Real-time quantification of nuclear RNA export using an intracellular relocation probe
Jie Shen, Jun Chen, Dong Wng, Zhengjie Liu, Gungmei Hn, Binhu Liu,Mingyong Hn, Ruilong Zhng,b,∗, Guoong Liu, Zhongping Zhng,b
a School of Chemistry and Chemical Engineering, Information Materials and Intelligent Sensing Laboratory of Anhui Province, and Institute of Physical Science and Information Technology, Anhui University, Hefei 230601, China
b Key Laboratory of Structure and Functional Regulation of Hybrid Materials (Anhui University), Ministry of Education, Hefei 230601, China
c Key Lab of Photovoltaic and Energy Conservation Materials, Institute of Solid State Physics, HFIPS, Chinese Academy of Sciences, Hefei 230031, China
d School of Life and Health Sciences, Anhui Science and Technology University, Chuzhou 233100, China
ABSTRACT Nuclear RNA export into the cytoplasm is one of the key steps in protein expression to realize biological functions.Despite the broad availability of nucleic acid dyes, tracking and quantifying the highly dynamic process of RNA export in live cells is challenging.When dye-labeled RNA enters the cytoplasm, the dye molecules are released upon degradation of the RNA, allowing them to re-enter the cell nucleus.As a result, the ratio between the dye exported with RNA into the cytoplasm and the portion staying inside the nucleus cannot be determined.To address this common limitation, we report the design of a smart probe that can only check into the nucleus once.When adding to cells, this probe rapidly binds with nuclear RNAs in live cells and reacts with intrinsic H2S.This reaction not only activates the fluorescence for RNA tracking but also changes the structure of probe and consequently its intracellular localization.After disassociating from exported RNAs in cytoplasm, the probe preferentially enters lysosomes rather than cell nucleus, enabling real-time quantitative measurement of nuclear RNA exports.Using this probe,we successfully evaluated the effects of hormones and cancer drugs on nuclear RNA export in live cells.Interestingly, we found that hormones inhibiting RNA exports can partially offset the effect of chemotherapy.
Keywords:RNA export Quantification Relocation probe Fluorescence imaging Hormone
RNA is one of the most critical substances in life, serving important roles in the physiological functions such as protein expression, signal transduction, and cellular proliferation and death [1–4].RNAs including messenger RNA (mRNA), ribosomal RNA, and long non-coding RNA are transcribed from DNA templates, and temporally stored in the cell nucleus [5].Upon needed, they are quickly transported into the cytoplasm through the nuclear pore complexes (NPC) [1,5].Therefore, nuclear RNA transport through the nuclear membrane is one of the first indicators for cell behaviors.For example, in response to drug stimulations [6] or stresses[5,7], RNA export can accelerate or decelerate resulting in a series of biological events and sometimes even diseases.Therefore,the understanding, monitoring, and measuring of this indicator of great interest in both cell biology research and drug discovery, but challenging to achieve because of the fast dynamics of RNA export[1,5,8].
Currently, nuclear RNA exports are analyzed by separating the cell nucleus and cytoplasm from cell lysates and to quantify the cytoplasmic RNA levels [9].Another widely accepted method is to measure the proteins corresponding to the transcripts, an indirect measurement [10].A common and fundamental limitation of these approaches is their incapability in tracking the fast dynamics of RNA export, not to mention the loss of spatial information.To address this issue, a variety of RNA fluorescent dyes have been made in recent years to label nuclear RNA in live cells [11–16].These dyes are often bright and activable upon binding to RNAs but still fall short for quantitative measurement of nuclear RNA export.This is because when the fluorescently labeled nuclear RNA entering the cytoplasm and eventually gets degraded, the released dye molecules return to the nucleus due to their preferential nuclear localization.This recycling process skews the dyes’nucleuscytoplasm distribution.
