupled Hairpin RT-PCR assay should be used as a potential clinical molecular diagnostic assay for CHIKV in acute-phase patient serum samples.
2. Epidemiology and spread of CHIKV
Chikungunya fever was first reported in 1952 from Makonde plateaus, along the borders between Tanzania and Mozambique[6,43]. Chikungunya virus was first isolated by Ross in 1953 from the serum of a febrile human during an epidemic in Newala district of Tanzania[44].
The virus probably originated in Africa[1,12,45] where it maintained in ‘sylvatic cycle’ involving wild primates and forest dwelling mosquitoes[11,12,14,46].
Chikungunya was introduced subsequently in Asia where it has been transmitted from human to human mainly by Aedes aegypti and, to a lesser extent by Aedes albopictus through an urban transmission cycle[45,46].
Since Tanzania outbreak in 1952, chikungunya virus caused outbreaks (emerging and re-emerging) between the 1960s and 1990s in East Africa (Uganda)[1,47], in Zimbabwe[48], in West Africa (Senegal)[13,49], and in Central Africa (Central African Republic, Democratic Republic of the Congo and Cameroon)[11,46,48,50,51].
Chikungunya virus has also been reported from Portugal and Guinea[12]. Philippines, Malaysia, Mayotte and Reunion Island are commonly affected in Asia[52].
The chronological order of the documented outbreaks of CHIKV was described by Powers and Logue[54].
The first documented Asian outbreak was in 1958[46,54-56]. The outbreaks in Africa and Asia, were unpredictable, with interval of 7 to 20 years between two consecutive epidemics. In India there was a confirmed history of outbreaks during 1963-64 in Kolkata[57] (earlier known as Calcutta) and 1965 in Chennai (earlier known as Madras)[58,59]. The entry of chikungunya virus in India was unknown although Calcutta Sea and air roots are believed to be the probable entry
points[60].
Among the numerous large cities in South East Asia also Bangkok has been identified as a particularly active site of transmission and disease[49,61-63]. Outbreaks have been documented in Cambodia, Vietnam, Laos, Myanmar too[55]. In 2004, a large epidemic of CHIKV, sustained by the circulation of the East-Central-South African (ECSA) genotype, started on the coast of Kenya, followed in 2005 by sequential outbreaks on the Comoros, La Reunion, and other islands in the southwest Indian Ocean. On Reunion island alone there were approximately 266 000 cases (34% of the total island population)[5,64,65].
Then, an epidemic due to the same strain ravaged the Indian subcontinent in 2005- 2006, causing more than one and half million cases[66].
Interestingly, during the epidemic in La Reunion, the virus apparently mutated. The A226V mutation allowed the virus to better adapt to Aedes albopictus, the only competent vector present on the island[1]. Viral strains with the same mutation were also identified in India and caused an outbreak in North-eastern Italy[67,68]. In particular the outbreak reported in Emilia- Romagna Region (North-eastern Italy) in the summer of 2007 represented the first epidemic reported in a temperate area, probably related to the a high concentration of Aedes albopictus that affected this area[69].
Other cases were reported in Europe (UK, Belgium, Germany, Czech Republic, Norway, Spain and France), Hong Kong, Canada, Taiwan, Sri Lanka and the USA; these cases were directly associated with the return of travellers from India and affected islands of the Indian Ocean[53,70].
3. Phylogeny
Primarily phylogenetic analysis divided the CHIK virus isolates into three distinct genotypes based on geographical origins. The first clade consisted of the isolates from Senegal and Nigeria, forming the West Africa genotype (WAf). The remaining isolates formed two clades: one contained strains from central and eastern Africa (Central/East African genotype), while the other contained solely Asian isolates[71]. Genetic studies by Lanciotti et al[47] as well as the phylogenetic analyses presented by Powers[71] clearly demonstrate that ONN and CHIK viruses are genetically distinct. The paraphyletic grouping of the African CHIK viruses supports the historical evidence that the virus was introduced into Asia from Africa[45,71].
A recent study[72] investigated the CHIKV genotype in patients in Cambodia and analyzed the phylogenetic origin of the strains. The phylogenetic analysis performed by the authors showed that CHIKV was composed by three genotypes: West Africa, Asia and East/Central/South African (ECSA) (Figure 2). The viruses from Cambodia clustered with those isolated during the Indian Ocean outbreak and within the ECSA phylogenetic group (Figure 2).

