Antioxidant activity and free radical scavenging activities of Streptomyces sp. strain MJM 10778
Dong-Ryung Lee, Sung-Kwon Lee, Bong-Keun Choi, Jinhua Cheng, Young-Sil Lee, Seung Hwan Yang*, Joo-Won Suh1,*
1Division of Bioscience and Bioinformatics, Myongji University, Youngin-si, Gyeonggi-do 449728, Korea
2Center for Neutraceutical and Pharmaceutical Materials, Myongji University, Yongin-si, Gyeonggi-do 449728, Korea
Antioxidant activity and free radical scavenging activities of Streptomyces sp. strain MJM 10778
Dong-Ryung Lee1,2#, Sung-Kwon Lee2#, Bong-Keun Choi2, Jinhua Cheng2, Young-Sil Lee2, Seung Hwan Yang2*, Joo-Won Suh1,2*
1Division of Bioscience and Bioinformatics, Myongji University, Youngin-si, Gyeonggi-do 449728, Korea
2Center for Neutraceutical and Pharmaceutical Materials, Myongji University, Yongin-si, Gyeonggi-do 449728, Korea
Objective:To investigate the antioxidant activity of soil-borne actinobacteria.Methods:The total phenolic contents, the level of antioxidant potential by DPPH radical scavenging activity, NO scavenging activity, and ABTS radical scavenging activity in ethyl acetate extract were determined.Results:The 16S rDNA sequencing analysis revealed that Streptomyces sp. strain MJM 10778, which was isolated from Hambak Mountain, Korea, has 99.9% similarity to Streptomyces misionensis (S. misionensis) NBRC 13063. The physiological and the morphological test revealed that the strain MJM 10778 has different characteristics from the strain NBRC 13063. The entire antioxidant assay with the ethyl acetate extract displayed good radical scavenging activity. The IC50values of the strain MJM 10778 extract on DPPH, NO, and ABTS radicals were identified to be 92.8 μg/mL, 0.02 μg/mL, and 134.9 μg/mL, respectively. The ethyl acetate extract of the strain MJM 10778 showed an 81.50% of cell viability at 100 μg/mL in Raw264.7 cell viability assay. Conclusions:The results obtained suggest that the ethyl acetate extract of Streptomyces sp. strain MJM 10778 could be considered as a potential source of drug for the diseases that is caused by free radicals with its anti-oxidant activities and low cytotoxicity.
ARTICLE INFO
Article history:
Received 5 June 2014
Received in revised form 15 July 2014
Accepted 15 September 2014
Available online 20 December 2014
Streptomyces sp. strain MJM 10778
1. Introduction
In the soil micro-ecosystem, the actinomycetes is one of the major group[1] and a large number of actinomycetes have already been isolated and screened from soil[2]. Actinomycetes are making 75% of all known antibiotics, and among them, nearly 80% are produced by members of the genusStreptomyces[3-5].
Streptomycesis a genus of high G+C Gram-positive filamentous bacteria belonging to the phylum actinomycetes. A complex life cycle involving the multicellular development into an aerial hyphae that is developed into spores is a special trait ofStreptomycesspecies[6]. Moreover, they produce various extracellular enzymes that degrade complex biopolymers, such as chitin and lignocellulose. This feature makes them important in the nutrient recycling processes[7,8].
Previous studies demonstrated that a variety ofStreptomycesinhabit a wide range of plants as either symbionts or parasites[9,10]. They might play a crucial role in plant development and human health, because they could affect plant growth either by nutrient assimilation or through secondary metabolite productions.
Reactive oxygen species (ROS) production occurs during normal cell metabolism. Excessive amount of ROS increases oxidative stress, and it can cause deleterious effects such as atherosclerosis, reperfusion injury, cataractogenesis, rheumatoid arthritis, inflammatory disorders, and cancer[11]. In order to retard the oxidation process, many synthetic antioxidants such as butylated hydroxycanisole (BHA), butylated hydroxytoluene (BHT), and propyl gallate (PG) are being used in clinics. However, these synthetic antioxidantshave potential health hazards[12], so it have been attempted to screen alternative antioxidants from natural sources.
The novel compound, JBIR-94 and JBIR-125 that were found fromStreptomycessp. strain R56-07, organofluorine that was isolated fromStreptomycessp. strain TC1 have shown strong antioxidant activity [13,14].
In this study, we isolated oneStreptomycesstrain MJM 10778 from mountain forest soil in Korea and investigated the cultural characteristics, phylogenetic analysis, and antioxidant activity through a series ofin vitrotests such as total phenolics contents (TPC), reducing power measurement, nitric oxide (NO) scavenging activity, ABTS (2,2-azinobis (3-ethylbenzothiazoline-6-sulfonic acid) free radical scavenging activity, and DPPH (1,1-Diphenyl-2-picrylhydrazyl) radical scavenging activity.
