Identification of an AP2 gene related to open flowering in diploid wheat (Triticum monococcum)
ShunZong Ning , Ning Wang , Shun Sakuma , Mohammad Pourkheirandish ,Takato Koba , Takao Komatsuda *
1. National Institute of Agrobiological Sciences (NIAS), Plant Genome Research Unit, 2-1-2 Kannondai, Tsukuba,Ibaraki 305-8602, Japan
2. Graduate school of Horticulture, Chiba University, 648 Matsudo, Matsudo, Chiba 271-8510, Japan
1 Introduction
Cleistogamy, a trait which involves the shedding of pollen within an unopened flower, has been reported in some 693 angiosperm species, distributed over 228 genera and 50 families (Cully, 2002; Cully and Klooster,2007). In some cases, it can also be induced by poor environmental conditions, such as drought, low intensity light and low temperature (Langer and Wilson, 1964;Waller, 1979; Cully, 2002; Barthet al., 2006). The trait has some advantageous features in the context of crop plants, since it ensures genetic integrity and impedes gene flow via the pollen, the latter seen as a significant issue for genetically modified varieties (Daniell, 2002).
Non-cleistogamy in monocotyledonous plants generally relies on a functioning lodicule, a structure homologous with the dicotyledonous petal (Glover, 2007). The grass floret features a pair of lodicules lying between the lemma and the ovary base, and their rapid expansion around the time of anthesis pushes aside the rigid lemma and so allows for the extrusion of the anthers and stigma.When lodicule function is compromised, the result is cleistogamy (Heslop-Harrison and Heslop-Harrison,1996). In barley, lodicule function is determined by the allelic status at the major geneCly1; cleistogamy occurs when the recessive alleleCly1is present in the homozygous state. The isolation ofCly1hasdemonstrated that its sequence includes two domains which are characteristic forAPgenes, and that it also includes a putative targeting site for miR172, a non-coding microRNA (Nairet al., 2010). In theCly1allele, a synonymous single nucleotide change within the miR172 targeting site has been demonstrated as being responsible for the cleistogamous phenotype.
Wheat, and particularly its hexaploid form bread wheat (Triticum aestivum), is one of the world’s fore-most crop plants (Feldmanet al., 1995; Gustafsonet al.,2009). A cultivated form of diploid wheat (einkorn wheat,T. monococcum) is one of hulled wheat crop species which are underutilized in spite of their well-recognised genetic and agronomic potentials and still grown as a crop in a few marginal environments (Perrinoet al.,1996). Here, we describe the isolation and characterization of the einkorn wheatCly1/AP2orthologTmAP2.
2 Material and methods
2.1 Plant material and assessment of flowering phenotype
A sample of einkorn wheat (strain KT003-005) was obtained from National Bioresource Project, Kyoto University, Kyoto, Japan. The grain was autumn-sown in the field at Tsukuba, Japan. Three spikes still attached to peduncle and the flag leaf were sampled for analysis immediately prior to anthesis. The lemma was removed from the first floret of spikelet to facilitate imaging of the lodicules; these images were used to quantify lodicule width and depth with the aid of Makijaku v1.1 software (http://cse.naro.affrc.go.jp/iwatah/). Maintaining the spikes in 100 mg/L 2,4-D for 24 h at room temperature maintained the swollen state of the lodicules for an additional 1–2 days, a procedure which facilitated the assessment of lodicule size.
2.2 PCR primer design, amplification and sequencing
Genomic DNA was extracted from young leaves according to Komatsudaet al.(1998). Appropriate primers were designed using Oligo 6 software (W. Rychlick, National Bioscience, Plymouth, MN, USA) and DNAMAN v6.0 (Lynnon Biosoft, Quebec, Canada). The complete sequence ofTmAP2was amplified from genomic DNA template in the form of three overlapping fragments. The experimental details for the 50 μL reactions used are given in Table 1. The resulting amplicons were electrophoresed through agarose gels made with TBE buffer,and were visualized by ethidium bromide staining. Purification of the amplicons was achieved using a QIAquick PCR purification Kit (QIAGEN, Germantown,MD, USA), and the products were then subjected to cycle sequencing using a Big Dye Terminator kit (Applied Biosystem, Foster, CA, USA). The sequencing reactions were purified by Agencourt CleanSEQ (Beckman, Beverly, MA, USA), and processed through an ABI Prism 3130 genetic analyzer (Applied Biosystems). Sequence data were aligned using DNAMAN v6.0 software.
