Comprehensive profiling and evaluation of the alteration of RNA modifications in thyroid carcinoma by liquid chromatography-tandem mass spectrometry
Meng-Yuan Chen, Chu-Bo Qi, Xiao-Meng Tang, Jiang-Hui Ding, Bi-Feng Yuan,∗,Yu-Qi Feng
a School of Public Health, Wuhan University, Wuhan 430071, China
b Department of Chemistry, Sauvage Center for Molecular Sciences, Wuhan University, Wuhan 430072, China
c Jiangxi Provincial People’s Hospital Affiliated to Nanchang University, Nanchang 330006, China
ABSTRACT RNA molecules contain diverse modifications that display important functions in a variety of physiological and pathological processes.So far over 150 chemical modifications have been characterized to be present in various RNA species, such as in messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA).Previous studies revealed that certain RNA modifications were correlated to specific human diseases, indicating RNA modifications could serve as the potential indicator of human diseases.However,systemic investigation of the alteration of RNA modifications in different RNA species of carcinoma tissues are still lacked.Herein, we carried out the comprehensive profiling and evaluation of the alteration of RNA modifications in thyroid carcinoma by liquid chromatography-tandem mass spectrometry (LC-ESIMS/MS) analysis.The developed method allowed us to simultaneously detect 48 different types of RNA modifications.Using this method, we detected 10, 15, 14, and 25 modifications in mRNA, 18S rRNA, 28S rRNA and small RNA (< 200 nt), respectively.Compared to the normal tissues, we revealed a total of 14 RNA modification exhibited significant increase and 2 RNA modifications showed significant decrease in thyroid carcinoma tissues.Our study provided the first comprehensive profile as well as the alteration of modifications in different RNA species in thyroid carcinoma and matched tumor-adjacent normal tissues.The altered pattern RNA modifications may serve as the indicator of thyroid carcinoma.Moreover, this study may promote the in-depth understanding of the regulatory roles of RNA modifications in thyroid carcinoma.
Keywords:RNA modification Liquid chromatography-mass spectrometry Thyroid carcinoma Profiling
DNA cytosine methylation is a well-explored epigenetic modification that can regulate gene expression in mammals[1–5].Apart from DNA, RNA molecules also contain numbers of chemical modifications [6].Mirroring DNA epigenetics, RNA epigenetics is an emerging field and over 150 structurally distinct modifications have been identified in different RNA species, including messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA) [6,7].The nucleobases as well as the ribose of RNA can be chemically modified to form diverse modifications, such as 5-methylcytidine (m5C),N6-methyladenosine (m6A), 2′-O-methylation,N1-methyladenosine(m1A),N6,2′-O-dimethyladenosine (m6Am),N7-methylguanosine(m7G),N3-methylcytidine (m3C), andN4-acetylcytidine (ac4C) [8–13].
The modifications in RNA greatly diversify their functions.Many RNA modifications are reversible and they play critical roles in modulating both physiological and pathological processes [14,15].The recognition factors of modifications can delicately control and regulate RNA stability, folding, localization and maturation.Although the complete functional roles of RNA modifications have not been fully unveiled, the known physiological functions of RNA modifications have been demonstrated to be involved in carcinogenesis, adipogenesis, spermatogenesis, and cell differentiation[16,17].
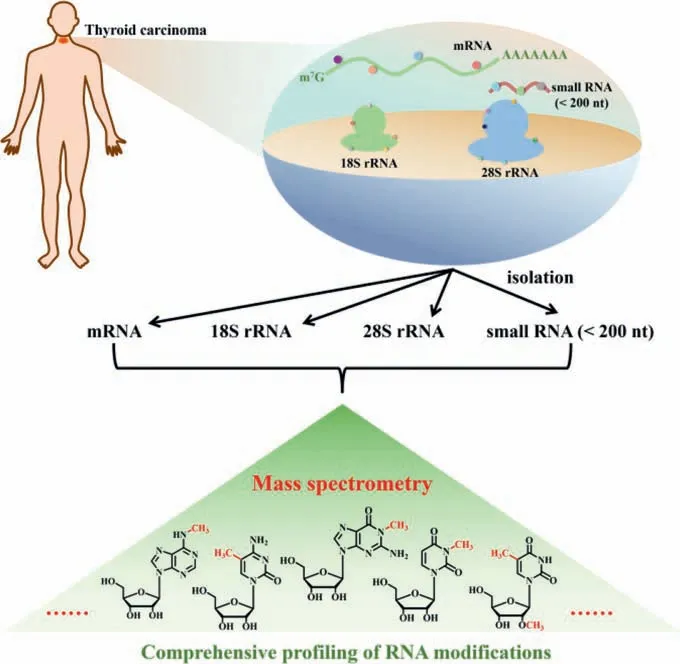
Fig.1.The schematic diagram of comprehensive profiling of RNA modifications in four RNA species from thyroid carcinoma by LC-ESI-MS/MS.Different RNA species(mRNA, 18S rRNA, 28S rRNA, and small RNA) were isolated from thyroid carcinoma and matched tumor-adjacent normal tissues.These RNA molecules were then enzymatically digested followed by LC-ESI-MS/MS analysis, which enables the comprehensive profiling and evaluation of the alteration of RNA modifications.