2. Materials and methods
2.1. Chemicals and reagents
The following reagents and solvents were purchased from Sigma-Aldrich: ascorbic acid, 1,1-Diphenyl-2-picrylhydrazyl (DPPH), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), 2,2-azinobis (3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), potassium ferricyanide, trichloroacetic acid, ferric chloride, sodium nitroprusside, suphanilamide, phosphoric acid, napthylethylene diamine dihydrobhloride, sodium nitrite, and Folin-Ciocalteu’s phenol.
2.2. Isolation of Streptomyces from mountain forest soil
In the present study, the soil sample was collected from Hambak mountain, Yongin city of Gyeonggi-do province, Korea (latitude 37˚ 12ˊ 51.00˝ N, longitude 127˚ 11ˊ 20.00˝ E), in September, 2007. The samples were dug up from a depth of 20 cm after removing approximately 3 cm of the soil surface. For each collected sample, 1 000 mg of the soil was suspended in 50 mL of deionized water, and then it was vortexed for 5 min. Mixtures were allowed to settle, serial dilutions of up to 10-4were prepared using sterile deionized water, and the diluted mixtures were agitated with the vortex. An aliquot of 0.2 mL of each dilution was taken and spread evenly over glucose-yeast extract-malt extract agar (GYM; medium 65; DSMZ) and Bennet’s agar (BN; medium 548; DSMZ). Plates were incubated at 28 ℃ for 7 days. Repeated streaking on GYM and BN agar plates led to purified bacterial colonies that showed an actinomyceteslike appearance.
2.3. Morphological identification of the strain MJM 10778
Morphological characteristics are the foundation for identifying actinobacteria. Isolated strain was identified according to the traditional morphological criteria including characteristics of colonies on the plate, the presence of aerial mycelium, spore mass color, distinctive reverse colony color, diffusible pigment, and morphological characteristics (sporophore and spore chain morphology)[15]. The cultural characteristics of the isolated strain were observed after incubation for 7 days in 4 different media.
2.4. Physiological characteristics
The utilization pattern of carbon sources by the strain was identified according to the methods of Gottlieb[16]. The isolated strain was grown on the basal medium, which was composed of the following: KH2PO4, 2.38 g; KH2PO4•3H2O, 5.65 g; MgSO4•7H2O, 1 g; (NH4)2SO4, 2.64 g; trace element solution, 6.25 mL; agar, 15 g; and distilled water, 1 L. The trace element solution contained the following: CuSO4•3H2O, 102 mg; FeSO4•7H2O, 176 mg; MnCl2•4H2O, 126 mg; ZnSO4•7H2O, 24 mg; and distilled water, 100 mL.
2.5. DNA isolation and 16S rDNA sequencing
Genomic DNA of the strain MJM 10778 was isolated using Bacterial Genomic DNA isolation kit (Corebio, Korea), following the manufacturer’s manual. PCR amplification of the 16S rDNA gene was carried out with primer set 27F/1492R[17]. Amplified products were sequenced using an automatic sequencer (ABI 3730XL; Applied Biosystems) at Solgent, Daejeon, Korea.
2.6. Phylogenetic analysis
For taxonomic positioning of the strain MJM 10778, 16S rDNA gene was amplified and sequenced (Figure 1). The 16S rDNA gene sequence (1 399 bp) of strain MJM 10778 was aligned with sequences of related type strains. The CLUSTAL_X program[18] was used for multiple alignments. Phylogenetic analysis was performed with MEGA software version 6.0 (MEGA6)[19]. Trees were generated by Neighbor-Joining[20] using Tajima-Nei model[21]. The topology of the phylogenetic tree was evaluated by bootstrap resampling method of Felsenstein with 500 replicates[22].