Figure 2. Phylogenetic tree based on the whole genome of CHIKV.Viruses were identified by using the GenBank accession number, country code, and year of isolation. Boldface indicates strains from Cambodia; circles indicate isolates from Preah Vihear Province; triangles indicate strains from Battambang Province. All 8 strains from Cambodia carried the A226V mutation. Numbers represent the bootstrap support obtained for respective branches (>70). The tree was rooted by o’nyongnyong virus (GenBank accession no. AF079456, UGA96-ONNV). ECSA, East Central South African genotype. Scale bars indicate nucleotide substitutions per site (Duong et al, 2012).
Another recent study[15] showed that the CHIKV phylogenetic trees included three distinct CHIKV clades, namely, ECSA (East/Central/South African), West Africa, and Asian, with the recent Indian Ocean basin outbreak forming a monophyletic lineage descendant from the ECSA clade. The divergence of each distinct lineage reflected, to some extent, the path of global transmission and occasional outbreaks. The authors showed that, excluding the 5’ and 3’ 20 nucleotides (nt) that were not sequenced, the genome length varied among and within geographic lineages, with those in the ECSA lineage being shorter (11 557 to 11 789 nt) than the WAf (11 843 to 11 881 nt) and Asian (11 777 to 11 999 nt) strains.
The authors[15] also found nucleotide differences in all genes, and the most variable genome regions included the 5’ and 3’ UTRs, as well as the 26S junction region. They observed that the ORFs were highly conserved, with occasional indels observed in high-passage strains. Because of the highly divergent UTRs made accurate alignments impossible the authors excluded the UTRs from the phylogenetic analyses and found poly(A) insertions in the 3’UTRs of two ECSA strains in addition to the Ross strain[73]. The insertion in the Senegal bat strain, which is located in the ECSA group, is in a different location from the other two, suggesting their independent generation.
A maximum parsimony, neighbour joining and maximum likelihood analysis was conducted on partial E1 gene sequences[71]; both the maximum-likelihood (ML) method and the Metropolis-coupled Markov Chain Monte Carlo (MCMCMC) method was peformed by Volk[15], they also estimated evolutionary rates and times to the most recent common ancestors on CHIKV genomic sequences. A recent phylogenetic study[45] analyzed CHIKV data set of partial E1 nucleotide sequences and estimated the evolutionary rates, the time-scaled phylogeny reconstruction and Bayesian phylogeography (Figure 3). Volk[15], showed that the overall nucleotide substitution rate was 4.33×10-4nucleotide substitutions per site per year (subs/nt/year) and that the rates estimated for each lineage exhibited considerable variation, with those for the epidemic lineages significantly higher than those estimated for the enzootic lineages. The authors showed that the Asian lineage exhibited a significantly higher substitution rate (i.e., non- overlapping HPD values) (4.16×10-4subs/nt/yr; 95% HPD: 3.26×10-4to 5.02×10-4subs/nt/year) than the WAf (2.39×10-4subs/nt/year; 95% HPD: 1.98×10-4to 2.84×10-4subs/nt/year) and ECSA lineages (2.30×10-4subs/nt/year; 95% HPD: 1.37×10-4to 3.24 ×10-4subs/nt/year). The Indian Ocean epidemic lineage yielded an even higher rate estimate (8.46×10-4subs/nt/year; 95% HPD: 5.81×10-4to 1.09×10-3subs/nt/year).

Figure 3. Significant non-zero rates for CHIKV E1 sequences. Only rates supported by a BF of > 3 were considered significant, and highlighted with arrows. In the Figure are showed the probable gene flows from the Origin (Kenya) to the other countries (Lo Presti et al, 2012).
CHIKV E1 evolutionary rate reported by a phylogeographic study[45] for the whole dataset was 1.4×10-3substitution/site/ year (95% HPD 6.4×10-4-2.5×10-3). The assessment of the evolutionary rate of the subset including only the strains involved in the recent Indian Ocean epidemic, gave an estimation of 2.2×10-3(95% HPD 9.6×10-4-3.8×10-3). The authors reconstructed the geographic spread of CHIKV during the last epidemic wave, which showed an eastward path from Africa to Indian Ocean Island to India, and from there to other South East Asian countries (Figure 3) [45].
Phylogenetic studies also showed that the Indian Ocean and the Indian subcontinent epidemics were caused by two different introductions of distinct strains of the ECSA genotype of CHIKV[45,74,75].
