First report on natural Leishmania infection of Phlebotomus sergenti due Leishmania tropica by high resolution melting curve method in Southeastern Iran
Aghaei Afshar A, Rassi Y, Sharifi I*, Vatandoost H, Mollaie HR, Oshaghi M.A, Abai MR, Rafizadeh S
1Leishmaniasis Research Center, Kerman University of Medical Sciences, Kerman, Iran
2Department of Medical Entomology & Vector Control, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran
3Department of Medical Virology, Tehran University of Medical Sciences, Tehran, Iran
4Department of Deputy of Strategic Planning, Reference Health Laboratories Research Center, Deputy of Treatment, Ministry of Health & Medical Education of Health, Tehran, Iran
First report on natural Leishmania infection of Phlebotomus sergenti due Leishmania tropica by high resolution melting curve method in Southeastern Iran
Aghaei Afshar A1, Rassi Y2*, Sharifi I1*, Vatandoost H2, Mollaie HR3, Oshaghi M.A2, Abai MR2, Rafizadeh S4
1Leishmaniasis Research Center, Kerman University of Medical Sciences, Kerman, Iran
2Department of Medical Entomology & Vector Control, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran
3Department of Medical Virology, Tehran University of Medical Sciences, Tehran, Iran
4Department of Deputy of Strategic Planning, Reference Health Laboratories Research Center, Deputy of Treatment, Ministry of Health & Medical Education of Health, Tehran, Iran
Objective: To identify the Leishmania species in infected sand flies by Real-time PCR coupled with HRM analysis. Methods: Real-time PCR coupled with HRM analysis targeting the first internal transcribed spacer (ITS1) of nuclear ribosomal DNA as the genetic marker was used to identify and distinguish Leishmania species in sand flies specimens. Results: Three out of 115 females of Phlebotomus sergenti (P. sergenti) (2.6%) were positive to Leishmania tropica (L. tropica).
Conclusions: This is the first report on P. sergenti as the main and proven vector of anthroponitic cutaneous leishmaniasis in Dehbakri County using Real-time PCR coupled with HRM analysis. This method is rapid, sensitive and specific for diagnosing of parasites in infected Sand flies and ideal for large scale genotyping projects.
ARTICLE INFO
Article history:
Received 9 October 2013
Received in revised form 19 December 2013
Accepted 15 January 2014
Available online 20 February 2014
Vector(s)
1. Introduction
Cutaneous leishmaniasis(CL) is endemic in many parts of the world and considered as a major public health problem[1]. The disease is one of the main health problem in Iran. Zoonotic cutaneous leishmaniasis (ZCL) due toLeishmaniamajor (L. major) is endemic in 50% of the 31 provinces of Iran, whereas anthroponotic cutaneous leishmaniasis (ACL) associated withLeishmania tropica(L. tropica) is an old endemic disease in 8 provinces, especially in South East Iran[2-6].
Two epidemiological forms of CL occur in Bam district, ACL is mainly limited to the city, with its main reservoir of host of human. Anthroponotic Cutaneous Leishmaniasis is a serious problem due to its difficulty of treatment and longer duration, hence potentially sever long-term complications[7-9]. However ZCL is the more prevalent form. After the massive earthquake of Bam in December 2003 the prevalence of CL cases increased among school children. It increased from 2% in 2005 to 5% in 2008 (unpublished data). The disease is outspread and a new emerging focus of cutaneous leishmaniasis was confirmed in Dehbakri County, adjacent to Bam. According to published data, this paper is the first report on identifying of responsible sand flies to transmitting of disease agent to human, employing Realtime PCR coupled with HRM analysis.
In many areas, however, despite considerable research on these diseases, the main ‘reservoir’ hosts and the species of sand fly responsible for most transmission have still to beidentified. In many foci of CL there is at least one species of sand fly that is sufficiently common and anthropophilic to be considered a probable vector, although good evidence to support this belief, such as the detection, in wild caught females of this species, of the parasites causing the CL, is lacking. The prevalence of infection in even the primary vector may be quite low, making the detection of sand fly infection, and particularly the detection of any temporal trends in the prevalence of sandfly infection, difficult, especially if dissection and microscopy constitute the detection method. Furthermore this traditional methods do not discriminate the organism in species level and often with variable and low sensitivity[10,11].
Molecular methods are currently being employed to detect Leishmania infection up to genus, complex or species level[12,13]. High-resolution melting (HRM) analysis is a relatively new technique that allows direct characterization of PCR amplicons in a closed system. It measures changes in the fluorescence intensity of a DNA intercalating dye during dissociation from double-stranded DNA to single-stranded DNA and can differentiate between single nucleotide polymorphisms (SNP)[14].