2.3 cDNA synthesis and quantitative real-time PCR(qRT-PCR)
Total RNA was extracted, using the TRIzol reagent(Invitrogen, Carlsbad, CA), from spikes sampled at the eight developmental stages defined by Kirby and Appleyard (1981), as well as from the leaf, shoot and root of young seedlings. The RNA represented the template for the synthesis of ss cDNA achieved via oligo (dT)priming, according to the Invitrogen RT-PCR first-strand synthesis protocol. Individual transcript abundances were obtained using the StepOne Real-Time PCR system (Applied Biosystems) in conjunction with THUNDERBIRD SYBR qPCR mix kit(Toyobo, Osaka), according to the manufacturers’ protocols. Each amplicon (relevant primer sequences given in Table 1) was inserted into pCR4-TOPO (Invitrogen),and used to generate a standard curve based on a dilution series of 5.1×10-7to 4.0×10-2ng plasmid forTmAP2, and 3.2×10-5to 2.5 ng for the reference gene wheatActin(NCBI accession number CJ932475). At least three independent biological replicates and at least two technical replicates per biological replicate were performed.
2.4 Mapping the miR172-guided cleavage site
Total spike RNA (extracted at the terminal spikelet stage) was subjected to an RNA-ligase mediated 5′RACE (Kasschauet al., 2003) reaction, employing a GeneRacer kit (Invitrogen). This developmental stage was chosen because of its analogy to the stamen primordium stage in barley, the stage when miR172-guided cleavage ofCly1was most clearly detectable (Nairet al., 2010). The dephosphorylation and decapping steps were both omitted, so that only the 5′ends of the truncated transcripts were ligated to the GeneRacer RNA oligomer. A nested PCR was based on a primer targeting the GeneRacer RNA oligomer, initially in combination with a gene-specific reverse primer, and subsequently with an internal gene-specific primer (Table 1). The resulting amplicons were electrophoresed through 1% agarose, inserted into the TA vector (TOPO TA Cloning Kit, Invitrogen), and thence intoE. coli(DH5α) competent cells. Randomly selected clones (without any prior size selection) were chosen for DNA sequencing.
3 Results
3.1 Flower gaping and lodicule swelling in einkorn wheat
The morphology of the pre-anthesis lodicules was normal (Figures 1a, 1c). As anthesis approached, the lodicules swelled (Figures 1b, 1d), forcing open the floret. Between the day prior to anthesis and anthesis itself, lodicule width expanded from 1.1 to 1.4 mm(Figures 1a, 1b) and its depth from 0.4 to 0.95 mm(Figures 1c, 1d).

?

Figure 1 Lodicule development in einkorn wheat prior to (a, c) and at (b, d) anthesis. The double headed arrows indicate the width(a, b), and the depth (c, d) of the lodicule. lo: lodicule. (Bar: 1 mm)
3.2 Isolation and structure of Cly1 homoeolog
The characteristics of theTmAP2sequence (Genbank accession No. AB778536) are given in Table 2.The length of its genomic DNA was 2,720 bp (compared to 2,691 bp forCly1) and its open reading frame(ORF) length was 1,437 bp (Cly1: 1,464 bp). Its predicted gene product comprised 478 residues (Cly1: 487),representing an estimated molecular weight of 50–52 kDa and predicted PI of approximately 7.2.The GC content in theTmAP2ORF (71.6%) was slightly greater than in the barley ORF (70.5%), and considerably greater than in the full genomic DNA (respectively,62.1% and 60.8%). Nucleotide identity between the einkorn and barley orthologs was 78.8% in the genomic DNA and 90.4% in the ORF, while at the peptide level,the extent of the homology reached 89.1%. A comparison of the predicted einkorn wheat and barley polypeptides (Figure 2) shows that their structural organization was identical and they shared several conserved sequence features, namely the central core of theAP2polypeptide includingAP2domains R1 and R2, the α-helix, the highly basic ten residue domain thought to be a nuclear localization signal, the AASSGF box and the motifs 1 through 3 (Jofukuet al., 1994; Tanget al.,2007).