A single E1-A226V mutation was sufficient to dramatically increase the ability of different strains of CHIKV to infect Ae. albopictus mosquitoes and this substitution required no additional adaptive mutations to gain intermolecular compatibility[76].
A study of strains obtained from the Democratic Republic of Congo during an urban outbreak in 1999-2000 demonstrated the close genetic relationship of these isolates with other strains from Central Africa[51] .
Since the major CHIK outbreak began in 2004, additional phylogenetic studies have been performed, most focusing on recent isolates[45] .
The MRCA for the recent epidemic which ravaged Indian Ocean islands and the Indian subcontinent in the years 2004-2007 was found to date in 2002[45], confirming the findings of a previous study[71]. Differently, a mathematical model by Cherian[77] estimated the progenitor of the 2005-2007 viruses to exist up to 9 years before. This appeared to be corroborated by the presence of a strain in India in 2000 that beared 99% identity with an Ugandan strain of 1982 and a high similarity with strains isolated during the recent CHIKV epidemic[74,78]. However, other studies did not support this finding that has been hypothesized to be the result of contamination[15] .
Asian lineage dated about 1952 and exhibited similar spread patterns of the recent Indian Ocean outbreak lineage, with successive epidemics detected along an eastward path[15]. Asian group splitted into two clades: an Indian lineage, which likely went extinct, and a south east lineage. The old Asia genotype has not been identified during the last epidemic in India. This was consistent with lack of sustainability of the human-mosquito cycle at a local scale in the absence of continued importation[15,79] .
Outbreaks of CHIKV fever in Asia had not been necessarily associated with outbreaks in Africa, which suggested an independent evolution of an African ancestor of CHIKV in Asia[45,80]. A new study on the phylogeny of Aedes albopictus and Aedes aegypti showed that Aedes albopictus probably spread in different times to the Indian Ocean islands starting from Madagascar, where these species where limited at the end of the XIX century[81].
On the contrary, Aedes albopictus was only recently introduced in Comores. These differences in the ecology of Aedes albopictus and Aedes aegypti could explain the simultaneous circulation of a wild type virus in the Comores and a mutated strain in La Reunion[81] .
4. Conclusion
Phylogenetic tools are important to reconstruct the geographic spread of CHIKV during the epidemics wave, because they can highlight a possible different way of virus diffusion i.e. the eastward path from Africa to Indian Ocean island to India, and from there to other South East Asian countries as also recently demonstrated[45] .
Some CHIKV cases outside tropical countries (ie. Italy) were directly associated with the return of travellers from India and from affected areas of the Indian Ocean and the major determinant of the outbreaks was the high density of the vector at the time of arrival of the index case.
Since the transmission of CHIKV is mediated by vectors, an issue to be considered and that has a fundamental role is the re-introduction and colonization of Aedes species in new areas and the problem of the vector control for disease containement.
The mosquito species Aedes albopictus originated in Southeast Asia, but has spread during the last 30-40 years to North, Central and Southern America, parts of Africa, northern Australia and several countries in Europe. The Aedes albopictus can colonise new geographical locations due to its ability to adapt to different climates[http://www. ecdc.europa.eu/en/healthtopics/vectors/mosquitoes/Pages/ aedes-albopictus.aspx].
Instead Aedes aegypti is an important invasive mosquitospecies that could potentially have an impact on European public health. It is found throughout tropical and subtropical regions of America, Africa and Asia, as well as southeastern US, Indian Ocean Islands and northern Australia. This species was established in Europe in the beginning of the 20th century and was also recently re-introduced in Europe[http://www.ecdc.europa.eu/en/healthtopics/vectors/ mosquitoes/Pages/aedes-aegypti.aspx].
Possible measures for vector control could be the use of fast-acting insecticides (synergised pyrethrins) for 3 days consecutively, to apply with a truck-mounted atomiser in public spaces and a backpack mist blower in private spaces. Antilarval measures using formulations of insect growth regulators. House-to-house interventions to eliminate breeding places, and to encourage community participation[67]. The occurrence of outbreaks of CHIKV infection in countries with a temperate climate highlighted that clinical and diagnostic capacities have to be developed where these vectors of exotic diseases already circulate.