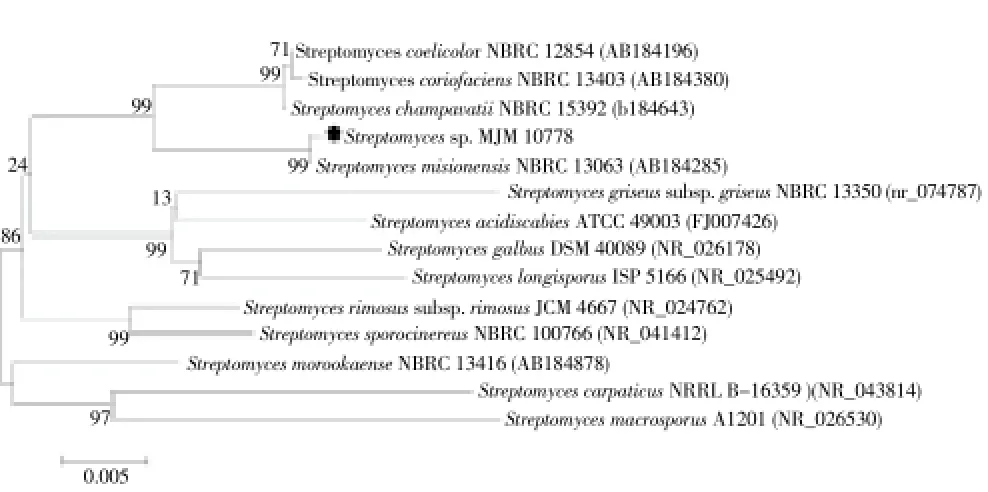
Figure 1. Phylogenetic tree based on the 16S rDNA sequences of the showing affiliation of the strain MJM 10778 with closely related members in GenBank.Phylogenetic trees were generated using MEGA version 6.0 with default parameters, Tajima-Nei model, and the Neighbor-Joining algorithm. The numbers at the branching prints are the percentages of occurrence in 500 bootstrapped tree.
2.7 Estimation of total phenolic content
The total phenolic content of the crude extract of MJM 10778 strain was measured using the Folin-Ciocalteau reagent. The crude extracts were diluted in water to obtain 125, 250, 500, and 1 000 μg/mL concentrations. The 50 μL of crude extract was mixed with 2.5 mL of Folin-Ciocalteau reagent (1/10 diluted in distilled water). The mixture was incubated at 45 ℃ for 15 min. The absorbance was measured at 765 nm. The Na2CO3solution (2 mL of 7.5% Na2CO3in 2.55 mL of distilled water) was used as blank. The results were expressed as gallic acid equivalence (GAE) in μg[23].
2.8. Extraction of the secondary metabolites
The spore suspensions of MJM 10778 were inoculated into 50 mL of BN medium and cultivated at 28 ℃ for 48 h as seed culture. Then, 500 μL of seed culture broth were collected and inoculated into 50 mL of BN medium and cultivated at 28 ℃ for 6 days. The culture broth was filtered through filter paper (Whatman No.1) and the filtered broth was extracted with two-volumes of ethyl acetate twice. The ethyl acetate extract was dried in vacuum at 38 ℃ and 200 mbar for various biological activity tests.
2.9. Antioxidant assays
2.9.1. Reducing power
The reducing power measurement was determined according to a method of Oyaizu[24] with slight modifications. The ethyl acetate (EtOAc) extract of MJM 10778 was diluted at various concentrations. One milliliter of each dilution was mixed with 0.1 mL of 1% potassium ferricyanide. The mixture was incubated at 50 ℃ for 20 min. After cooling, 0.1 mL of 1% trichloro acetic acid was added to the mixture, the upper layer of mixture was mixed with 0.1% ferric chloride. Absorbance was measured at 700 nm using spectrophotometer. The higher absorbance of the reaction mixture indicates increase in reducing power.
2.9.2. DPPH radicals scavenging activity
The DPPH radical scavenging assay was conducted according to the modified method of Diazet al[25]. EtOAc extracts of the strain MJM 10778 were diluted with ethanol, 10 μL of dilution was distributed into a 96-well plate. To each well, 190 μL of DPPH ethanol solution was added and allowed to react at room temperature for 30 min. The absorbance was measured at 550 nm by microplate reader (Tecan; Infinite pro 2000).
The DPPH radical scavenging capability was calculated by the following equation: DPPH radical scavenging activity (%) = [(A0- A1)/A0] × 100, where A0, and A1are the absorbance of control (blank) and the reaction system of sample or ascorbic acid.
2.9.3. Nitric oxide (NO) scavenging activity
The scavenging activity of MJM10778 EtOAc extract on nitric oxide was measured according to the method of Marcocciet al[26] with few modifications. One milliliter of extract was mixed with 1 mL of sodium nitroprusside (5 mM) and the mixture was incubated at 25 ℃ for 3 h. After the incubation, 50 μL of griess reagent (1% sulphanilamide, 2% phosphoric acid, and 0.1% of napthylene diaminedihydrochloride) was added to the sample solution. The nitric oxide scavenging was measured at 540 nm and referred to the absorbance of standard solutions of sodium nitrite salt treated in the same way with Griess reagent.
The NO radical scavenging capability was determined using the same equation that was used to calculate DPPH scavenging activity.
