转基因抗草甘膦棉花R1-3株系的分子特征鉴定
马燕斌,李换丽,文晋,周仙婷,2,秦欣,王霞,王新胜,李燕娥
转基因抗草甘膦棉花R1-3株系的分子特征鉴定
马燕斌1,李换丽1,文晋1,周仙婷1,2,秦欣1,王霞3,王新胜1,李燕娥1
1山西农业大学棉花研究所,山西运城 044000;2山西农业大学农学院,山西太谷 030800;3运城学院生命科学系,山西运城 044000
【目的】通过转化抗草甘膦除草剂基因获得抗草甘膦除草剂的棉花转基因株系,对其进行分子特征鉴定分析,为今后棉花育种利用该株系提供必要的分子依据。【方法】利用农杆菌介导法,在草甘膦筛选条件下,通过组织培养获得棉花再生株系,利用Western blot对转基因棉花株系不同组织的外源蛋白表达进行检测;通过Southern blot确定株系中外源整合位点的拷贝数;利用TAIL-PCR扩增外源基因整合位点侧翼序列,并通过NCBI BLAST工具比较分析其定位的染色体位置。【结果】通过草甘膦筛选,利用组织培养获得R1-3棉花再生株系;Western blot结果表明,外源基因在叶、苞叶、花、茎中均可正常表达,其蛋白大小约为46 kDa,与预期目标条带一致;基因组DNA酶切后杂交结果显示,R1-3株系外源序列的整合位点为单拷贝插入,其中,Ⅰ酶切的杂交条带位于约6 557 bp处,RⅠ酶切的2条杂交带位于略大于4 316 bp处;侧翼序列比对结果显示,外源序列融合到陆地棉A或D基因组的第11号染色体上,且在交换插入的过程中,左右边界的融合位点分别位于该染色体47 525 303和47 525 449处。另外,利用特异引物进行PCR鉴定,可知左侧融合位点可扩增出约300 bp的预期特定目标条带,右侧融合位点可扩增出约600 bp的预期特定目标条带。【结论】获得具有稳定遗传特征的R1-3转基因棉花株系,不同组织中外源基因编码的蛋白均有表达,提高了该株系对草甘膦的高抗性。含有的外源序列为单个位点插入,融合位点位于陆地棉A或D基因组第11号染色体上,该融合位点处缺失约146 bp的核苷酸序列。
陆地棉;抗草甘膦棉花;拷贝数;融合位点;分子特征
0 引言
【研究意义】田间杂草可导致作物减产,利用除草剂进行田间杂草去除是较为简便的管理方式。抗草甘膦除草剂棉花种植可有效提高棉田杂草的去除,达到降低人工投入和简化种植的目标。从近几十年来抗除草剂作物发展趋势可知,抗草甘膦除草剂的玉米、大豆、棉花及油菜等作物具有经济效益显著的商业化价值,研究培育抗草甘膦棉花等作物有助于应对今后我国农业发展在国际市场的竞争。同时,在转化获得抗草甘膦棉花的基础上,进一步鉴定转基因棉花外源基因相关分子的特征,对满足转基因生物安全评价中环境释放条件等具有重要的意义。【前人研究进展】草甘膦具有广谱除草特性,可抑制5-烯醇式丙酮酸莽草酸-3-磷酸合成酶(5-enolpyruvylshikimate 3-phosphate synthase,EPSPS)活性,阻断芳香族氨基酸的合成,最终导致植株死亡[1-3]。利用过表达抗草甘膦基因改良作物的抗草甘膦特性是重要的途径之一,国内外抗草甘膦等除草剂作物研究较多[4-5]。近年来,科研人员从不同生物克隆、人工设计等途径获得了编码EPSPS酶的基因,经不同方式验证,这些基因对草甘膦均具有较好的耐受性[6-9],且利用基因编辑技术替换不同位点的氨基酸获得具有草甘膦抗性的木薯和水稻[10-11]。