The good management of patients with acute CHIKV infection is essential for public health in susceptible areas with current Aedes spp activity. Chikungunya fever is diagnosed based on symptoms, physical findings (e.g., joint swelling), laboratory testing, and the possibility of exposure to infected mosquitoes (http://www.cdc.gov/chikungunya/). The early diagnosis of CHIKV infection remains difficult because the clinical picture of CHIKV infection is similar to that of other viral infections, which results in frequent diagnostic uncertainty[82]. In areas with current Aedes spp activity, most health authorities recommend prompt suspicion of imported or autochthonous cases, adequate use of diagnostic tools, isolation of suspect patients, rapid contact with the local health department, and sometimes mandatory case declaration. The final aim is to avoid epidemics spreading around the new cases[33]. In conclusion, the present review gave a critical appraisal of the epidemiology, clinics and phylogenesis of CHIKV and reinforces the need to monitor the geographic spread of CHIKV, and the vectors.
Conflict of interest statement
We declare that we have no conflict of interest
Acknowledgments
The authors thank: Laurence Dupuis-Maguiraga, Marion Noret, Sonia Brun, Roger Le Grand, Gabriel Gras, Pierre Roques (authors of the article entitled “Chikungunya Disease: Infection-Associated Markers from the Acute to the Chronic Phase of Arbovirus-Induced.
“Arthralgia” published in Plos Neglected Tropical Diseases [6(3):e1446] to have kindly provided and given permission to re-use and to publish their Figure (Figure 1 of their article). The authors also thank the journal Plos Neglected Tropical Diseases for the same reason.
The authors also thank: Veasna Duong, Anne-Claire Andries, Chantha Ngan, Touch Sok, Beat Richner, Nima Asgari-Jirhandeh, Steve Bjorge, Rekol Huy, Sovann Ly, Denis Laurent, Bunheng Hok, Maria Concepcion Roces, Sivuth Ong, Meng Chuor Char, Vincent Deubel, Arnaud Tarantola, and Philippe Buchy [authors of the article entitled “Reemergence of Chikungunya Virus in Cambodia”published in Emerg Infect Dis. 2012; 18(12):2066-9. doi: 10.3201/eid1812.120471.] to have kindly provided and given permission to re-use and to publish their Figure (Figure 2 of their article). The authors also thank the journal Emerging Infectious Diseases (http://wwwnc.cdc.gov/eid/; http:// wwwnc.cdc.gov/eid/article/18/12/12-0471_article.htm) for the same reason.
[1] Schuffenecker I, Iteman I, Michault A, Murri S, Frangeul L, Vaney MC, et al. Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med 2006; 3: e263.
[2] Deller JJ Jr, Russell PK. An analysis of fevers of unknown origin in American soldiers in Vietnam. Ann Intern Med 1967; 66: 1129-1143.
[3] McGill PE. Viral infections: alpha-viral arthropathy. Baillieres Clin Rheumatol 1995; 9: 145-150.
[4] Adebajo AO. Rheumatic manifestations of tropical diseases. Curr Opin Rheumatol 1996; 8: 85-89.
[5] Ligon BL. Reemergence of an unusual disease: the chikungunya epidemic. Semin Pediatr Infect Dis 2006; 17: 99-104.
[6] Robinson MC. An epidemic of virus disease in southern province, Tanganyika territory, in 1952-53. Trans R Soc Trop Med Hyg 1955; 49: 28-32.
[7] Higashi N, Matsumoto A, Tabata K, Nagatomo Y. Electron microscope study of development of Chikungunya virus in green monkey kidney stable (VERO) cells. Virology 1967; 33(1): 55-69.
[8] Simizu B, Yamamoto K, Hashimoto K, Ogata T. Structural proteins of Chikungunya virus. J Virol 1984; 51: 254-258.
[9] Powers AM, Brault AC, Shirako Y, Strauss EG, Kang W, Strauss JH, et al. Evolutionary relationships and systematics of the alphaviruses. J Virol 2001; 75(21): 10118-10131.
[10] Khan AH, Morita K, Parquet Md, Mdel C, Hasebe F, Mathenge EG, et al. Complete nucleotide sequence of chikungunya virus and evidence for an internal polyadenylation site. J Gen Virol 2002; 83(12): 3075-3084.
[11] Barrett ADT, Weaver SC. Arboviruses: alphaviruses, flaviviruses and bunyaviruses. In: Greenwood D, Slack RCB, Peutherer JF (editors). Medical microbiology. 16 edn. London: Churchill Livingstone; 2002, p. 484-501.