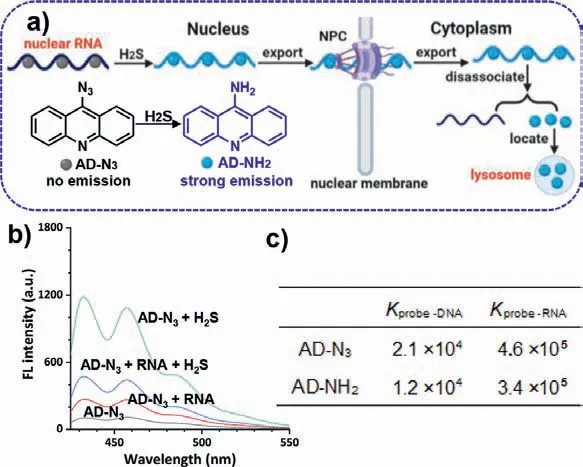
Fig.1.(a) Schematic illustration of the probing strategy using AD-N3.(b) The fluorescent response of AD-N3 (10 μmol/L) to H2S (50 μmol/L) and RNA (0.5 mg/mL)under the excitation wavelength of 405 nm.(c) The binding constants (K) of AD-N3 and AD-NH2 with nucleic acids.
In this context, we have designed a new class of RNA imaging probe, azido acridine (AD-N3), that changes its intracellular localization preference after the mission of measuring nuclear RNA export is completed (Fig.1a).This probe features an acridine group that provides the capability of cell nucleus localization and specific recognition of RNAs through hydrogen bonds andπ-πinteractions similar to common nuclear RNA dyes [17,18], and a reactive -N3group.As soon as AD-N3encounters intrinsic H2S that has a typical concentration range of ∼50 μmol/L in the nucleus [19,20], the -N3group is immediately converted to amino acridine (AD-NH2).This quick and simple reaction serves two important purposes.First, it transforms the non-emissive AD-N3into a highly fluorescent label for nuclear RNA in live cells [21].Second, it changes the acridine probe’s affinity from the nucleus to lysosomes due to the alkalinity of -NH2and the acidity of lysosomes [22], preventing the fluorescent probe from going back to cell nucleus.Although in most fluorescent assays, recycling fluorescent probes are desirable, we designed AD-N3capable of only checking into the nucleus once,making real-time quantification of nuclear RNA export feasible.
The synthetic route and characterizations of probe AD-N3were shown in Scheme S1 and Figs.S1-S3 (Supporting information), respectively.After confirmation of its chemical structure, we first examined the AD-N3probe’s fluorescence in response to H2Sin vitro.The original probe was nearly non-fluorescent but was immediately activated to emit bright blue fluorescence upon the addition of H2S (excitation at 405 nm, Fig.S4 in Supporting information).The fluorescence increased with increasing concentrations of H2S,and the enhancement peaked at 8.2-fold at 50 μmol/L H2S (Fig.1b).To confirm the reaction mechanism, the reaction product, ADNH2, was characterized by1H NMR and HR-MS (Figs.S5 and S6 in Supporting information).In the presence of RNA (no H2S), the intact probe AD-N3could also emit weak fluorescence, suggesting that the probe interacts with RNA mainly through the hydrogen bonds andπ-πinteractions.Further addition of H2S could also significantly enhance the probe’s fluorescence, showing that probe molecules bound to RNA are still reactive to H2S (Fig.1c).The measured binding constants (K) indicate that AD-N3and AD-NH2have strong affinities to nucleic acids, but the affinity to RNA is ∼20-fold stronger than that to DNA (inset of Fig.1c) [23].Additional selectivity tests indicated that only H2S can enhance the probe fluorescence, whereas other common compounds found in cells have no significant effects (Fig.S7 in Supporting information).The MTT assay for both AD-N3and AD-NH2showed that the probe was almost nontoxic to cells in the concentration range of 0–30 μmol/L(Fig.S8 in Supporting information).