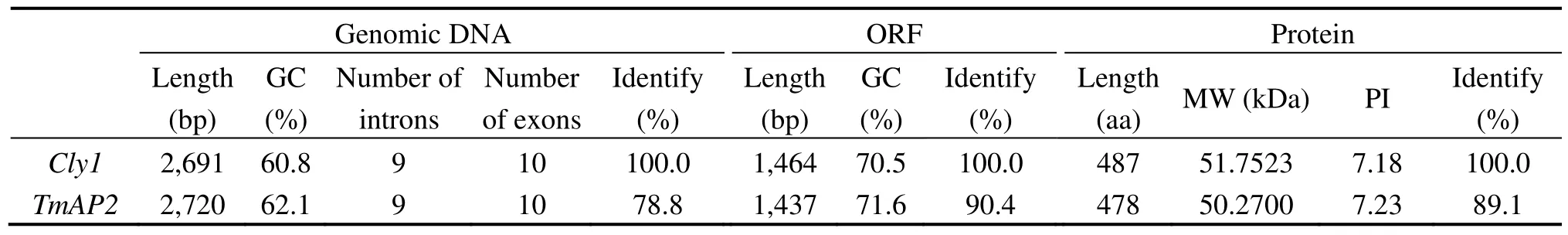
Table 2 Homology between Cly1 and its einkorn wheat ortholog TmAP2
3.3 Transcription profiling of TmAP2
qRT-PCR analysis revealed thatTmAP2was transcribed throughout spike development (from the glume primordium stage until anthesis) (Figure 3). The abundance of the transcript increased as the spike developed.TmAP2transcript was also detectable in leaf, shoot and root tissue, although its abundance was markedly lower than in the spike (Figure 3).
3.4 miR172-guided cleavage of TmAP2 transcript
TheTmAP2sequence shared the miR172 binding site sequence carried by the dominant allele atCly1, except for an A/U polymorphism at the 5' end (Figure 4);this polymorphism also occurs within barley, but it has no effect on flowering habit (Nairet al., 2010). The modified 5' RACE experiment was expected to generate 134 bp fragments from theTmAP2transcript, and this was the case for 83 of the 90 clones analysed; cleavage occurred between the A and U nucleotides (Figure 4), as is also the situation inCly1(Nairet al., 2010). The remaining seven clones represented 3' untranslated region sequence consistent with random mRNA breakage, again as has been observed in barley (Nairet al., 2010).