[12] Brooks GF, Butel JS, Morse SA. Human arboviral infections. In: Jawetz. Melnick and Adelberg’s Medical microbiology. 23rd edn. Singapore: Mc Graw Hill, 2004, p. 514-524.
[13] Diallo M, Thonnon J, Traore Lamizana M, Fontenille D. Vectors of Chikungunya virus in Senegal: current data and transmission cycles. Am J Trop Med Hyg 1999; 60: 281-286.
[14] Vanlandingham DL, Hong C, Klingler K, Tsetsarkin K, McElroy KL, Powers AM, et al. Differential inactivities of o’nyong-nyong and Chikungunya virus isolates in Anopheles gambiae and Aedes aegypti mosquitoes. Am J Trop Med Hyg 2005; 72: 616-621.
[15] Volk SM, Chen R, Tsetsarkin KA, Adams AP, Garcia TI, Sall AA, et al. Genome-scale phylogenetic analyses of chikungunya virus reveal independent emergences of recent epidemics and various evolutionary rates. J Virol 2010; 84: 6497-6504.
[16] Yadav P, Gokhale MD, Barde PV, Singh DK, Mishra AC, Maurya DT. Experimental transmission of Chikungunya virus by Anopheles stephensi mosquitoes. Acta Virol 2003; 47: 45-47.
[17] Talarmin F, Staïkowsky F, Schoenlaub P, Risbourg A, Nicolas X, Zagnoli A, et al. Skin and mucosal manifestations of chikungunya virus infection in adults in Reunion Island. Med Trop (Mars) 2007; 67: 167-173.
[18] Robin S, Ramful D, Zettor J, Benhamou L, Jaffar-Bandjee MC, Rivière JP, et al. Severe bullous skin lesions associated with Chikungunya virus infection in small infants. Eur J Pediatr 2010; 169: 67-72
[19] Schwartz O, Albert ML. Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol 2010; 8: 491-500
[20] Dupuis-Maguiraga L, Noret M, Brun S, Le Grand R, Gras G, Roques P. Chikungunya disease: infection-associated markers from the acute to the chronic phase of arbovirus-induced arthralgia. PLoS Negl Trop Dis 2012; 6(3): e1446.
[21] Morrison JG. Chikungunya fever. Int J Dermatol 1979; 18: 628-629.
[22] Bodenmann P, Genton B. Chikungunya: an epidemic in real time. Lancet 2006; 368: 258.
[23] Mourya DT, Mishra AC. Chikungunya fever. Lancet 2006; 368: 186-187.
[24] Yazdani R, Kaushik VV. Chikungunya fever. Rheumatology (Oxford) 2007; 46: 1214-1215.
[25] Lemant J, Boisson V, Winer A, Thibault L, André H, Tixier F, et al. Serious acute chikungunya virus infection requiring intensive care during the Reunion Island outbreak in 2005-2006. Crit Care Med 2008; 36(9): 2536-2541.
[26] Sissoko D, Malvy D, Ezzedine K, Renault P, Moscetti F, Ledrans M, et al. Post-epidemic chikungunya disease on Reunion Island: course of rheumatic manifestations and associated factors over a 15-month period. PLoS Negl Trop Dis 2009; 3: e389.
[27] Manimunda SP, Vijayachari P, Uppoor R, Sugunan AP, Singh SS, Rai SK, et al. Clinical progression of chikungunya fever during acute and chronic arthritic stages and the changes in joint morphology as revealed by imaging. Trans R Soc Trop Med Hyg 2010; 104: 392-399.
[28] Borgherini G, Poubeau P, Jossaume A, Gouix A, Cotte L, Michault A, et al. Persistent arthralgia associated with Chikungunya virus: a study of 88 adult patients on Reunion Island. Clin Infect Dis 2008; 47: 469-475.
[29] Simon F, Parola P, Grandadam M, Fourcade S, Oliver M, Brouqui P, et al. Chikungunya infection: an emerging rheumatism among travelers returned from Indian Ocean islands. Report of 47 cases. Medicine (Baltimore) 2007; 86: 123-137.
[30] Mizuno Y, Kato Y, Takeshita N, Ujiie M, Kobayashi T, Kanagawa S, et al. Clinical and radiological features of imported chikungunya fever in Japan: a study of six cases at the National Center for Global Health and Medicine. J Infect Chemother 2011; 17(3): 419-423.