Real-time PCR offers several advantages over traditional PCR, including faster processing time, higher sensitivity and decreased contamination risk. A previous study used HRM PCR assay for Old WorldLeishmaniabased on gene 7SLHRM that can be used to differentiate betweenL. tropica,L. major, and theLeishmania donovanicomplex[15].
PCR processing. Nowadays, the HRM has mostly been used in human clinical studies[16-20]. However, the application of the HRM technique to the diagnosis of parasitic organisms has been rather limited and the method has mostly been applied in molecular studies of parasitic protozoa such as the old worldLeishmaniaspp[14]. The aim of the present study was to evaluate the potential use of ITS1 gene for identifying Old WorldLeishmaniaspecies by HRM method based real-time PCR. The method was successfully applied to identifyLeishmaniaspecies in sand flies collected from Dehbakri county, Bam, South eastern Iran.
2. Material and methods
2.1. Study area
This study was carried out in Dehbakri county, Bam district, Kerman province during summer of 2011. Dehbakri is located 60 km west of Bam city. This area has a semidesert climate temperate in summer and cold at winter. The mean of monthly maximum and minimum temperatures were 40 ℃ and -5 ℃ in July and Dec, respectively. The total annual rainfall was 220 mm. The minimum and maximum of monthly relative humidity were 45% and 92%, respectively in July and January.
2.2. Sample collection and identification
Sand flies were caught using aspirator from 8.00 pm till 2.00 am during summer 2011 where cases of ACL had been reported. The caught sand flies were transfered to the entomological cage and then were kept by wet towel and transported to laboratory research center for identification and Detection of Leishmania species. All the collected sand flies were mounted separately using Puri’s media for species identification. Male and female were considered separately[21-23].
2.3. DNA extraction
Sand flies were washed in 1% detergent (washing-up liquid) solution for 5 min, Heads and last abdominal segments were kept for morphological identification based on the keys described by Legeret al., 1986[24]. Individual code names, consisting of a letter(s) taken from the collection area name followed by a number were assigned. DNA was extracted by maceration in a micro tube using a plastic pestle, followed by addition of 35 μL of lysis buffer (100 mM TRIS-HCl, 100 mM NaCl, 25 mM EDTA, 0.5% SDS, pH 8.0) and another maceration step. The test samples were digested overnight at 37 ℃ by proteinase K (1.25 μL of a 10 mg/mL solution) and the DNA extracted by phenol-chloroform. The DNA pellet was resuspended in 20 μL of TE (10 mM TRISHCl pH 8.0, 1 mM EDTA). Ten μL were used to estimate the DNA concentration and purity at 280 and 260 nm in a spectrophotometer, then discarded. The remaining 10 μL were stored at -20 ℃ until use. Double distilled water and DNA fromL. majorandL. tropicaprovided to the Iran Institute of Pasteur by the World Health Organization were used as negative and positive controls.
2.4. HRM-real-time PCR assay
We designed two primers AGCTGGATCATTTTCCGATG and ATCGCGACACGTTATGTGAG using the software Beacon as forward and reverse respectively. Real-time PCR was performed in a total reaction mixture of 20 mL containing 10 mL of HRM Master Mix, 10 pmole of each primer, approximately 10 ng/mL of genomic DNA and sterile deionized water using a 7500 fast real-time PCR system. Infection was detected by internal transcribed spacer 1 (ITS1) real-time PCR and high-resolution melt analysis (ITS1-HRM) PCR. All samples were tested in duplicates and results were compared with those from HRM analysis of positive controls for each assay. Positive samples also were verified by kDNA PCR.
3. Results
3.1. Sand fly identification
A total of 1 261 sand flies were captured and identified to the species level. Fourteen species including, 5 species of Phlebotomus genus and 9 species ofSergentomyiagenus were identified. They werePhlebotomus sergenti (P. sergenti),Phlebotomus papatasi(P. papatasi), Phlebotomus monglensis, Phlebotomus major, Phlebotomus longiductus,Sergentomyia sintoni, Sergentomyia baghdadis, Sergentomyia sumbarica, Sergentomyia africana,Sergentomyia dentata, Sergentomyia squemipleuris, Sergentomyia grekovi, Sergentomyia mervynaeandSergentomyia antennata. Two species ofP. sergenti(67.8%) andP. papatasi(19.4) were the dominant specimens respectively.