2.9.4. ABTS free radical scavenging assay
ABTS free radical scavenging assay was done using the method by Zhishenet al[27]. with modifications. ABTS, 2,2′-azinobis (3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (Sigma-Aldrich, USA) was dissolved in water to a 7 mM concentration. ABTS radical cation (ABTS•+) was produced by reacting ABTS solution with 2.45 mM potassium persulfate (K2S2O8) and storing in the dark at room temperature for 12-16 h. The ABTS radical solution was diluted to an absorbance of (0.7±0.2) at 734 nm. The 10 μL of MJM 10778 EtOAc extracts were distributed into 96-well plate. To each well, 190 μL of ABTS radical solution was added and incubated for 3 min at room temperature. The absorbance was measured at 734 nm by microplate reader (Tecan; Infinite pro 2000). ABTS•+scavenging activity was determined using the same equation that was used to calculate DPPH scavenging activity.
2.10. Cytotoxicity assay
Cytotoxicity of the EtOAc extract was assessed by MTT cell viability assay, following the method by Mosmanet al[28]. Raw 264.7 cells (5×104cells/well) were pated in a 96-well plate, and then incubated at 37 ℃ in 5% CO2. After 24 h, cells were treated with MJM 10778 EtOAc extracts (final concentration: 25, 50, and 100 μg/mL) and incubated for 24 h. Then, the cells were treated with MTT solution (5 mg/mL in PBS) for 4 h. The medium was removed and the remaining cells were dissolved in DMSO. The optical density at 570 nm was measured by microplate reader (Tecan; Infinite pro 2000). The control (untreated) optical densities of cells were taken as 100% of viability.
2.11. Statistical analysis
The data of all experiments were represented as Means±SD and were analyzed with Sigmaplot (version 12.5). Differences were considered significantly at P<0.05.
3. Results
3.1. Morphological identification of the isolated strain MJM 10778
The strain MJM 10778 formed grayish brown-colored and lite grayish colored aerial mass on oatmeal agar (ISP-3)and salt-starch agar (ISP-4) medium, respectively. Color in media: soluble pigments were not formed in oatmeal agar, salt-starch agar, and glycerol-asparagine agar (ISP-5). In yeast extract-malt extract agar, orange brown exopigment was found (Table 1).
3.2. Carbon sources utilization of the isolated strain MJM 10778
Eight carbon sources (glucose, arabinose, rhamnose, fructose, raffinose, mannitol, sucrose, and inositol) were test in this study, and the strain MJM 10778 efficiently utilized glucose and arabinose for growth. The utilization of rhamnose, fructose, raffinose, mannitol, sucrose, and inositol was doubtful.
3.3. Molecular identification of the isolated strain MJM 10778
The phylogenetic analysis demonstrated that the strain MJM 10778 belongs to the genusStreptomyces. The rooted phylogenetic tree for MJM 10778 and related and representative type strains of the genusStreptomycesindicated that this strain is most closely related toStreptomyces misionensisNBRC 13063 (GenBanK accession no. AB184285) (Figure 1). The 16S rDNA gene sequence similarity between the strain MJM 10778 and the type strain NBRC 13063 was 99.9%.
3.4. Total phenolic content and reducing power
Total phenolic content of the strain MJM 10778 extract was expressed as gallic acid exultance (GAE) in μg. The total phenolic compound was found to be (8.8±0.2) μg GAE/g dry weights. The reducing power is associated with antioxidant activity and may serve as a significant reflection of the antioxidant activity[29]. The extract can serve as a significant indicator of potential antioxidant because of its electron donating ability. In this assay, the green and blue color displays the reducing power of the extract. Figure 2a shows the reductive capabilities of theStreptomycessp. strain MJM 10778 extract, and the highest reducing power was found to be 500 μg.
3.5. DPPH radical scavenging activity
The DPPH radical has been widely used to investigate the scavenging activities of natural compounds. It was scavenged by antioxidants via the donation of hydrogen, forming the non-radical DPPH. The color changed from purple to yellow after the reaction. The EtOAc extract showed the scavenging activities with increasing concentrations of extract as follows: (28.9±0.1)% at 15 μg/mL, (41.9±3.2)% at 30 μg/mL, (53±3)% at 60 μg/mL, (64.6±4.0)% at 125 μg/mL, (77.9±2.4)% at 250μg/mL, (82.6±0.4)% at 500 μg/mL, and (81.2±0.2)% at 1 000 μg/mL. The IC50value was found to be 92.8 μg/mL (Figure 2b).
3.6. NO scavenging activity
The strain MJM 10778 extract showed NO scavenging activities with increasing concentration of extract as follows: (89.6±0.2)% at 31.25 μg/mL, (90.8±0.2)% at 62.5 μg/mL, (91 ±0.2)% at 125 μg/mL, (92.4±0.2)% at 250 μg/mL, (92.6±0.6)% at 500 μg/mL, and (95.4±0.1)% at 1 000 μg/mL. The IC50value was found to be 0.02 μg/mL (Figure 2c).