[31] Chhabra M, Mittal V, Bhattacharya D, Rana UVS, Lal S. Chikungunya fever: A re-emerging viral infection. Indian J Med Microbiol 2008; 26(1): 5-12.
[32] Chikungunya Fever Fact Sheet - CDC Division of Vector Borne Infectious Diseases.[Online]. Availiable from: http://www.cdc.gov/ ncidod/chikungunya/chickfact.htm [Accessed on 2006].
[33] Simon F, Javelle E, Oliver M, Leparc-Goffart I, Marimoutou C. Chikungunya virus infection. Curr Infect Dis Rep 2011; 13: 218-228.
[34] Panning M, Hess M, Fischer W, Grywna K, Pfeffer M, Drosten C. Performance of the RealStar Chikungunya virus real-time reverse transcription-PCR kit. J Clin Microbiol 2009; 47: 3014-3016.
[35] Parola P, de Lamballerie X, Jourdan X, Rovery C, Vaillant V, Minodier P, et al. Novel chikungunya virus variant in travelers returning from Indian Ocean islands. Emerg Infect Dis 2006; 12: 1493-1499.
[36] Niedrig M, Zeller H, Schuffenecker I, Drosten C, Emmerich P, Rumer L, et al. International diagnostic accuracy study for the serological detection of chikungunya virus infection. Clin Microbiol Infect 2009; 15: 880-884.
[37] Oliver M, Grandadam M, Marimoutou C, Rogier C, Botelho-Nevers E, Tolou H, et al. Persisting mixed cryoglobulinemia in Chikungunya infection. PLoS Negl Trop Dis 2009; 3: e374.
[38] Jaffar-Bandjee MC, Das T, Hoarau JJ, Krejbich Trotot P, Denizot M, Ribera A, et al. Chikungunya virus takes centre stage in virally induced arthritis: possible cellular and molecular mechanisms to pathogenesis. Microbes Infect 2009; 11: 1206-1218.
[39] Carletti F, Bordi L, Chiappini R, Ippolito G, Sciarrone MR, Capobianchi MR, et al. Rapid detection and quantification of Chikungunya virus by a one-step reverse transcription polymerase chain reaction real-time assay. Am J Trop Med Hyg 2007; 77(3): 521-524.
[40] Chiam CW, Chan YF, Loong SK, Yong SS, Hooi PS, Sam IC. Real-time polymerase chain reaction for diagnosis and quantitation of negative strand of chikungunya virus. Diagn Microbiol Infect Dis 2013; 77: 133-137.
[41] Agarwal A, Singh AK, Sharma S, Soni M, Thakur AK, Gopalan N, et al. Application of real-time RT-PCR in vector surveillance and assessment of replication kinetics of an emerging novel ECSA genotype of Chikungunya virus in Aedes aegypti. J Virol Methods 2013; 193: 419-425.
[42] Chen H, Takei F, Koay ES, Nakatani K, Chu JJ. A novel DANP-coupled hairpin RT-PCR for rapid detection of Chikungunya virus. J Mol Diagn 2013; 15: 227-233.
[43] Lumbsden WHR. An epidemic of virus disease in southern province of Tanganyika territory, in 1952-53; II General description and epidemiology. Trans R Soc Trop Med Hyg 1955; 49(1): 33-55.
[44] Ross RW. The Newala epidemic III; the virus: isolation, pathogenic properties and relationship to the epidemic. J Hyg 1956; 54: 177-191.
[45] Lo Presti A, Ciccozzi M, Cella E, Lai A, Simonetti FR, Galli M, et al. Origin, evolution, and phylogeography of recent epidemic CHIKV strains. Infect Genet Evol 2012; 12(2): 392-398.
[46] Jupp PG, McIntosh BM. Chikungunya disease. In: Monath TP editor. The arboviruses: epidemiology and ecology. Boca Raton (Florida): CRC Press; 1988, p. 137-57.
[47] Lanciotti RS, Ludwig ML, Rwaguma EB, Lutwama JJ, Kram TM, Karabatsos N, et al. Emergence of epidemic O’nyong-nyong feverin Uganda after a 35-year absence: genetic characterization of the virus. Virology 1998; 252: 258-268.
[48] Williams MC, Woodall JP, Corbet PS, Gillett JD. O’nyong-nyong fever: an epidemic virus disease in East Africa and virus isolations from Anopheles mosquitoes. Trans R Soc Trop Med Hyg 1965; 59: 300-306.