3.2. Leishmania species identification
We tested 164 specimens of sand flies including 115 Ph. sergenti and 49P. papatasiby ITS1-HRM PCR. The overallLeishmaniainfection rate forP. sergentiwas 2.6% (3/115) and allP. papatasisamples were negative. Positive specimens by ITS1-HRM PCR were also positive by kDNA PCR and produced a 750 bp kDNA product. Infected sand flies had been collected from indoors places with empty position of abdomen. This is the first report on natural infection ofP. sergentitoL. tropicausing high resolution melting curve (HRM) method in Iran. According our results this species was the only infected sand fly and it seems was responsible to transmitting ofL. tropicaamong human.

Figure 1. Differentiation L. tropica positive control and L. major positive control using HRM method.
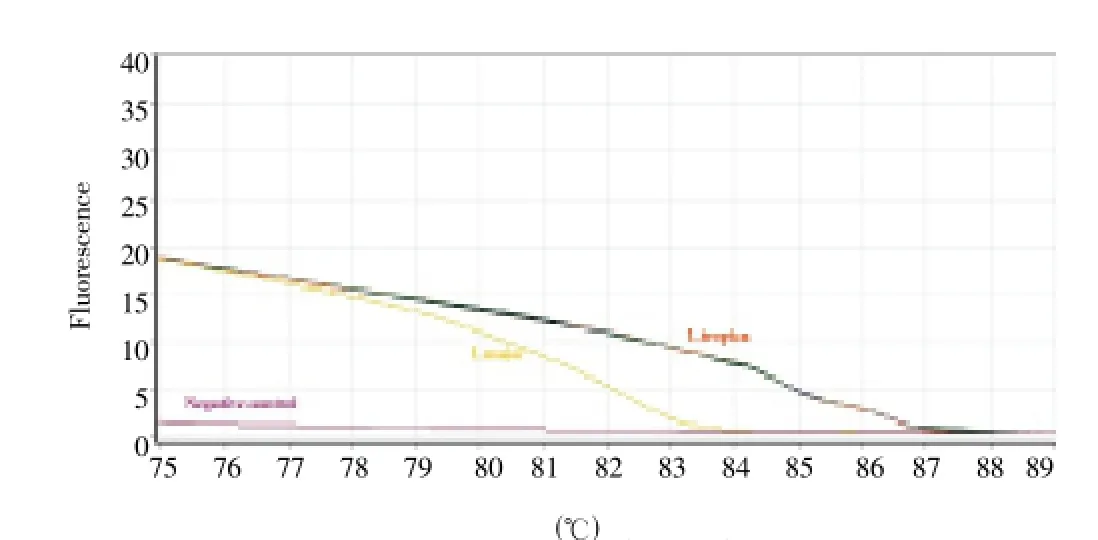
Figure 2. Representative curve of the melting curves of ITS-1 amplicons for infected P. sergenti species to L. tropica, Dehbakri County, Southeastern Iran, 2011.
4. Discussion
Control of leishmaniasis need to understand on ecology and epidemiology of disease in endemic areas. There is a major problem for epidemiologists both in identification of reservoir hosts and in detection of vectors. Therefore, finding naturally infected sand flies is essential in identifying a species as a vector ofLeishmaniaand in studying infection rates in areas of endemicity[25, 26].
The applicability of kDNA for the detection and identification ofLeishmaniawithin sand flies by DNA hybridization have been shown previously[27]. In recent years, several molecular techniques, mainly those based on PCR have been developed for the specific identification and characterization ofLeishmaniaspecies[28-30]. Although other techniques are sensitive and specific for identification ofLeishmaniaspecies, they are laborious and time consuming, especially the post-PCR processing steps. In addition, there is also a higher risk of contamination, more expensive (e.g., DNA sequencing) and the techniques only provide qualitative information. A highly sensitive method is needed to determine sand fly infection ofLeishmaniaparasite. The choice of the gene used for HRM analysis is important in developing a successful assay, since it can take advantage of small differences in melting curves to distinguish between organisms with highly homologous sequences[14].
The ITS1 gene can be used for diagnosis of human leishmaniasis, as well as epidemiological studies on potential reservoir hosts, sand fly vectors, and parasite genotypes[12]. In the present study, we have successfully utilized HRM analysis along with real-time PCR for the rapid detection and species identification of Leishmania species in sand flies samples. To the best of our knowledge, this is the first report on the utilization of the HRM approach for rapid detection and discrimination of sand flies species by employing the ITS1 of nuclear ribosomal DNA as a genetic marker in Iran. We reportL. tropicainfections inP. sergenti(2.6%) specimens, the only proven vector ofL. tropicain Iran.