除此之外,研究发现在水稻自然进化中存在比野生型更具抗草甘膦特性的突变体[12]。但也有在抗虫棉花中直接转入抗草甘膦基因的研究,在保留抗虫特性的同时达到便捷实现棉花的抗草甘膦特性[13],同时,国内在玉米、棉花等作物中也获得了具有极高耐受草甘膦除草剂的转基因作物[14-15]。综上所述,目前,抗草甘膦基因及相关抗除草剂作物的研究取得了较大进展,抗草甘膦转基因等作物在今后的农业生产中可能具有重要的应用潜力。【本研究切入点】抗草甘膦除草剂棉花品种培育有助于促进棉花的轻简化栽培,具有自主知识产权的优良的抗草甘膦棉花品种亟待培育。研究人员在获得高抗草甘膦的转基因棉花材料后,为满足国内转基因作物环境释放要求,抗性材料的遗传稳定性、单个位点插入及整合位点侧翼序列等是需要提交的分子特征,同时也是国内自主培育作物产权的要求之一。【拟解决的关键问题】本研究利用农杆菌介导法转化获得的抗草甘膦棉花材料R1-3,检测不同组织中外源蛋白的表达,利用Southern blot、TAIL-PCR等方法分别鉴定转基因棉花R1-3材料中外源序列的拷贝数、侧翼序列,以及分析融合位点处序列的特征,为后续该转基因材料环境释放、育种利用及生物安全检测等方面提供较详实的分子依据。
1 材料与方法
1.1 植物表达载体及棉花受体材料
棉花受体为山西省农业科学院棉花所选育材料陆地棉R15(L.),植物表达载体为含抗草甘膦基因的pCAMBIA 1300(图1)。
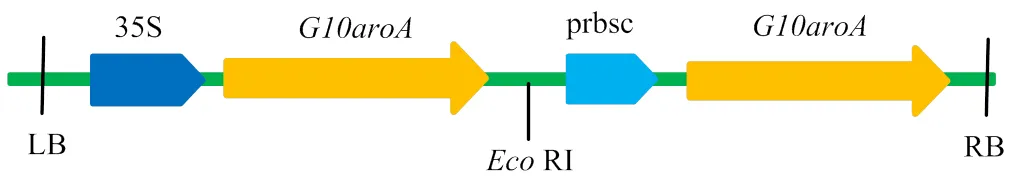
图1 G10aroA的pCAMBIA1300载体构建示意图
1.2 外源G10aroA的棉花转化及再生株系R1-3的获得
LB液体培育含有植物转化载体的农杆菌菌株LBA4404,培养基中加入抗生素卡那霉素和利福平,28 ℃摇床悬浮过夜培养,离心倒去上清液收集农杆菌,沉淀用30 g·L-1的葡萄糖重新悬浮,调整OD值为0.6,浸染棉花下胚轴切段后,22 ℃黑暗共培养48 h,在草甘膦浓度为2.5 mmol·L-1的MS培养基上进行愈伤诱导,按照常规转化方法进行棉花转基因[14,16],获得再生植株后,PCR检测确定阳性植株,自交后收获种子,多年自交繁育获得纯合稳定的转基因抗草甘膦株系R1-3。
1.3 棉花DNA提取与Southern blot分析
试验转基因材料种植于隔离棚内。取幼嫩棉花叶片放入离心管内,液氮快速冷冻后,-80 ℃保存备用。采用改良的CTAB法[17]提取转基因棉花植株等试验样品DNA,采用碱裂解法提取质粒DNA,利用内切酶RⅠ和Ⅰ分别进行酶切,电泳转膜后,采用地高辛标记法进行Southern blot分析,探针为扩增特异片段序列后标记,扩增引物为779F:5′-GCGTGGATATGTCCTGCGGG-3′;1481R:5′-TCT ACACAGCCATCGGTCCAG-3′。
1.4 外源基因融合插入位点鉴定
利用TAIL-PCR进行扩增,根据pCAMBIA 1300植物转化载体左右边界序列设计特异引物后扩增。随机引物设计及扩增程序参照Liu等[18],引物见表1。
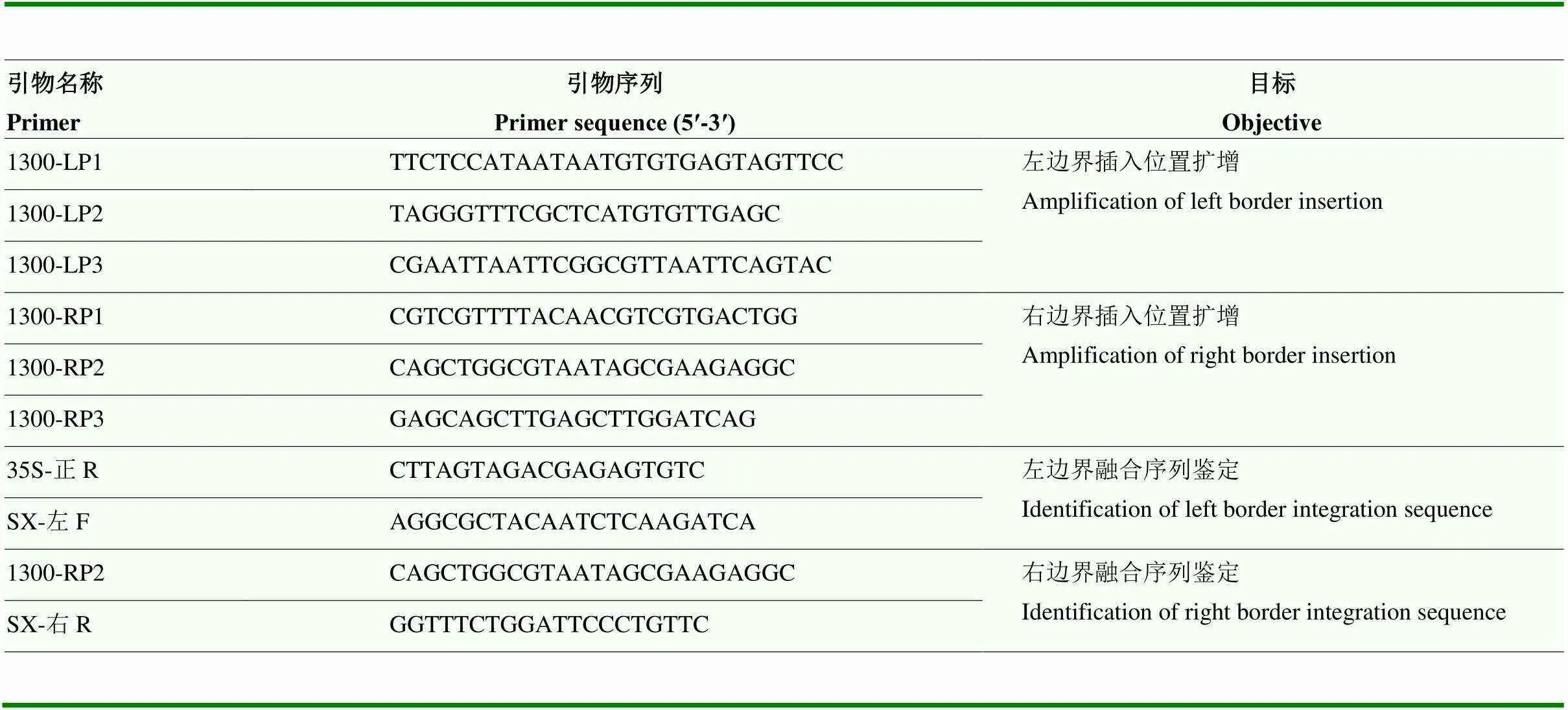