3.7. ABTS free radical scavenging activity
The antioxidant capacity of EtOAc extract was evaluated according to the ABTS decolorization method (Figure 2d). The extract showed the scavenging activities with increasing concentrations of extract (Figure 2d) as follows: (2.7±0.4)% at 31.25 μg/mL, (16.1±1.5)% at 52.5 μg/mL, (38±1.3)% at 125 μg/mL, (71.2±1.3)% at 250 μg/mL, (82.9±0.1)% at 500 μg/mL, and (82.6±0.3)% at 1 000 μg/mL. The IC50value was found to be 134.9 μg/mL.
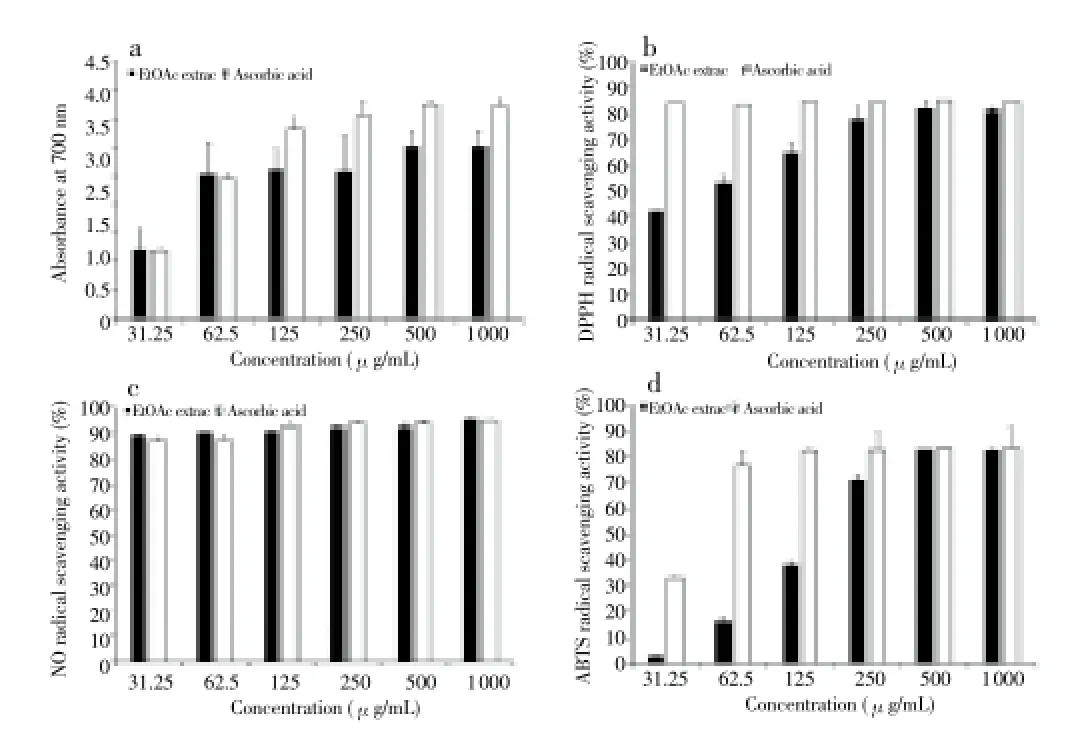
Figure 2. Antioxidant results of the EtOAc extract from Streptomyces sp. strain MJM 10778 on (a) reducing power assay (b) DPPH radical scavenging assay, (c) nitric oxide scavenging assay, (d) ABTS free radical scavenging assay. Results represent the average of three replicates (n=3). Error bars represent standard deviation.

Table 1 Comparison of culture characteristics between the isolated strain MJM 10778 and the strain ATCC 14991 on the different medium.
3.8. MTT cell viability test
The cytotoxicity of the EtOAc extract at various concentrations on Raw 264.7 cells was determined by the MTT assay. The EtOAc extract showed cell viabilities of (100 ±2.1)% at control (untreated), (83.20±3.80)% at 25 μg/mL, (82.18±5.15)% at 50 μg/mL, and (81.50±2.09)% at 100 μg/mL, respectively (Figure 3).