In conclusion, using HRM assay for identifying of Old WorldLeishmaniaspecies is rapid, sensitive, specific, and simple and can be used to directly diagnose parasites in the sand flies vector with a minimum of operator post-PCR manipulation and interpretation.
Conflict of interest statement
We declare that we have no conflict of interest.
Acknowledgments
This investigation was supported by the Iranian Center of Diseases Management, Ministry of Health and MedicalEducation. We wish to express our sincere thanks for providing facilities needed for this study. The authors would like to thank the Leishmaniasis Research Center, Kerman University of Medical Sciences and the School of Public Health, Tehran University of Medical Sciences for their close collaboration. This study was financially supported by School of Public Health, Tehran University of Medical Sciences, and Leishmaniasis Research Center, Kerman University of Medical Sciences, project No.10487
[1] Desjeux P. Leishmaniasis: current situation and new perspectives. Comp Immunol Microbiol Infect Dis 2004; 27: 305-318.
[2] Rassi Y, Javadian E, Jalali M, Motazedian MH, Vatandoost H. Investigation on zoonotic cutaneous leishmaniasis, southern Iran. Iran J Public Health 2004; 33(1): 31-35.
[3] Rassi Y, Javadian E, Amin M, Rafizadeh S, Vatandoost H, Motazedian H. Meriones libycus is the main reservoir of zoonotic cutaneous leishmaniasis in south Islamic Republic of Iran. East Med Health 2006; 12(3-4): 475-457.
[4] Rassi Y, Gassemi MM , Javadian E, Rafizadeh S, Motazedian H, Vatandoost H. Vectors and reservoirs of cutaneous leishmaniasis in Marvdasht district, southern Islamic Republic of Iran. Est Mediterr Health J 2007; 3(3): 686-693.
[5] Rassi Y, Oshaghi MA, Mohammadi Azani S, Abaie MR, Rafizadeh S, et al. Molecular detection of Leishmania infection due to Leishmania major and Leishmania turanica in the vectors and reservoir host in Iran. Vector-borne Zoonotic Dis 2011; 11: 145-150.
[6] Nadim A. Epidemiology of leishmaniasis in Iran, leishmania parasites and leishmaniases. Tehran, Iran: University press; 2008, p. 191-211.[In Persian]
[7] Aghaei Afshar A, Rassi Y, Sharifi I, Abai MR, Vatandoost H, Oshaghi MA, et al. Susceptibility status of Phlebotomus papatasi and P. sergenti (Diptera: Psychodidae) to DDT and Deltamethrin in a focus of cutaneous leishmaniasis after earthquake strike in Bam, Iran. J Arthropod-Borne Dis 2011; 5(2): 32-41.
[8] Sharifi I, Zemani F, Aflatoonian MR, Fekri AR. An epidemic of cutaneous leishmaniasis in Baft district in Kerman Province and its probable causative risk factors. Iranian J Epidemiol 2008; 4(1): 53-58.
[9] Sharifi I, Poursmaelian S, Aflatoonian MR, Ardakani RF, Mirzaei M, Fekri AR, et al. Emergence of a new focus of anthroponotic cutaneous leishmaniasis due to Leishmania tropica in rural communities of Bam district after the earthquake, Iran. Trop Med Int Health 2011; 16(4): 510-513.
[10] Hajjaran H, Vasigheh F, Mohebali M, Rezaei S, Mamishi S, Charedar S. Direct diagnosis of Leishmania species on serosity materials punctured from cutaneousleishmaniasis patients using PCR-RFLP. J Clin Lab Anal 2011; 25(1): 20-24.
[11] Shahbazi F, Shahabi S, Kazemi B, Mohebali M, Abadi AR, Zare Z. Evaluation of PCR assay in diagnosis and identification of cutaneous leishmaniasis: a comparison with the parasitological methods. Parasitol Res 2008; 103(5): 1159-1162.
[12] Bensoussan-Hermano E, Nasereddin A, Jonas F, Schnur LF, Jaffe CL. Comparison of PCR assays for diagnosis of cutaneous leishmaniasis. J Clin Microbiol 2006; 44: 1435-1439.
[13] Nasereddin A, Bensoussan-Hermanoe, Schoniane G, Baneth G, Jaffe CL. Molecular diagnosis of Old World cutaneous leishmaniasis and species identification by use of a reverse line blot hybridization assay. J Clin Microbiol 2008; 46: 2848-2855.