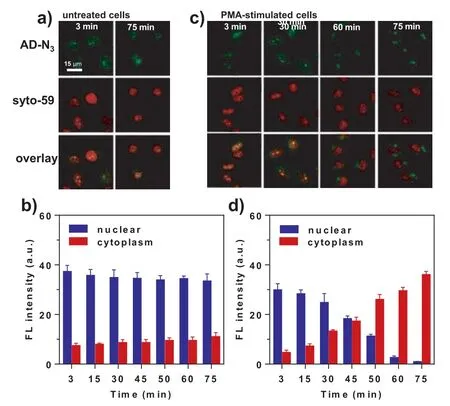
Fig.2.Fluorescent imaging of untreated and PMA-stimulated cells.(a) HeLa cells were treated with 5 μmol/L AD-N3 alone for 3 min, followed by incubation in culture media.(b) Time-dependent fluorescent intensity of (a).(c) HeLa cells were first treated with 2 μg/mL PMA for 1 h and then incubated with 5 μmol/L AD-N3 for 3 min.(d) Time-dependent fluorescent intensity of (c).The error bar represents the standard deviation (± SD).
When HeLa cells were treated with AD-N3, the fluorescent signal was detected within only 3 min (Fig.2a and Fig.S9 in Supporting information), showing fast reaction kinetics of converting AD-N3to AD-NH2.The addition of a well-established commercial nuclear-RNA dye, Syto-59, exhibited complete colocalization, suggesting that our probe can rapidly bind with nuclear RNA and become activated by H2S.Periodic imaging of the cells (once every 15 min) revealed virtually no change in fluorescence intensity (3–75 min, Fig.2b), indicating the chemical stability of AD-NH2.It was also noted that the cytoplasm exhibited negligible fluorescence,likely because the probe entered the nucleus very quickly and did not have time to react with cytoplasmic H2S.
In contrast, when the cells were stimulated by an apoptosisinducing drug, phorbol 12-myristate 13-acetate (PMA), the fluorescence in the cytoplasm gradually increased, while the fluorescence in the nucleus gradually decreased, suggesting that apoptosis accelerates the export of nuclear RNAs into the cytoplasm (Figs.2c and d).In the case of Syto-59, the red fluorescence in the cell nucleus also became weaker (likely due to photobleaching), but almost no signal was detected in the cytoplasm due to its recycling back to the nucleus.
To further verify the mechanism of our imaging probe, additional detailed imaging experiments were carried out.First, we performed a dual-color imaging experiment using our probe and a commercial Lyso-tracker and observed colocalization of the two colors in the cytoplasm, confirming that the cytoplasmic fluorescence was from lysosomes.This result shows that the fluorescent AD-NH2dissociated from the exported RNA was concentrated in lysosomes (Fig.S10 in Supporting information).Separately, when the cells were treated directly with AD-NH2(pre-activated), the fluorescence signal was also detected in lysosomes rather than the cell nucleus (Fig.3a).Combined, these images confirmed that the activated probe can not check back into the cell nucleus.The results in Fig.3 also proved that the probe was carried into the cytoplasm by nuclear RNA export, instead of free diffusion.When the AD-N3-treated cells were fixed, the fluorescence of the probe remained in the nucleus and did not leak into the cytoplasm (Fig.3b).This can be attributed to the strong affinity between the probe and nuclear RNA, preventing probe leaching.To further confirm this observation, we performed two additional control experiments(Figs.3b and c).First, live cells were incubated with PMA at 37°C for 2 h followed by treatment with the probe at 4 °C.No fluorescence enhancement was detected in the cytoplasm, showing that the entry of AD-NH2into the cytoplasm was through active transportation with energy consumption.Second, cells were pretreated with leptomycin B (LMB) which is a classic inhibitor of nuclear RNA export [24].Addition of PMA and the probe only resulted in fluorescence in the nucleus (not in lysosomes).Taken together,these characterization experiments validated our probe for imaging and quantification of nuclear RNA export, enabled by its intracellular relocation from the nucleus to lysosomes.