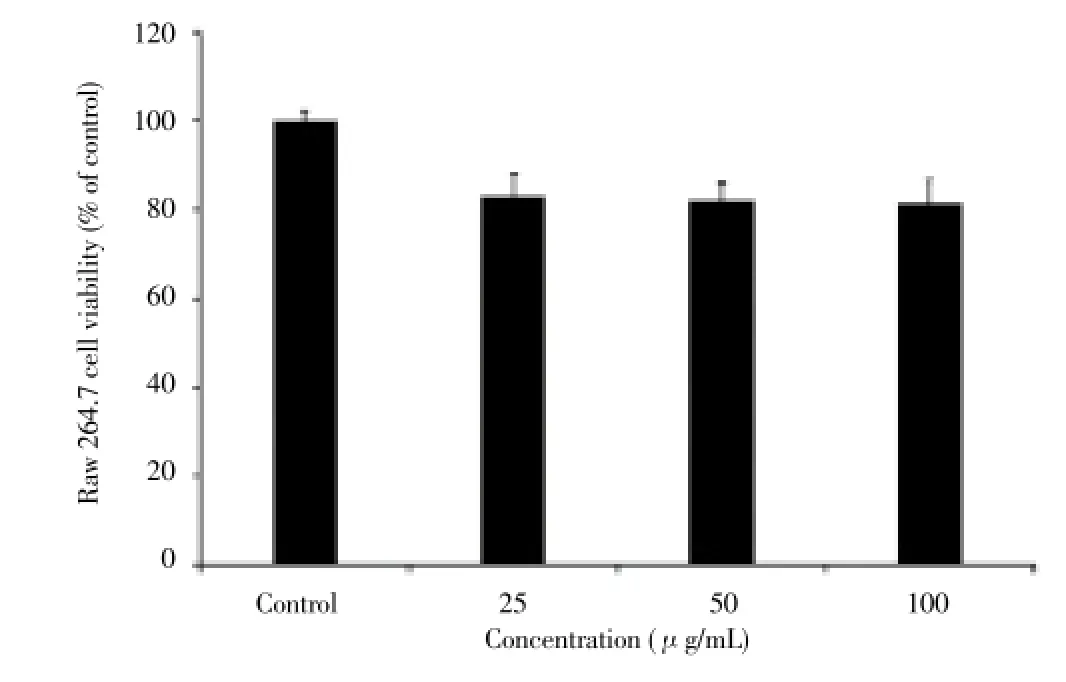
Figure 3. Cytotoxicity of EtOAc extract from Streptomyces sp. strain MJM 10778 on Raw 264.7 cells. Results represent the average of three replicates (n=3). Error bars represent standard deviation.
4. Discussion
Recent studies have revealed that free oxygen radicals have the critical roles in various diseases such as cancer, autoimmune disorders, cardiovascular and neurodegenerative diseases[11,30-33]. These clinical hazards led to investigation of novel and potent antioxidant compounds from actinobacteria and someStreptomycesstrains, which were isolated from soil or ocean and have been reported to have prominent antioxidant activity[34-37]. In order to find the novel antioxidant compounds, we isolated actinomycetes from mountain forest soil in Korea. The soil-borne actinobacteria,Streptomycessp. strain MJM 10778, exhibited potential antioxidant activity against DPPH, NO, and hydrogen peroxide radicals. The 16S rDNA sequencing was conducted to trace bacterial phylogeny and determine the taxonomy. Hence, the potential strain was identified asStreptomycessp. strain MJM 10778 and it shows 99.9% similarity withStreptomyces misionensisNBRC 13063. However, the strain MJM 10778 showed differences in morphological and physiological properties. The previous study reported that the aerial mass color of strain NBRC 13063 is light grayish brown or light grayish reddish brown on ISP-2, ISP-3, ISP-4, and ISP-5 agar, and this strain did not produce exopigments in ISP-2, ISP-3, ISP-4, and ISP-5 agar[38]. Even though the strain MJM 10778 shows high 16S rDNA sequence homology with the strain NBRC 13063, its aerial mass color is white, grayish brown, and light grayish brown in ISP-2 and 5, ISP-3, and ISP-4 agar, respectively. Moreover, the strain MJM 10778 produced orange brown colored pigment in ISP-2 agar. These phenetic results support the classification of the isolate MJM 10778 as a new strain.
The reducing power assay showed that the EtOAc extract of the strain MJM 10778 possess significant antioxidant activity. In order to understand the antioxidant mechanisms of the strain MJM 10778 EtOAc extract, several antioxidant tests were conducted, such as donating hydrogen to radicals, reducing power, free radical scavenging activity, and quenching singlet oxygen. The antioxidant tests of the strain MJM 10778 EtOAc extract demonstrated that it possess the significant DPPH free radical scavenging at 500 μg/mL (81.6%), nitric oxide free radical scavenging at 1 000 μg/mL (95.4%), and ABTS free radical scavenging at 500 μg/mL (83.4%).
In all antioxidant tests, the ascorbic acid was used as a positive control and it showed 96.6% of DPPH free radical scavenging at 31.25 μg/mL, 94.6% of nitric oxide free radical scavenging at 1 000 μg/mL, and 83.4% of ABTS free radical scavenging at 500 μg/mL. These results demonstrate that the EtOAc extract of the strain MJM 10778 has similar antioxidant activity to ascorbic acid in the free radical scavenging of DPPH, nitric oxide, and hydrogen peroxide.