[14] Talmi-Frank D, Nasereddin A, Schnur LF, Schonian G, Töz SO. Detection and identification of Old World Leishmania by high resolution melt analysis. PLoS Negl Trop Dis 2010; 4(1): e581.
[15] Nasereddin A, Jaffe CL. Rapid diagnosis of Old World Leishmaniasis by high-resolution melting analysis of the 7SL RNA gene. J Clin Microbiol 2010; 2240-2242.
[16] Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High resolution genotyping by amplicon melting analysis using LC Green. Clin Chem 2003; 49: 853-860.
[17] Zhou L, Vandersteen J, Wang L, Fuller T, Taylor M. Highresolution DNA melting curve analysis to establish HLA genotypic identity. Tissue Antigens 2004; 64: 156-164.
[18] Liew M, Pryor R, Palais R, Meadows C, Erali M. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin Chem 2004; 50: 1156-1164.
[19] Radvansky J, Resko P, Surovy M, Minarik G, Ficek A. Highresolution melting analysis for genotyping of the myotonic dystrophy type1 associated Alu insertion/deletion polymorphism. Anal Biochem 2010; 398: 126-128.
[20] Saitsu H, Kato M, Okada I, Orii KE, Higuchi T. STXBP1 mutations in early infantile epileptic encephalopathy with suppression-burst pattern. Epilepsia 2010; 51: 2397-2405.
[21] Rassi Y, Hanafi-Bojd AA. Phlebotomine sand flies, vectors of Leishmaniases: Morphology, biology, ecology, and field and laboratory methods with pictorial key of Iranian sand flies. Noavaran-Elm Publication 2006. (In Persian)
[22] Lewis DJ. A taxonomic review of the genus Phlebotomus. Bull Br Mus 1982; 45: 121-209.
[23] Theodor O, Mesghali A. On the phlebotominae of Iran. J Med Entomol 1964; 1: 285-300.
[24] Leger NB, Pesson G, Madulo-Leblond G. Les phlebotomes de Grece. Biol Gallo-Hellenica 1986; 11: 165-192.
[25] Killick-Kendrick R. Phlebotomine vectors of the leishmaniases: a review. J Med Vet Entomol 1990; 4: 1-24.
[26] Sharma U, Singh S. Insect vectors of Leishmania: distribution, physiology and their control. J Vector Borne Dis 2008; 45: 255-272.
[27] Ready PD, Lainson R, Shaw JJ, Souza AA. DNA probes for distinguishing Psychodopygus wellcomi from Psychodopygus complexus (Diptera: Psychodidae). Memrias do Instituto Oswaldo Cruz 1991; 86: 41-49.
[28] Nicolas L, Milon G, Prina E. Rapid differentiation of Old World Leishmania species by LightCycler polymerase chain reaction and melting curve analysis. J Microbiol Methods 2002; 51(3): 295-299.
[29] Nicolas L, Prina E, Lang T, Milon G. Real-time PCR for detection and quantitation of Leishmania in mouse tissues. J Clin Microbiol 2002; 40(5): 1666-1669.
[30] Rotureau B, Joubert M, Clyti E, Djossou F, Carme B. Leishmaniasis among gold miners, French Guiana. Emerg Infect Dis 2006; 12(7): 1169-1170.
*Corresponding author: Professor Yavar Rassi, Department of Medical Entomology & Vector Control, School of Public Health, Tehran University of Medical Sciences, Tehran, Iran.
E-mail: rassiy@tums.ac.ir
Professor Iraj Sharifi, Leishmaniasis Research Center, Kerman University of Medical Sciences, Kerman, Iran.
E-mail: iraj.sharifi@yahoo.com
Leishmania tropica
Real-time PCR
Iran
 Asian Pacific Journal of Tropical Medicine2014年2期
Asian Pacific Journal of Tropical Medicine2014年2期
- Asian Pacific Journal of Tropical Medicine的其它文章
- Bond strength analysis of the bone cement- stem interface of hip arthroplasties
- Hepatic effect of NAC on sevear acute pancteatise of rats
- Comparative analysis of different cyclosporine A doses on protection after myocardial ischemia/reperfusion injury in rat
- Comparison on serum biomarkers for anovulatory and ovulatory dysfunctional uterine bleeding in Lizu females
- Preparation of novel biodegradable pHEMA hydrogel for a tissue engineering scaffold by microwave-assisted polymerization
- Mathematical modeling for selecting center locations for medical and health supplies reserve in Hainan Province