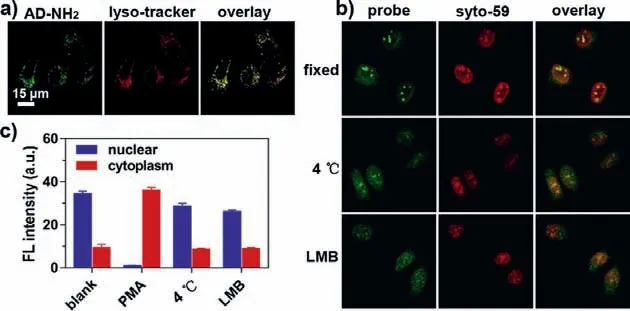
Fig.3.Fluorescent imaging for the certification of AD-N3 real-time measuring nuclear RNA export.(a) Fluorescent imaging of AD-NH2 in lysosomes.HeLa cells were incubated with 5 μmol/L AD-NH2 for 20 min, then treated with 1 μmol/L Lysotracker Deep Red.The overlay coefficient is 0.88.(b) Top row: HeLa cells incubated with 5 μmol/L AD-N3 for 5 min followed by fixation by a mixture of methanol and acetic acid (v:v=3:1) for 15 min.Middle row: cells incubated with 2 μg/mL PMA at 37 °C for 1 h, followed by treatment with AD-N3 at 4 °C for 75 min.Bottom row:cells simultaneously treated with 2 μg/mL PMA and 6 ng/mL leptomycin B (LMB)for 1 h, incubated with 5 μmol/L AD-N3 for 5 min, and imaged after 75 min.(c) Fluorescence intensities corresponding to the imaging results in (b).The error bars represent the standard deviation (± SD).
To demonstrate the application of this new probe, we evaluated the effect of hormones on nuclear RNA export.It is well-documented that hormones are critical regulators to activate or inhibit the expression of specific proteins, which are achieved through controlling the export of nuclear RNAs [25].Seven common hormones, tetraiodothyronine (T4), adrenaline,17α-methyltestosterone (MT), diethylstilbestrol (DES), dopamine,insulin, and melatonin, were selected to treat cells at the dosages in their normal physiological ranges.We found that among these hormones, T4 and adrenaline promoted nuclear RNA export, while the other five did not have a significant effect (Figs.4a and b).To further investigate the reason for the absence of RNA export for the five hormones, cells were simultaneously treated with PMA and each of the five hormones (Figs.4c and d), followed by incubation with our probe.Fluorescence imaging revealed that these five hormones suppressed the export of nuclear RNAs.
Very interestingly, we also discovered that these five hormones(MT, DES, dopamine, insulin, and melatonin) also effectively inhibited apoptosis induced by PMA.Flow cytometry showed that the cell viability was clearly elevated in the presence of these hormones (Fig.4e).Further immunofluorescence study also showed that the expression of apoptosis hallmark protein, caspase-3, was significantly inhibited (Fig.4f and Fig.S11 in Supporting information) [26,27].These results suggest that the five hormones are inhibitors of nuclear RNA export related to cell apoptosis, and can be suitable for the study of cellular responses to chemotherapeutics since anti-tumor drugs are often associated with hormone fluctuations in humans [28].
We proceeded to use the probe to test the correlation between hormones and a popular anti-tumor drug, paclitaxel, in affecting nuclear RNA export as well as cell killing.Paclitaxel kills tumor cells through binding with the microtubules to prevent the formation of spindle fiber [29].Surprisingly, we found that the use of paclitaxel alone promoted the export of nuclear RNA in HeLa cells (Figs.5a and b) and cell death (viability dropped to ∼21%),while joint using of paclitaxel with any of the five hormones reduced the paclitaxel’s effect on RNA export and cell killing (viability only dropped to 40%−50%, Fig.5c).These results show that the anti-tumor effect of chemotherapy could be partially offset by some human hormones, which deserves future investigation as a possible remedy to alleviate chemotherapy side effects.