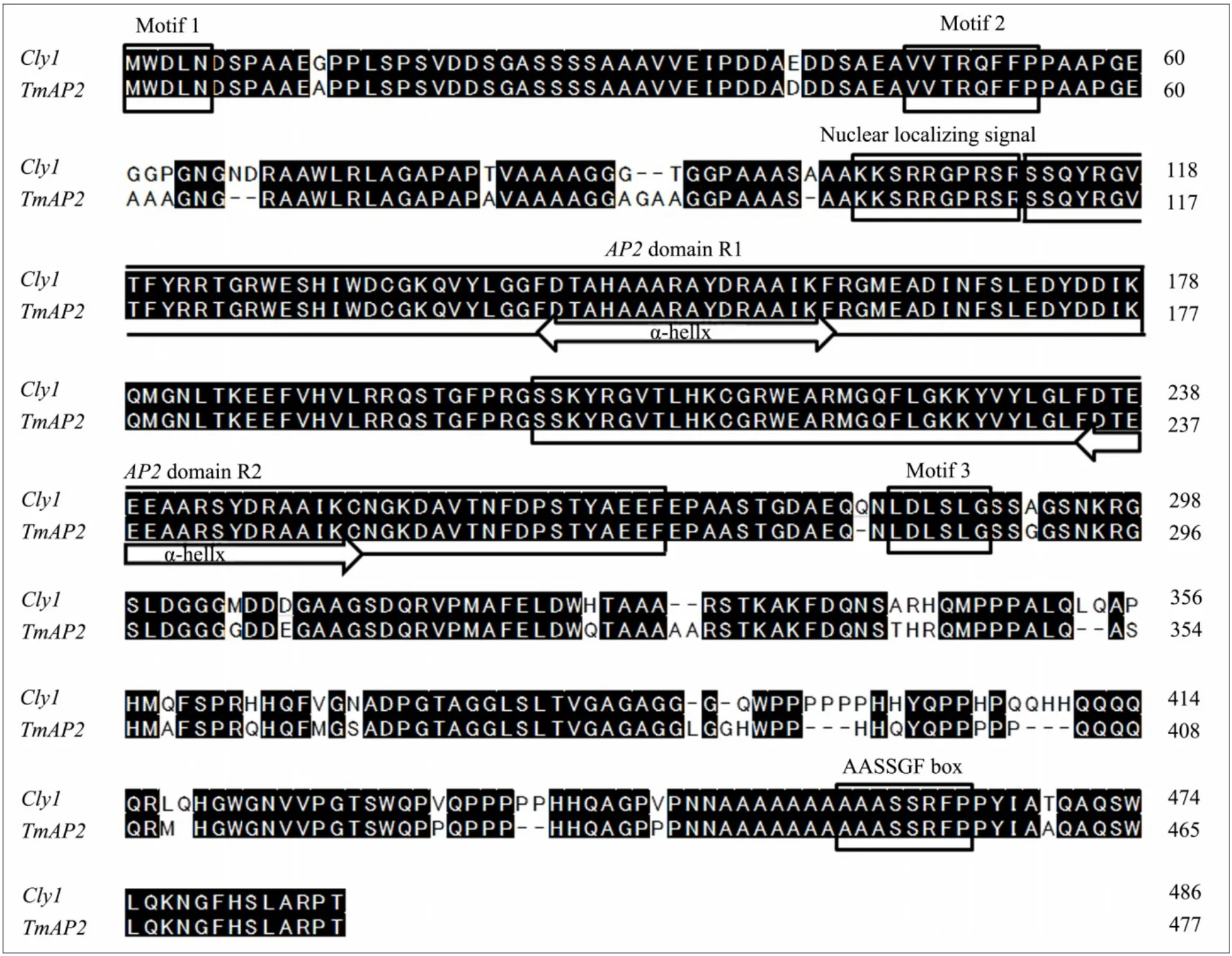
Figure 2 Alignment of the predicted TmAP2 protein from einkorn wheat and Cly1 (ACY29532). The characteristic features motifs 1–3, the nuclear localization signal, the two AP2 domains R1 and R2 and the AASSGF box are shown boxed.The α-helical structure in the core of region of each AP2 domain is shown delimited by arrows

Figure 3 qRT-PCR-based transcription profiles of TmAP2 in the developing spike. 1: glume primordium stage; 2: lemma primordium stage; 3: floret primordium stage; 4: terminal spikelet stage; 5: white anther stage; 6: green anther stage;7: yellow anther stage; 8: anthesis, and in non-floral organs. Actin was used as the reference gene.Values represent the mean ± S.E. (n=3)
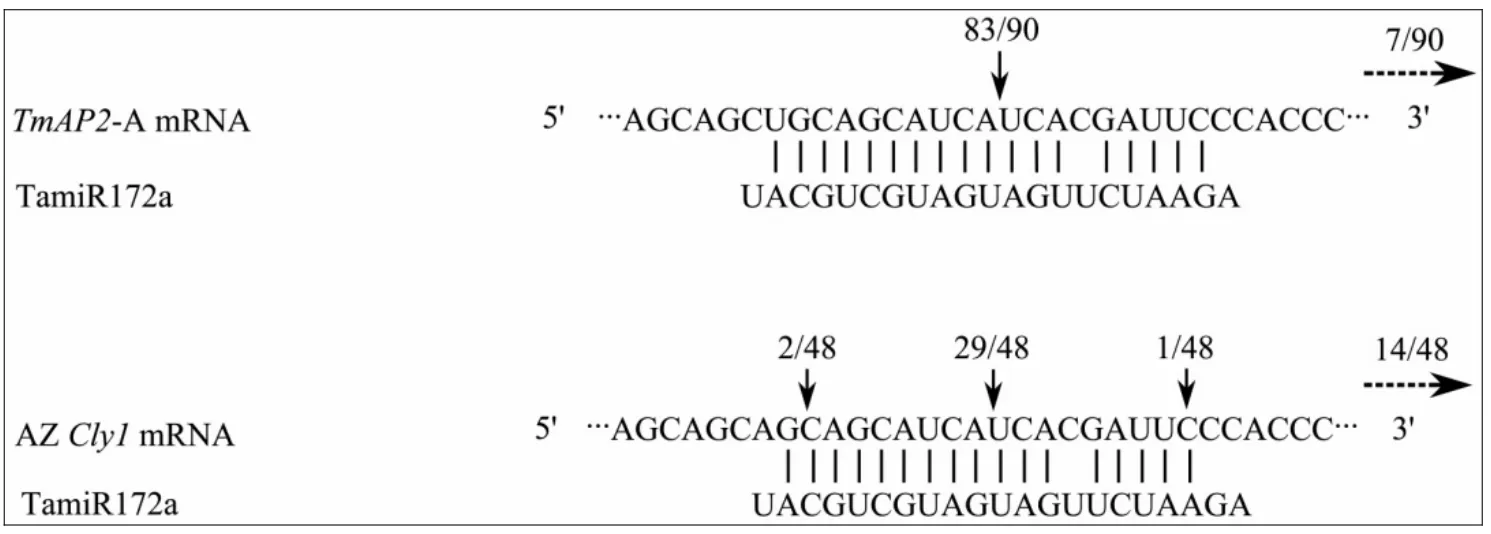
Figure 4 miR172-guided mRNA cleavage of TmAP2 mRNA. The 5' termini of the cleaved products were identified using a modified 5' RACE approach. Vertical arrows indicate the inferred 5' termini of miR172-guided cleavage, and the number above each arrow gives the proportion of clones containing that site. The horizontal arrows refer to cleavage downstream of the miR172 site. The miR172-guided mRNA cleavage of Cly1 mRNA was taken from Nair et al. (2010)
4 Discussion