表1 R1-3转基因棉花R1-3整合位点的TAIL-PCR与侧翼序列扩增特异引物
LP:左边界特异引物;RP:右边界特异引物 LP: specific primers for left boundary; RP: specific primers for right boundary
1.5 侧翼序列分析
通过对TAIL-PCR克隆的序列进行测序,利用DNAMAN软件,将测序结果分别与引物、载体边界序列进行比较,结果相匹配后,利用NCBI网站Blast分析工具,输入测序结果进行融合位点预测分析。
1.6 Western blot免疫印迹分析
取R1-3棉花株系的不同组织,液氮研磨样品,取约0.1 g样品加磷酸缓冲液后转入2 ml灭菌离心管中,离心提取上清液后,用分光光度计检测提取的样品蛋白浓度,浓度均一化处理。蛋白电泳上样前加入1×loading buffer,样品煮沸变性10 min,取等量样品进行SDS-PAGE电泳,电泳结束后,经转膜、封闭、抗体杂交,二抗为编码的EPSPS蛋白特异抗体[15],直接显影后对杂交膜拍照。
2 结果
2.1 外源G10aroA的转化及转基因植株的获得
用农杆菌菌液浸染切好的棉花无菌苗茎段,经共培养后,在含有草甘膦的培养基上进行愈伤诱导(图2-A),持续诱导待形成块状愈伤(图2-B)。块状愈伤单独分离后培养增殖,转入分化培养基诱导后继续诱导形成胚性愈伤(图2-C)。持续培养后转入分化培养基再生并生成完整幼苗植株(图2-D—E)。选取生长正常的幼苗嫁接成活后移入盆中温室种植(图2-F),待转基因植株开花自交棉铃成熟后收获种子。

A:农杆菌侵染后的棉花下胚轴外植体愈伤诱导;B:下胚轴诱导的愈伤;C:愈伤增殖;D:胚性愈伤分化;E:分化后生成的幼苗;F:再生植株
2.2 外源蛋白在不同组织中表达分析
通过对转基因植株喷施草甘膦进行抗性筛选和多个世代的分离纯化,获得纯合转基因R1-3等转基因植株,为进一步鉴定优良的转基因抗性植株,提取转基因株系R1-3的叶、苞叶、花、茎等不同组织中的总蛋白,利用特异抗体对外源EPSPS蛋白表达进行鉴定分析。Western blot杂交结果表明,与阴性对照相比,在转基因株系R1-3的不同组织中,均检测到EPSPS蛋白的表达(图3),表明外源基因在叶、苞叶、花、茎中均可正常翻译表达,其中,外源蛋白预期大小约为46 kDa。因此,可确定外源抗草甘膦基因在棉花株系R1-3中可正常表达。
2.3 外源基因在棉花基因组插入拷贝数鉴定
根据外源基因序列设计特异探针引物,扩增特异序列后,利用地高辛标记探针,提取转基因植株等试验材料的基因组DNA纯化后,分别加入内切酶RⅠ和Ⅰ进行酶切消化,其中,前期分析已知抗草甘膦基因中不含有该内切酶位点;电泳转膜后进行Southern blot杂交,结果(图4)表明,受体R15无杂交条带信号,pCAMBIA1300载体阳性质粒杂交在小于23 130 bp的位置有明显的杂交条带,表明该特异探针杂交效果可满足目标基因检测;同时,为选择具有单拷贝的转基因植株,对转基因株系R1-3进行杂交分析,结果显示,杂交条带清晰,条带位置约介于Marker标记的4 316—23 130 bp;其中,由于构建载体的2个序列之间存在RⅠ酶切位点,杂交结果显示,有紧密相邻的2条杂交带,而Ⅰ酶切的R1-3杂交条带显示为单拷贝插入,这些条带分别位于Marker标记约4 316 bp和略小于6 557 bp位置处。