[49] Halstead SB, Scanlon JE, Umpaivit P, Udomsakdi S. Dengue and Chikungunya virus infection in man in Thailand, 1962-1964, epidemiological study in Bangkok metropolitan area. Am J Trop Med Hyg 1969; 18: 997-1021.
[50] Padbidri VS, Gnaneswar TT. Epidemiological investigations of chikungunya epidemic at Barsi, Maharashtra state, India. J Hyg Epidemiol Microbiol Immunol 1979; 23(4): 445-451.
[51] Pastorino B, Muyembe-Tamfum JJ, Bessaud M, Tock F, Tolou H, Durand JP, et al. Epidemic resurgence of Chikungunya virus in Democratic Republic of the Congo: identification of a new central African strain. J Med Virol 2004; 74: 277-282.
[52] WHO-Disease outbreak news. 2006. Chikungunya and dengue in the southwest Indian Ocean, 17 March 2006.[Online]. Geneva: Available from: http://www.who.int/csr/don/2006_03_17/print. html [Accessed on May 01, 2006].
[53] Powers AM, Logue CH. Changing patterns of chikungunya virus: re-emergence of a zoonotic arbovirus. J Gen Virol 2007; 88: 2363-2377.
[54] Mathew T, Tiruvengadam KV. Further studies on the isolate of Chikungunya from the Indian repatriates of Burma. Indian J Med Res 1973; 61(4): 517-520.
[55] Laras K, Sukri NC, Larasati RP, Bangs MJ, Kosim R, Djauzi, et al. Tracking the re-emergence of epidemic chikungunya virus in Indonesia. Trans R Soc Trop Med Hyg 2005; 99: 128-141.
[56] Lam SK, Chua KB, Hooi PS. Chikungunya infection: emerging disease in Malasiya. Southeast Asian J Trop Med Pub Hlth 2001; 32: 447-451.
[57] Shah KV, Gibbs CJ Jr, Banerjee G. Virological investigation of the epidemic of haemorrhagic fever in Calcutta: isolation of three strains of Chikungunya virus. Indian J Med Res 1964; 52: 676-683.
[58] Jadhav M, Namboodripad M, Carman RH, Carey DE, Myers RM. Chikungunya disease in infants and children in Vellore: a report of clinical and haematological features of virologically proved cases. Indian J Med Res 1965; 53: 764-776.
[59] Thiruvengadam KV, Kalyanasundaram V, Rajgopal J. Clinical and pathological studies on Chikungunya fever in Madras City. Indian J Med Res 1965; 53: 729-744.
[60] Lahariya C, Pradhan SK. Emergence of chikungunya virus in Indian subcontinent after 32 years: A review. J Vector Borne Dis 2006; 43: 151-160.
[61] Pavri KM. Presence of chikungunya antibodies in human sera collected from Calcutta and Jamshedpur before 1963. Indian J Med Res 1964; 52: 698-702.
[62] Sarkar JK, Chatterjee SN, Chakravarty SK, Mitra AC. The causative agent of Calcutta haemorrhagic fever: chikungunya or dengue. Bull Calcutta Sch Trop Med 1965; 13: 53-54.
[63] Burke DS, Nisalak A, Nimmannitya S. Disappearance of Chikungunya virus from Bangkok. Trans R Soc Trop Med Hyg 1985; 79: 419-420.
[64] Chastel C. Chikungunya virus: its recent spread to the southern Indian Ocean and Reunion Island (2005-2006). Bull Acad Natl Med 2005; 189: 1827-1835.
[65] Paganin F, Borgherini G, Staikowsky F, Arvin-Berod C, Poubeau P. Chikungunya on Reunion Island: chronicle of an epidemic foretold. Presse Med 2006; 35: 641-646.
[66] Pialoux G, Gaüzère BA, Jauréguiberry S, Strobel M. Chikungunya, an epidemic arbovirosis. Lancet Infect Dis 2007; 7: 319-327.
[67] Rezza G, Nicoletti L, Angelini R, Romi R, Finarelli AC, Panning M, et al. CHIKV study group. Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet 2007; 370: 1840-1846.
[68] Kumar NP, Joseph R, Kamaraj T, Jambulingam P. A226V mutation in virus during the 2007 chikungunya outbreak in Kerala, India. J Gen Virol 2008; 89: 1945-1948.