The einkorn wheat ortholog ofCly1shares the same ten exon structure as the barley gene, and also includes the 21 nt miR172 targeting site (Table 2). Both genes belong to theAP2family of putative transcription factors,the characteristic feature of which is the presence of two DNA binding motifs consisting of approximately 68 conserved residues, referred to as theAP2domain(Jofukuet al., 1994). The two genes share the same sequence in motifs 1, 2 and 3, the nuclear localization signal, the twoAP2-domains and the AASSGF box. The einkorn wheat gene is transcribed in the leaf, shoot, root,but most abundantly in the spike throughout development, particularly at anthesis (Figure 3). This pattern of transcription is similar to that ofCly1(Nairet al., 2010).TheTmAP2sequence at the miRNA targeting site is the same as that of the non-cleistogamous allele in barley(Figure 4). Its transcripts were consequently cleaved,mainly between the A and U nucleotides (Figure 4), as again in barleys carryingCly1(Nairet al., 2010).Around anthesis, the lodicules expanded to reach more than double the volume which they occupied at the green anther stage, allowing them to function as a lever to push the lemma aside and permit anther extrusion,just as in non-cleistogamous barley (Nairet al., 2010).Thus the evidence is strong thatTmAP2is intimately involved in lodicule development and the determination of cleistogamy.