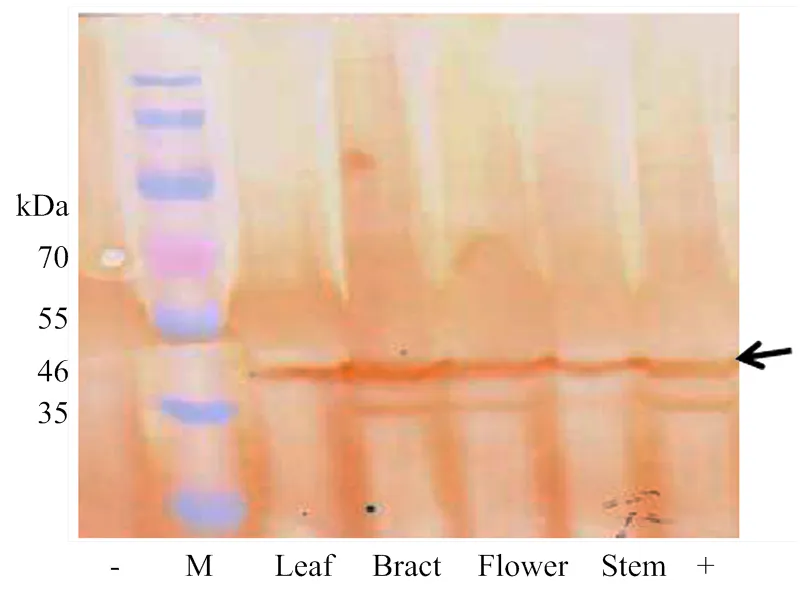
-:阴性对照R15;M:蛋白Marker,46 kDa为R1-3棉花株系中EPSPS蛋白条带大小

左侧2条带分别为R1-3被EcoRⅠ和KpnⅠ酶切的杂交结果;-:R15阴性对照结果,内切酶EcoRⅠ消化;M:DNA Marker;+:内切酶EcoRⅠ消化的含外源基因G10aroA的pCAMBIA1300载体,箭头指示杂交条带的位置
2.4 外源基因在棉花基因组融合位点及侧翼序列鉴定
提取R1-3株系DNA,分别利用转基因载体左右边界序列设计特异引物和基因组随机引物进行TAIL-PCR扩增,进行三轮扩增后,选择特异条带回收并克隆转化,测序后得到核苷酸序列信息。首先利用DNAMAN软件比较测序结果与引物、载体边界序列的一致性,结果显示,克隆的部分测序序列可与扩增引物和载体边界序列相匹配。利用NCBI数据库对测序序列进行Blast分析,结果显示,与载体边界相融合的克隆序列,分别与棉花TM1参考基因组A11和D11染色体基因组序列相一致。由于陆地棉A、D基因组相似性较高,以A基因组序列参考分析可知(图5-A),载体左边插入位置为棉花A(D)基因组第11号染色体47 525 303处(图5-B,L位置),载体右边界插入位置为染色体47 525 449处(图5-B,R位置),由序列比较可知,左右边界序列插入染色体位置缺失约146 bp核苷酸序列。同时,从插入边界的序列分析可知,外源基因插入染色体编码方向为由L到R。另外,利用融合插入位点附近比较一致的序列分别设计左右两端的特异引物(表1),PCR扩增验证结果表明,左侧融合位点可扩增出约300 bp特异目标条带(图5-C),右侧融合位点可扩增出约600 bp特异目标条带(图5-D)。
3 讨论
3.1 抗除草剂棉花具有潜在的种植优势
抗除草剂作物的种植在田间杂草防治、生产管理简化、防止作物减产等方面均具有突出的优势[19-20]。棉花是世界上重要的纤维类经济作物之一,当前除新疆棉区外其他棉区均面临棉花种植面积逐渐减少的趋势。因此,提高棉花种植经济效益和简化种植模式是当前棉花产业发展的亟需要求。同时,为满足潜在的棉花种植轻简化发展需求,培育具有自主知识产权的抗除草剂棉花是理想的途径之一。
3.2 培育具有自主产权的抗草甘膦除草剂棉花有利于国内棉花产业发展
世界上种植转基因作物80%以上面积为单一抗草甘膦作物,其中,这些作物抗草甘膦基因受专利保护,被利用最多的是[21],潜在的利用价值也促进了不同单位对新的抗草甘膦基因的研究[22-23]。本研究在通过草甘膦筛选的基础上获得一批抗草甘膦转基因棉花株系。通过自交选育和喷施草甘膦分离鉴定获得纯合的转基因植株后,选育抗性和农艺性状优良的株系用于后续的分子鉴定。通过Western blot杂交分析R1-3植株不同组织外源蛋白表达可知,各组织中均可以检测到编码的EPSPS外源蛋白,这与田间喷施草甘膦R1-3植株叶、花、蕾等各组织可正常生长发育的表现相一致,不同组织对草甘膦的高耐受性也是转基因植株所必需的性状[15, 24],在种植时处理田间杂草会达到较好的杀灭效果。另外,该外源基因编码蛋白在转基因植株中具有明显的杂交条带,受体植株R15检测不到该蛋白的表达,因此,在转基因检测中利用抗体检测也是识别该转基因植株特异性的方法之一。此外,从国内抗草甘膦棉花的相关研究脉络可知,不同单位从较早的抗性基因克隆、转基因植株获得、田间抗性鉴定到生物安全评价等各方面均有较多研究[25-26];这些自主研究的积累将有助于促进我国农业生产的国际竞争力。