[69] Moro ML, Gagliotti C, Silvi G, Angelini R, Sambri V, Rezza G, et al. Chikungunya Study Group. Chikungunya virus in North-Eastern Italy: a seroprevalence survey. Am J Trop Med Hyg 2010; 82: 508-511.
[70] Centers for Disease Control and Prevention (CDC). Chikungunya fever diagnosed among international travelers--United States, 2005-2006. MMWR Morb Mortal Wkly Rep 2006; 55: 1040-1042. [71] Powers AM, Brault AC, Tesh RB, Weaver SC. Re-emergence of Chikungunya and O’nyong-nyong viruses: evidence for distinct geographical lineages and istant evolutionary relationships. J Gen Virol 2000; 81: 471-479.
[72] Duong V, Andries AC, Ngan C, Sok T, Richner B, Asgari-Jirhandeh N, et al. Reemergence of Chikungunya virus in Cambodia. Emerg Infect Dis 2012; 18(12): 2066-2069.
[73] Arankalle VA, Shrivastava S, Cherian S, Gunjikar RS, Walimbe AM, Jadhav SM, et al. Genetic divergence of Chikungunya viruses in India (1963-2006) with special reference to the 2005-2006 explosive epidemic. J Gen Virol 2007; 88: 1967-1976.
[74] Brisse S, Iteman I, Schuffenecker I. Chikungunya outbreaks. N Engl J Med 2007; 356: 2650-2652.
[75] Charrel RN, de Lamballerie X, Raoult D. Chikungunya outbreaks the globalization of vectorborne diseases. N Engl J Med 2007; 356: 769-771
[76] Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S. A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 2007; 3: e201.
[77] Cherian SS, Walimbe AM, Jadhav SM, Gandhe SS, Hundekar SL, Mishra AC, et al. Evolutionary rates and timescale comparison of Chikungunya viruses inferred from the whole genome/E1 gene with special reference to the 2005-07 outbreak in the Indian subcontinent. Infect Genet Evol 2009; 9: 16-23.
[78] Yergolkar PN, Tandale BV, Arankalle VA, Sathe PS, Sudeep AB, Gandhe SS, et al. Chikungunya outbreaks caused by African genotype. Indian Emerg Infect Dis 2006; 12: 1580-1583.
[79] Chevillon C, Briant L, Renaud F, Devaux C. The chikungunya threat: an ecological and evolutionary perspective. Trends Microbiol 2008; 16: 80-88.
[80] Ng LC, Hapuarachchi HC. Tracing the path of chikungunya virus -evolution and adaptation. Infect Genet Evol 2010; 10: 876-885.
[81] Delatte H, Bagny L, Brengue C, Bouetard A, Paupy C, Fontenille D. The invaders: phylogeography of dengue and chikungunya viruses Aedes vectors, on the South West islands of the Indian Ocean. Infect Genet Evol 2011; 11: 1769-1781.
[82] Carey DE. Chikungunya and dengue: a case of mistaken identity? J Hist Med Allied Sci 1971; 26(3): 243-262.
ment heading
10.1016/S1995-7645(14)60164-4
*Corresponding author: Department of Infectious, Parasitic and Immunomediated Diseases, Epidemiology Unit, Reference Centre on Phylogeny, Molecular Epidemiology and Microbial Evolution (FEMEM), Istituto Superiore di Sanita`, Viale Regina Elena 299, 00161, Rome, Italy.
Tel: +39-06-49903187
E-mail: massimo.ciccozzi@iss.it
Epidemiology
Phylogeny
 Asian Pacific Journal of Tropical Medicine2014年12期
Asian Pacific Journal of Tropical Medicine2014年12期
- Asian Pacific Journal of Tropical Medicine的其它文章
- Induction of deletion mutation on ompR gene of Salmonella enterica serovar Typhi isolates from asymptomatic typhoid carriers to evolve attenuated strains for vaccine development
- Evaluation of protective effect of IL-22 and IL-12 on cutaneous leishmaniasis in BALB/c mice
- Immunogenic potential and protective efficacy of formalin inactivated circulating Indian strain of West Nile virus
- Genetic diversity and gene structure of mitochondrial region of Anopheles minimus (Diptera: Culicidae) - major malaria vector of North east India
- Fumigant and repellent properties of sesquiterpene-rich essential oil from Teucrium polium subsp. capitatum (L.)
- Antioxidant activity and free radical scavenging activities of Streptomyces sp. strain MJM 10778