Two naturally occurring cleistogamous alleles are known in barley, both of which are believed to be of relatively recent origin (Nairet al., 2010). The recessive nature of these alleles implies that their equivalents in wheat will not produce a cleistogamous phenotype unless present in the homozygous state. Mutagenesis could represent a good option for creating a cleistogamous einkorn wheat and the tilling approach (Henikoffet al.,2004) provides an alternative to conventional phenotypic selection for identifying mutants. A further possibility for generating novel alleles is provided by site-specific nucleases based on the fusion between the DNA cleavage domain ofFokI and a custom-designed DNA binding domain, such as the C2H2 zinc-finger motif for zinc finger nucleases (ZFNs) (Urnovet al., 2010) and the truncated transcription activator-like effector (TALE)domain for TALE nucleases (Milleret al., 2011).
The identification ofTmAP2as the controlling gene for cleistogamy provides a starting point for developing a breeding program based on the cleistogamous flowering type in diploid wheat. Although known in barley, the trait has not been documented as yet in wheats at any of its three ploidy levels. The major advantage of cleistogamy is that pollen-mediated gene flow is minimized,thus assuring the maintenance of genetic integrity, and in the case of genetically modified varieties, protecting against transgene escape. A further advantage of the trait lies in improving the level of resistance against certainFusariumspp.pathogens, which enter the plant through the gaping floret (Gilsingeret al., 2005; kuboet al.,2010). Some suggestion has been made that cleistogamy can impart a degree of drought tolerance to plants at flowering time. In Collomia grandiflora, for example,moisture stress has been shown to increase the proportion of cleistogamous flowers produced, an adaptation interpreted as representing a means of increasing reproductive success in the face of drought (Minter, 1981).The non-cleistogamous flowers of wheat provide no protection to the stigma and the anther from dehydration,so it is possible that seed set under drought stress may be higher in cleistogamous than in non-cleistogamous types.
We thank G. Chen for crucial help and advice in publication. This research was funded by the Japanese Ministry of Agriculture, Forestry and Fisheries (Genomics for Agricultural Innovation Grants No. TRG1004) to T.K.S.N. appreciates the award of a Japanese Government(Monbukagakusho: MEXT) scholarship.
Barth C, Tullio MD, Conklin PL, 2006. The role of ascorbic acid in the control of flowering time and the onset of senescence. Journal of Experimental Botany, 57(8): 1657–1665.
Cully TM, 2002. Reproductive biology and delaying selfing inViola pubescens(Violaceae), an understory herb with chasmogamous and cleistogamous flowers. International Journal of Plant Sciences,163(1): 113–122.
Cully TM, Klooster MR, 2007. The cleistogamous breeding system: a review if its frequency, evolution, and ecology in angiosperms.Botanical Review, 73(1): 1–30.
Daniell H, 2002. Molecular strategies for gene containment in transgenic crops. Nature Biotechnology, 20(6): 581–586.
Feldman M, Lupton FGH, Miller TE, 1995. Wheats,Triticumspp.(Gramineae-Triticinae). In: Smartt J, Simmonds NW (eds.). Evolution of Crop Plants. Longman Scientific and Technical, Harlow, pp.84–192.
Gilsinger J, Kong L, Shen X, Ohm H, 2005. DNA markers associated with low Fusarium head blight incidence and narrow flower opening in wheat. Theoretical and Applied Genetics, 110(7):1218–1225.
Glover B, 2007. Understanding Flowers and Flowering. Oxford, New York, pp. 227.
Gustafson P, Raskina O, Ma X, Nevo E, 2009. Wheat evolution, domestication, and improvement. In: Carver BF (ed.). Wheat: Science and Trade. Wiley, Danvers, pp. 5–30.
Henikoff S, Till BJ, Comai L, 2004. TILLING. Traditional mutagenesis meets functional genomics. Plant Physiology, 135(2): 630–636.
Heslop-Harrison Y, Heslop-Harrison JS, 1996. Lodicule function and filament extension in the grasses: potassium ion movement and tissue specialization. Annals of Botany, 77(6): 573–582.