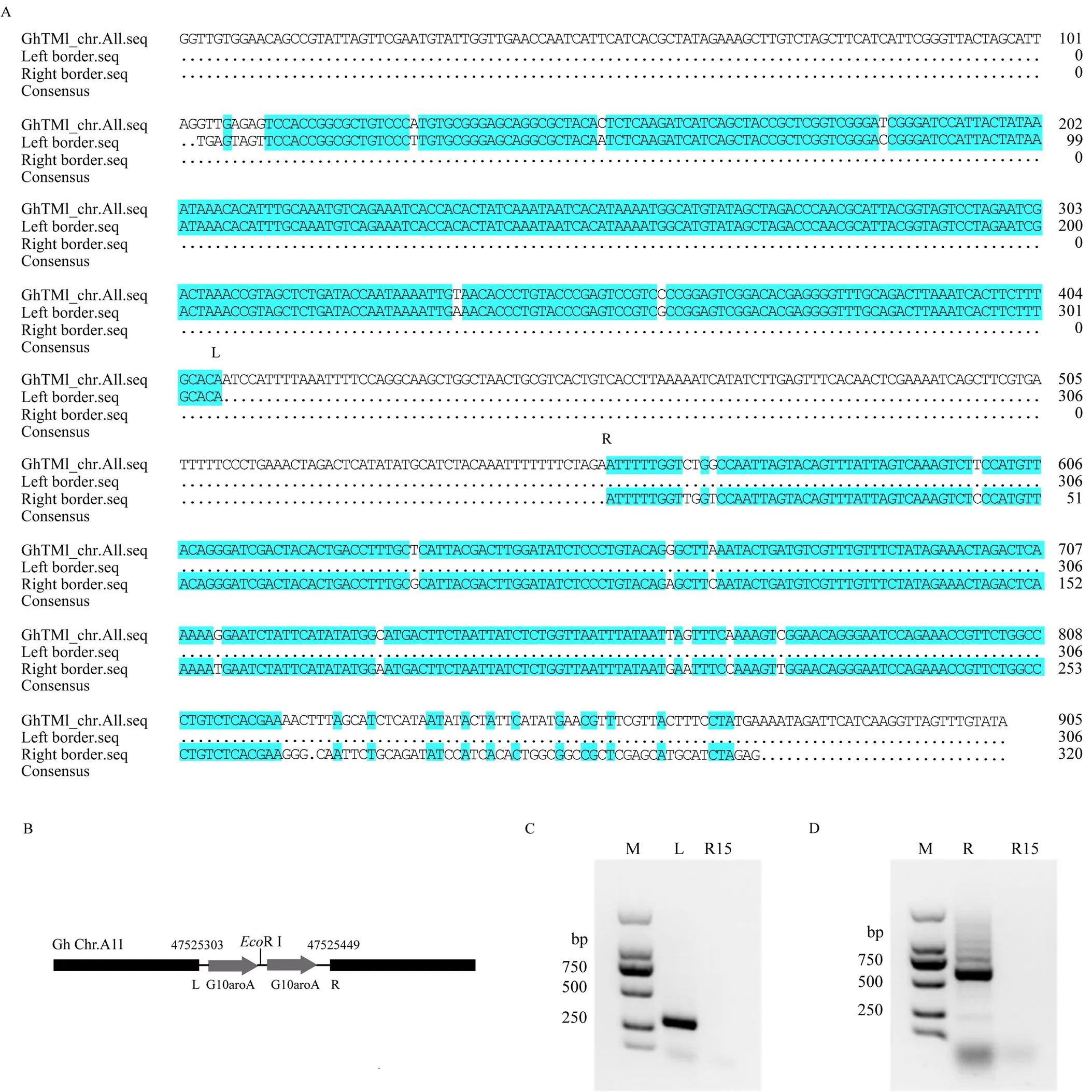
A:R1-3株系中外源序列的融合位点两侧侧翼序列经TAIL-PCR扩增后测序结果与TM-1基因组的一致性比较,L为pCAMBIA 1300载体左边界插入位置,R为pCAMBIA 1300载体右边界插入位置;B:外源序列整合到R15基因组中的示意图;C:左边界融合侧翼序列的特异PCR扩增;D:右边界融合侧翼序列的特异PCR扩增
3.3 转基因植株分子信息鉴定是后续安全评价利用重要参考数据
本研究在抗草甘膦除草剂中间试验的基础上,鉴定提供可靠的R1-3植株中外源基因融合位点、外源序列拷贝数以及侧翼序列等特征是该材料安全利用的必要科学证据。从基因拷贝数分析结果看,RⅠ和Ⅰ酶避开了外源基因内插入区域的酶切位点,整合到染色体上后,RⅠ酶切的R1-3条带与构建载体酶切后的杂交条带均为2条,而Ⅰ酶切条带为单一条带,这两个酶切结果可相互验证表明,该外源基因区域的插入染色体为单一融合位点,该融合位点具有稳定遗传特性。另外,T-DNA位点的分子特征信息为公共风险评估和监测所提供了必要的依据,同时也用于分析相关内源性基因的表达是否受影响[27-28]。本研究根据整合位点序列比对结果预测为棉花A基因组第11号染色体,由于陆地棉为异源四倍体,含有A、D 2个染色体组且具有高度的核苷酸序列一致性,Blast序列比较时结果显示,与D染色体组第11号染色体也具有极高的一致性,因此,从结果上不排除该外源基因整合到D11染色体的可能性,后续有待进一步验证。在此基础上,分析外源基因整合位点侧翼序列可知,整合位点区及上下游区域附近的基因组核苷酸序列没有任何假定或注释的基因,表明该外源基因插入对棉花基因组表达的影响较小,后续该侧翼序列的分子特征将为验证利用提供必要的科学依据。
4 结论
R1-3转基因棉花植株的不同组织中均可检测到约46 kDa大小的目标蛋白。含有抗草甘膦基因的外源序列为单个位点插入,左边界融合位点位于棉花基因组A或D的第11号染色体47 525 303处,载体右边界融合插入位置为染色体47 525 449处。表明获得外源序列为单个整合位点的、可稳定遗传的高抗草甘膦转基因棉花植株。
[1] Duke S O. The history and current status of glyphosate. Pest Management Science, 2018, 74(5): 1027-1034.
[2] Powell H A, KerBby N W, Rowell P. Natural tolerance of cyanobacteria to the herbicide glyphosate. New Phytologist, 1991, 119(3): 421-426.