The cytotoxicity of the strain MJM 10778 EtOAc extract was determined by MTT assay on RAW 264.7 cells, and it was measured as low cytotoxicity (81.50% of cell viability at 100 μg/mL).
From the present findings, it was revealed that the EtOAc extract ofStreptomycessp. strain MJM 10778, which was isolated from mountain forest soil, has strong antioxidant capacity on DPPH, nitric oxide, and hydrogen peroxide free radicals. The low cytotoxicity of the extract gives a possibility of being used in the clinical setting as a therapeutic agent of diseases that are caused by free oxygen radicals. Further studies are being carried out for the isolation of single compound with antioxidant activities from the EtOAc extract ofStreptomycessp. strain MJM 10778.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgements
This work was supported by a grant from next-generation BioGreen 21 project (No. PJ009643), RDA Korea.
[1] Kuster E. The actinomycetes. In: Burges A, Raw F, eds. London: Academic Press; 1968, p.111-124.
[2] Muiru WM, Mutitu EW, Mukunya DM. Identification of selected actinomycetes isolates and characterization of their antibiotic metabolites. J Biol Sci 2008; 8: 1021-1026.
[3] Kieser T, Bibb MJ, Chater KF, Hopwood DA. Practical Streptomyces genetics. UK: John Innes Foundation; 2000.
[4] Tiwari K, Gupta RK. Rare actinomycetes: a potential storehouse for novel antibiotics. Crit Rev Biotechnol 2012; 32(2): 108-132.
[5] Thakur D, Bora TC, Bordoloi GN, Mazumdar S. Influence of nutrition and culturing conditions for optimum growth and antimicrobial metabolite production by Streptomyces sp. 201. J Med Mycol 2009; 19: 161-167.
[6] Ohnish Y, Seo JW, Horinouchi S. Deprogrammed sporulation in Streptomyces. FEMS Microbiol Lett 2002; 216(1): 1-7.
[7] McCarthy AJ, Williams ST. Actinomycetes as agents of biodegradation in the environment-a review. Gene 1992; 115(1-2): 189-192.
[8] Brown ME, Chang MC. Exploring bacterial lignin degradation. Curr Opin Chem Biol 2014; 19C: 1-7.
[9] Schrey SD, Tarkka MT. Friends and foes: streptomycetes as modulators of plant disease and symbiosis. Antonie Van Leeuwenhoek 2008; 94(1): 11-19.
[10] Pornthip R, Akira Y, Saisamorn L. Characterization of Streptomyces strain CMU-MH021, a nematicidal actinomycetes isolated from plant-parasitic nematode-infested soil in northern Thailand. Ann Microbiol 2012; 62(4): 1447-1452.
[11] Valko M, Leibfritz D, Moncol J, Cronin MT, Mazur M, Telser J. Free radicals and antioxidants in normal physiological functions and human disease. Int J Biochem Cell Biol 2007; 39: 44-84.
[12] Tiwari AK. Imbalance in antioxidant defence and human diseases: Multiple approach of natural antioxidant therapy. Curr Sci 2001; 9: 1179-1186.
[13] Kawahara T, Izumikawa M, Otoguro M, Yamamura H, Hayakawa M, Takagi M, et al. JBIR-94 and JBIR-125, antioxidative phenolic compounds from Streptomyces sp. R56-07. J Nat Prod 2012; 75(1): 107-110.
[14] Nanjundan J, Chokkalingam Uvarani, Ramasamy R, Devedasan V, Ponnusamy M. Natural occurrence of organofluorine and other constituents from Streptomyces sp. TC1. J Nat Prod 2014; 77(1): 2-8.
[15] Goodfellow J, Cross T. Classification. In: The biology of the Actinomycetes. Goodfellow M, Mordarski M (Eds.). London: Academic Press; 1984, p. 7-164.
[16] Gottlieb D. An evaluation of criteria and procedures used in the description and characterization of Streptomyces: A cooperative study. Appl Microbiol 1961; 9: 55-65.
[17] Lane DJ. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M. Nucleic acid techniques in bacterial systematics. London: Wiley; 1991.
[18] Thomson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 1997; 25: 4876-4882.
[19] Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 2013; 30: 2725-2729.
[20] Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4: 406-425.
[21] Tajima F, Nei M. Estimation of evolutionary distance between nucleotide sequences. Mol Biol Evol 1984; 1: 269-285.
[22] Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 1985; 39(4): 783-791.
[23] Guha G, Rajkumar V, Ashock Mumar R, Mathew L. Aqueous extract of Phyllanthus amarus inhibits Chromium (VI)-induced toxicity in MDA-MB-435S cells. Food Chem Toxicol 2010; 48: 396-401.