Jofuku KD, den Boer BG, Montagu MV, Okamuro JK, 1994. Control of Arabidopsis flower and seed development by the homeotic geneAPETALA2. Plant Cell, 6(9): 1211–1225.
Kasschau KD, Xie Z, Allen E, Llave C, Chapman EJ, Krizan KA,Carrington JC, 2003. P1/HC-Pro, a viral suppressor of RNA silencing, interferes with Arabidopsis development and miRNA function. Developmental Cell, 4(2): 205–217.
Kirby EJM, Appleyard M, 1981. Cereal Development Guide. National Agricultural Center Cereal Unit, Stoneleigh. Warwickshire, UK.
Komatsuda T, Nakamura I, Takaiwa F, Oka S, 1998. Development of STS markers closely linked to thevrs1locus in barley,Hordeum vulgare. Genome, 41(5): 680–685.
Kubo K, Kawada N, Fujita M, Hatta K, Oda S, Nakajima T, 2010.Effect of cleistogamy on Fusarium head blight resistance in wheat.Breeding Science, 60(4): 405–411.
Langer RHM, Wilson D, 1964. Environmental control of cleistogamy in prairie grass (Bromus unioloides H.B.K.). New Phytologist,64(1): 80–85.
Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X,Paschon DE, Leung E, Hinkley SJ, Dulay GP, Hua KL, Ankoudinova I, Cost GJ, Urnov FD, Zhang HS, Holmes MC, Zhang L,Gregory PD, Rebar EJ, 2011. A TALE nuclease architecture for efficient genome editing. Nature Biotechnology, 29(2): 143–148.
Minter TC, 1981. Cleistogamyin Collomiagrandiflora: comparative floral development and effects of water stress, abscisic acid, and gibberellic acid on flower production. M.S. thesis. University of California, Riverside.
Nair SK, Wang N, Turuspekov Y, Pourkheirandish M, Sinsuwongwat S,Chen G, Sameri M, Tagiri A, Honda I, Watanabe Y, Kanamori H,Wicker T, Stein N, Nagamura Y, Matsumoto T, Komatsuda T, 2010.Cleistogamous flowering in barley arises from the suppression of microRNA-guidedHvAP2mRNA cleavage. Proceedings of the National Academy of Sciences of the United States of America,107(1): 490–495.
Perrino P, Laghetti G, D’Antuono LF, Al-Ajlouni M, Kanbertay M,Szabó AT, Hammer K, 1996. Ecogeographical distribution of hulled wheat species. In: Padulosi S, Hammer K, Heller J (eds.).Hulled Wheats. Proceedings of the First International Workshop on Hulled Wheats. Castelvecchio Pascoli, Tuscany, pp. 101–119.
Tang M, Li G, Chen M, 2007. The phylogeny and expression pattern ofAPETALA2-like genes in rice. Journal of Genetics and Genomics,34(10): 930–938.
Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD, 2010.Genome editing with engineered zinc finger nucleases. Nature Reviews Genetics, 11(9): 636–646.
Waller DM, 1979. The relative costs of self- and cross-fertilized seeds in Impatiens capensis (Balsaminaceae). American Journal of Botany,66(3): 313–320.
 Sciences in Cold and Arid Regions2013年6期
Sciences in Cold and Arid Regions2013年6期
- Sciences in Cold and Arid Regions的其它文章
- Involvement of anti-oxidative enzymes, photosynthetic pigments and flavonoid metabolism in the adaptation of Reaumuria soongorica to salt stress
- Spatial coupling relationships of gas hydrate formation in the Tibetan Plateau
- The morphological characteristics of glacial deposits during the Last Glaciation, taking the Parlung Zangbo River Basin as an example
- The change of Ningchan River Glacier No. 3 at Lenglongling, east Qilian Mountain, China
- The influence of human activity and precipitation change on mid-long term evolution of landslide and debris flow disasters
- Seasonal variation in soil microbial biomass carbon and nitrogen in an artificial sand-binding vegetation area in Shapotou, northern China