[3] Funke T, Han H, Healy-Fried M L, Fischer M, SchÖnbrunnE. Molecular basis for the herbicide resistance of roundup ready crops. Proceedings of the National Academy of Sciences of the United States of America, 2006, 103(35): 13010-13015.
[4] Comai L, Facciotti D, Hiatt W R, Thompson G, Rose R E, Stalker D M. Expression in plants of a mutant aroA gene fromconfers tolerance to glyphosateNature, 1985, 317(6039): 741-744.
[5] Ye G N, Hajdukiewicz P T J, Broyles D, Rodriguez D, Xu C W, Nehra N, Staub J M. Plastid-expressed 5-enolpyruvylshikimate -3-phosphate synthase genes provide high level glyphosate tolerance in tobacco. The Plant Journal, 2001, 25(3): 261-270.
[6] Yi S Y, Cui Y, Zhao Y, Liu ZD, Lin YJ, Zhou F. A novel naturally occurring class I 5-enolpyruvylshikimate-3-phosphate synthase from Janibacter sp. confers high glyphosate tolerance to rice.Scientific Reports, 2016, 6: 19104.
[7] Liu F, Cao Y P. Cloning and characterization of 5-enopyruvylshikimate-3-phosphate synthase fromsp. Genetics and Molecular Research, 2015, 14(4): 19233-19241.
[8] Cui Y, Huang S Q, Liu Z D, Yi S Y, Zhou F, Chen H, Lin Y J. Development of novel glyphosate-tolerantrice lines: a step toward commercial release.Frontiers in Plant Science, 2016, 7: 1218.
[9] Han J, Tian YS, Xu J, Wang LJ, Wang B, Peng RH, Yao QH. Functional characterization of aroAfromwith significant glyphosate tolerance in transgenic. Journal of Microbiology and Biotechnology, 2014, 24(9): 1162-1169.