[24] Oyaizu M. Studies on products on browning reaction prepared from glucose amine. Japanese J Nut 1986; 44: 307-315.
[25] Diaz P, Jeong SC, Lee S, Khoo C, Koyyalamudi SR. Antioxidant and anti-inflammatory activities of selected medicinal plants and fungi containing phenolic and flavonoid compounds. Chin Med 2012; 7(1): 26.
[26] Marcocci L, Maguire JJ, Droy-Lefaix MT, Packer L. The nitric oxide-scavenging properties of Ginkgo biloba extract EGb 761. Biochem Biophys Res Commun 1994; 15: 748-755.
[27] Zhishen J, Mengcheng T, Jianming W. The determination of flavonoid contents on mulberry and their scavenging effects of superoxide radical. Food Chem 1999; 64: 555-559.
[28] Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J Immunol Methods 1983; 65: 55-63.
[29] Oktay M, Gülçin İ, Küfrevioğlu Öİ. Determination of in vitro antioxidant activity of fennel (Foeniculum vulgare) seed extracts. Lebensm Wiss Technol 2003; 36: 263-271.
[30] Chen AF, Chen DD, Daiber A, Faraci FM, Li H, Rembold CM, et al. Free radical biology of the cardiovascular system. Clin Sci (Lond) 2012; 123(2): 73-91.
[31] Jomova K, Valko M. Importance of iron chelation in free radicalinduced oxidative stress and human disease. Curr Pharm Des 2011; 17(31): 3460-3473.
[32] Brieger K, Schiavone S, Miller FJ Jr, Krause KH. Reactive oxygen species: from health to disease. Swiss Med Wkly 2012; 142: w13659.
[33] Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic Biol Med 2010; 49(11): 1603-1616.
[34] He F, Yang Y, Yang G, Yu L. Components and antioxidant activity of the polysaccharide from Streptomyces virginnia H03. Z Naturforsch C 2008; 63:181-188.
[35] Thenmozhi M, Sindhura S, Kannabiran K. Characterization of antioxidant activity of Streptomyces species VITTK3 isolated form Puducherry Coast, India. J Adv Sci Res 2010; 1: 46-52.
[36] Kawamura T, Izumikawa M, Otoguro M, Yamamura H, Hayakawa M, Takagi M, et al. JBIR-94 and JBIR-125, Antioxidative phenolic compound from Streptomyces sp. R56-07. J Nat Prod 2012; 75: 107-110.
[37] Dharmik PG, Gomashe AV. isolation, identification and antioxidant activity of melanin pigment from Actinomycete (Streptomyces species) isolated from garden soil, Nagpur District, India. Int J Pure Appl Sci Technol 2013; 18: 69-72.
[38] Shirling EB, Gottlieb D. Cooperative description of type cultures of Streptomyces. IV. Species descriptions from the second, third and fourth studies. Int J Syst Bacteriol 1969; 19: 391-512.
ment heading
10.1016/S1995-7645(14)60170-X
#These authors contributed equally to this work.
*Corresponding author: J.-W. Suh, PhD, Professor, Division of Bioscience and Bioinformatics, Myongji University, Youngin-si, Gyeonggi-do 449728, Korea.
Tel: 82-31-330-6881
E-mail: jwsuh@mju.ac.kr
S. H. Yang, PhD, Professor, Center for Neutraceutical and Pharmaceutical Materials, Myongji University, Yongin-si, Gyeonggi-do 449728, Korea.
Tel: +82-31-330-6880
E-mail: ymichigan@mju.ac.kr
Foundation project: This work was supported by a grant from next-generation BioGreen 21 project (No. PJ009643), RDA Korea.
Antioxidant activity
DPPH radical scavenging assay
NO radical scavenging assay
 Asian Pacific Journal of Tropical Medicine2014年12期
Asian Pacific Journal of Tropical Medicine2014年12期
- Asian Pacific Journal of Tropical Medicine的其它文章
- Chikungunya virus, epidemiology, clinics and phylogenesis: A review
- Induction of deletion mutation on ompR gene of Salmonella enterica serovar Typhi isolates from asymptomatic typhoid carriers to evolve attenuated strains for vaccine development
- Evaluation of protective effect of IL-22 and IL-12 on cutaneous leishmaniasis in BALB/c mice
- Immunogenic potential and protective efficacy of formalin inactivated circulating Indian strain of West Nile virus
- Genetic diversity and gene structure of mitochondrial region of Anopheles minimus (Diptera: Culicidae) - major malaria vector of North east India
- Fumigant and repellent properties of sesquiterpene-rich essential oil from Teucrium polium subsp. capitatum (L.)