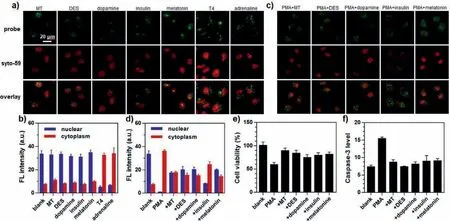
Fig.4.The evaluation of hormones regulating nuclear RNA export.(a) HeLa cells were treated with various hormones (MT: 0.2 μg/mL; DES: 0.2 μg/mL; dopamine: 1 μmol/L;insulin: 20 μg/mL; melatonin: 0.2 μg/mL; T4: 70 ng/mL; or adrenaline: 0.3 μg/mL) for 1 h, incubated with 5 μmol/L AD-N3 for 3 min, and imaged after 75 min.(b) Fluorescence intensity corresponding to the images in (a).(c) The cells were simultaneously treated with 2 μg/mL PMA plus individual hormones (MT: 0.2 μg/mL; DES: 0.2 μg/mL;dopamine: 1 μmol/L; insulin: 20 μg/mL; or melatonin: 0.2 μg/mL), incubated with 5 μmol/L AD-N3 for 3 min, and imaged after 75 min.(d) Fluorescence intensity corresponding to the images in (b).(e) Cell viability under the simultaneous treatment with PMA and the individual hormones assessed by flow cytometry.(f) The immunofluorescence intensity of caspase-3 in cells co-treated with PMA and individual hormones.The error bars represent the standard deviation (± SD).
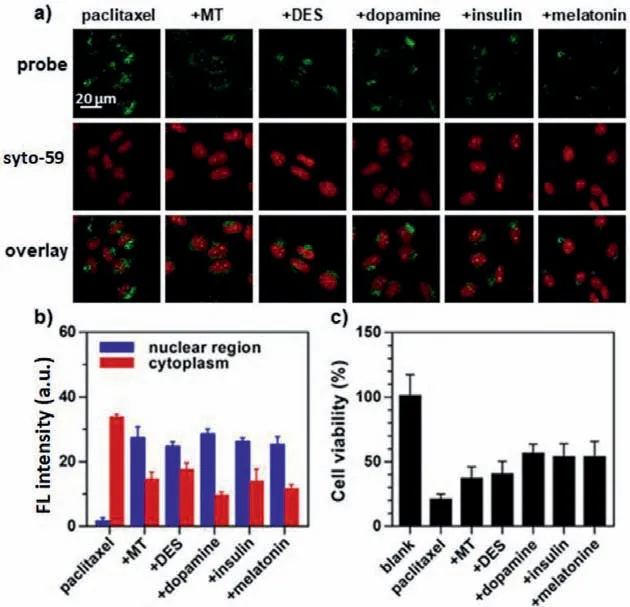
Fig.5.The anti-tumor effect of paclitaxel in the presence of nuclear RNA exportinhibiting hormones.(a) Cells were simultaneously treated with 1 μmol/L paclitaxel and the individual hormones (MT: 0.2 μg/mL; DES: 0.2 μg/mL; dopamine: 1 μmol/L;insulin: 20 μg/mL; or melatonin: 0.2 μg/mL), incubated with 5 μmol/L for 5 min, and imaged after 75 min.(b) Fluorescence intensity corresponding to the images in (a).(c) Cell viability after co-treatments with paclitaxel and the individual hormones assessed by flow cytometry.The error bars represent the standard deviation (±SD).
In summary, we have developed a novel molecular probe to address the lack of means for studying nuclear RNA export.We designed a probe that responds to intracellular H2S to activate its fluorescence while changing its affinity from the nucleus to lysosomes.This relocation from the nucleus feature is similar to a hotel stay: once the guest checks out of the hotel, this person can no longer check back in.The one-time stay allows real-time quantitative imaging of nuclear RNA exports, a highly useful indicator for drug screening and discovery.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos.21775001, 21874137, 21974001, 21976183 and 22074001), Natural Science Foundation of Anhui Province (No.1808085MB32) and Nature and Science Foundation from Anhui Province Ministry of Education (No.KJ2019A0011).
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2021.10.032.
 Chinese Chemical Letters2022年8期
Chinese Chemical Letters2022年8期
- Chinese Chemical Letters的其它文章
- Adsorptive removal of PPCPs from aqueous solution using carbon-based composites: A review
- A review on hollow fiber membrane module towards high separation efficiency: Process modeling in fouling perspective
- Recent advances in DNA glycosylase assays
- Chiral pillar[n]arenes: Conformation inversion, material preparation and applications
- Recent progress in carbon-based materials boosting electrochemical water splitting
- Working principle and application of photocatalytic optical fibers for the degradation and conversion of gaseous pollutants