[10] Hummel AW, Chauhan RD, Cermak T, Mutka AM, Vijayaraghavan A, Boyher A, Starker CG, Bart R, Voytas DF, Taylor NJ. Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnology Journal, 2018, 16(7): 1275-1282.
[11] Li J, Meng X B, Zong Y, Chen K L, Zhang H W, Liu J X, Li J Y, Gao C X. Gene replacements and insertions in rice by intron targeting using CRISPR-Cas9.Nature Plants, 2016, 2: 16139.
[12] Yu Q, Jalaludin A, Han H P, Chen M, Sammons R D, Powles S B. Evolution of a double amino acid substitution in the 5-enolpyruvylshikimate-3-phosphate synthase in Eleusine indica conferring high-level glyphosate resistance.Plant Physiology, 2015, 167(4): 1440-1447.
[13] Latif A, Rao A Q, Khan M A, Shahid N, Bajwa K S, Ashraf M A, Abbas M A, Azam M, Shahid A A, Nasir I A, Husnain T. Herbicide-resistant cotton () plants: an alternative way of manual weed removal. BMC Research Notes, 2015, 8: 453.
[14] 王霞, 马燕斌, 吴霞, 沈志成, 林朝阳, 李朋波, 孙璇, 王新胜, 李燕娥, 李贵全. 转G10aroA棉花株系的获得及分子生物学鉴定. 中国农业科学, 2014, 47(6): 1051-1057.
Wang X, Ma Y B, Wu X, Shen Z C, Lin C Y, Li P B, Sun X, Wang X S, Li Y E, Li G Q. Molecular biology identification of transgenic cotton lines expressing exogenous G10aroA gene. Scientia Agricultura Sinica,2014, 47(6): 1051-1057. (in Chinese)
[15] Liu M M, Zhang X J, Gao Y, Shen Z C, Lin C Y. Molecular characterization and efficacy evaluation of a transgenic corn event for insect resistance and glyphosate tolerance. Journal of Zhejiang University Science B, 2018, 19(8): 610-619.
[16] 李燕娥, 朱祯, 陈志贤, 吴霞, 王伟, 李淑君, 朱玉, 焦改丽, 吴家和, 徐鸿林, 范小平, 孟晋红, 肖桂芳, 李向辉. 豇豆胰蛋白酶抑制剂转基因棉花的获得. 棉花学报, 1998, 10(5): 237-243.
Li Y E, Zhu Z, Chen Z X, Wu X, Wang W, Li S J, Zhu Y, Jiao G L, Wu J H, Xu H L, Fan X P, Meng J H, Xiao G F, Li X H. Obtaining transgenic cotton plants with cowpea trypsin inhibitor gene. Acta Gossypii Sinica,1998, 10(5): 237-243. (in Chinese)
[17] Paterson A H, Brubaker C L, Wendel J F. A rapid method for extraction of cotton (spp.) genomic DNA suitable for RFLP or PCR analysis.Plant Molecular Biology Reporter,1993, 11(2): 122-127.
[18] Liu Y G, Chen Y L. High-efficiency thermal asymmetric interlaced PCR for amplification of unknown flanking sequences. Biotechniques, 2007, 43(5): 649-650, 652, 654 passim.
[19] ZABLOTOWICZ R M, REDDY K N. Impact of glyphosate on thesymbiosis with glyphosate-resistant transgenic soybean: A minireview. Journal of environmental quality,2004, 33(3): 825-831.
[20] Heard M S, Hawes C, Champion G T, Clark S J, Firbank L G, Haughton A J, Parish A M, Perry J N, Rothery P, Scott R J, Skellern M P, Squire G R, Hill M O. Weeds in fields with contrasting conventional and genetically modified herbicide-tolerant crops: I. Effects on abundance and diversity. Philosophical Transactions of the Royal Society B, 2003, 358(1439): 1819-1832.
[21] Bonny S. Genetically modified herbicide-tolerant crops, weeds, and herbicides: Overview and impact. Environmental Management, 2016, 57(1): 31-48.
[22] Tan X L, Othman R Y, Teo C H. Isolation and functional characterization of 5-enolpyruvylshikimate 3-phosphate synthase gene from glyphosate-tolerantstrains FY43 and FY47. 3 Biotech, 2020, 10(4): 183.
[23] Ghaderitabar H, Mousavi A, Hatef S A, Hadi F. Novel aroA of Glyphosate-tolerant bacterium Pseudomonas sp. strain HA-09 isolated from roundup-contaminated garden soils in iran. Iranian journal of biotechnology, 2020, 18(3): e2597.
[24] WENDY A P, ANDREW J P, JOHN W W, KEITH L E, RANDY W. Absorption and translocation of glyphosate in glyphosate-resistant cotton as influenced by application method and growth stage.Weed Science, 2001, 49(4): 460-467.
[25] 赵福永, 谢龙旭, 田颖川, 徐培林. 抗草甘膦基因aroAM12及抗虫基因Bts1m的转基因棉株. 作物学报, 2005, 31(1): 108-113.
ZHAO F Y, XIE L X, TIAN Y C, XU P L. Glyphosate-resistant and bollworm-resistant transgenic cotton plants with the aroAM12 and Bts1m genes. Acta Agronomica Sinica, 2005, 31(1): 108-113. (in Chinese)
[26] 刘锡娟, 刘昱辉, 王志兴, 王旭静, 张永强. 转5-烯醇式丙酮酰莽草酸-3-磷酸合酶(EPSPS)基因抗草甘膦烟草和棉花的获得. 农业生物技术学报, 2007, 15(6): 958-963.
Liu X J, Liu Y H, Wang Z X, Wang X J, Zhang Y Q. Generation of glyphosate-tolerant transgenic tobacco and cotton by transformation with a 5-enolpyruvyl-shikimate-3-phosphate synthase (EPSPS) gene. Journal of Agricultural Biotechnology, 2007, 15(6): 958-963. (in Chinese)
[27] Chen X H, Dong Y, Huang Y L, Fan J M, Yang M S, Zhang J. Whole-genome resequencing using next-generation and Nanopore sequencing for molecular characterization of T-DNA integration in transgenic poplar 741. BMC Genomics, 2021, 22(1): 329-342.
[28] Tang G X, Zhong X B, Hong W, Li J F, Shu Y, Liu L L. Generation and identification of the number of copies of exogenous genes and the T-DNA insertion site in SCN-resistance transformation event ZHs1-2. International Journal of Molecular Sciences, 2022, 23(12): 6849.
Identification of Molecular Characterizations for Transgenic Cotton R1-3 Line of Glyphosate Tolerance

1Institute of Cotton Research, Shanxi Agriculture University, Yuncheng 044000, Shanxi;2College of Agronomy, Shanxi Agriculture University, Taigu 030800, Shanxi;3Department of Life Science, Yuncheng University, Yuncheng 044000, Shanxi
【Objective】To obtain the transgenic cotton by-mediated method was the purpose using the new genewith high glyphosate tolerance in our laboratory. Meanwhile, it was necessary to provide molecular characteristics of genomic integration of the exogenous gene for breeding utilization in future. 【Method】The-mediated method was used for transgenic cotton plants obtained via tissue culture with glyphosate herbicide. Western blot was utilized to detect the expression of exogenous proteins in different organs of transgenic cotton R1-3. The number of loci in cotton genomes were evaluated by Southern blot for detecting integration of the exogenous sequence from the pCAMBIA1300construct. The flanking sequence near the insertion site was amplified by TAIL-PCR, which the extractions of DNA were cloned and sequenced. The location of the chromosome for the flanking sequences were compared and analyzed on the website of NCBI blast. 【Result】Regenerated R1-3 cotton plants were successfully obtained by tissue culture depending on glyphosate screening. the specific protein coded by exogenousgene could be detected normally via Western blotting in the leaves, bracts, flowers and stems separately, and the size of the exogenous protein around 46 kDa were also observed in this experiment. In addition, the result of Southern-blot confirmed that the exogenous fragment containingsequences was single integration in the genome of transgenic cotton R1-3, in which the bands digested severally byⅠ andRⅠ endonucleases were distinctly observed near the position of 6 557 bp and 4 316 bp strips on the nylon membrane respectively. The analysis of flanking sequence alignment for the integration site was predicted to be located on the 11th chromosome of either cotton A or D genome, and the left and right boundaries of the insertion site were further located between 47 525 303 and 47 525 449 of the chromosomes. In addition, the specific identification for the fusion site showed that the target band of approximately 300 bp for testing left border junction, and a specific target band can be amplified about 600 bp for testing the right border fusion site.【Conclusion】In this study, we obtained R1-3 transgenic cotton plants that also have exhibited stable genetic characteristics of glyphosate resistance during the process of self-crossing breeding. The protein coded bygene was about the size of 46 kDa that could be detected in different tissues of transgenic cotton R1-3 plant. Furthermore, the exogenous fragment includinggene was identified with a single location by southern blot in the cotton genomes, and the integration site was located at the 11th chromosome. The results of comparative analysis were predicted that a nucleotide sequence about 146 bp length was deleted at the integration of the genome.
L.; glyphosate-tolerance cotton; copy number; integration site; molecular characterization
10.3864/j.issn.0578-1752.2023.17.004
2023-02-01;
2023-07-05
转基因生物新品种培育重大专项(2016ZX08010-003)、山西省应用基础研究面上项目(202103021224145)、山西省农业科学院农业科技创新研究课题(YGJPY2007)
通信作者马燕斌,E-mail:myb0517@163.com
(责任编辑 李莉)