Due to the vital roles of RNA modifications in regulating cell fate decision, the dysregulation of RNA modifications, in terms of their levels and distributions, may contribute to carcinogenesis.In fact, the accumulated lines of evidence have established the links between specific RNA modifications and cancer onset and evolution [18].Several RNA modifications have been characterized for their effects on promoting cancer cell proliferation or invasiveness [19].For example, the level of pseudouridine (Y) is increased in malignant prostate compared to normal tissue [20].N6,2′-Odimethyladenosine (m6Am) can promote chemoresistance in colorectal cancer cells [21].A few studies have shown that RNA modification pathways can regulate cell metabolism; conversely, cancerspecific metabolic changes have the potential to affect RNA modifications [22].These studies highlight the importance of RNA modification pathways in tumorigenesis.
Considering we are in the early stage of RNA epigenetics, the effects of RNA modifications on cancer hallmark are still poorly understood.Nevertheless, deciphering the alteration of the levels of RNA modification from carcinoma tissues and normal tissues should provide valuable information for in-depth understanding of carcinogenesis as well as for disease diagnosis.Thyroid carcinoma is one of the most prevalent human malignancies [23].However,there are few studies investigating the correlation of RNA modifications in different RNA species with thyroid carcinoma.In this respect, herein we systematically carried out the comprehensive profiling and evaluation of the alteration of RNA modifications in different RNA species of thyroid carcinoma and matched tumoradjacent normal tissues by liquid chromatography-tandem mass spectrometry (LC-ESI-MS/MS) analysis (Fig.1).
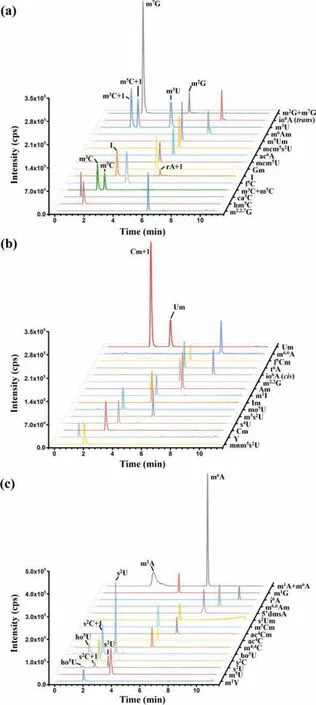
Fig.2.The extraction-ion chromatograms of 48 RNA modifications.The extraction-ion chromatograms of (a) m2G, m7G, io6A (trans), m3U, m6Am,m5Um, mcm5s2U, ac6A, mcm5U, Gm, I, f5C, m3C, m5C, ca5C, hm5C, m2,2,7G,(b) Um, m6,6A, f5Cm, t6A, io6A (cis), m2,2G, Am, m1I, Im, mo5U, m5s2U, s4U,Cm, Y, mnm5s2U, (c) m1A, m6A, m1G, i6A, m6,6Am, 5′dmsA, s2Um, m5Cm,ac4Cm, ac4C, m4,4C, ho5U, s2C, s2U, m5U and m1Y.m2G, N2-methylguanosine;m7G, N7-methylguanosine; io6A (trans), trans-zeatin-riboside; m3U, 3-methyluridine; m6Am, N6,2′-O-dimethyladenosine; m5Um, 5,2′-O-dimethyluridine;mcm5s2U, 5-methoxycarbonylmethyl-2-thiouridine; ac6A, N6-acetyladenosine;mcm5U, 5-methoxycarbonylmethyluridine; Gm, 2′-O-methylguanosine; I, inosine; f5C, 5-formylcytidine; m3C, N3-methylcytidine; m5C, 5-methylcytidine;ca5C, 5-carboxycytidine; hm5C, 5-hydroxymethylcytidine; m2,2,7G, N2,N2,7-trimethylguanosine; Um, 2′-O-methyluridine; m6,6A, N6,N6-dimethyladenosine;f5Cm, 5-formyl-2′-O-methylcytidine; t6A, N6-threonylcarbamoyladenosine;io6A (cis), cis-Zeatin-riboside; m2,2G, N2,N2-dimethylguanosine; Am, 2′-Omethyladenosine; m1I, 1-methylinosine; Im, 2′-O-methylinosine; mo5U, 5-methoxyuridine; m5s2U, 5-methyl-2-thiouridine; s4U, 4-thiouridine; Cm, 2′-Omethylcytidine; Y, pseudouridine; mnm5s2U, 5-methylaminomethyl-2-thiouridine;m1A, N1-methyladenosine; m6A, N6-methyladenosine; m1G, 1-methylguanosine;i6A, N6-isopentenyladenosine; m6,6Am, N6,N6,2′-O-trimethyladenosine; 5′dmsA,5′-deoxy-5′-methylthioadensine; s2Um, 2-thio-2′-O-methyluridine; m5Cm, 5,2′-Odimethylcytidine; ac4Cm, N4-acetyl-2′-O-methylcytidine; ac4C, N4-acetylcytidine;m4,4C, N4,N4-dimethylcytidine; ho5U, 5-hydroxyuridine; s2C, 2-thiocytidine; s2U,2-thiouridine; m5U, 5-methyluridine; m1Y, 1-methylpseudouridine.
A LC-ESI-MS/MS method to simultaneously detect 48 RNA modifications was first established (Fig.2, Fig.S1 and Table S1 in Supporting information).Among these modifications, there are eight groups of isomeric RNA modifications (m1A and m6A; io6A (cis)and io6A (trans); m1G, m2G, and m7G; m3C and m5C; f5C and m4,4C; s2U, s4U and ho5U; m3U and m5U; mo5U and m5s2U) and eight groups of RNA modifications with similar molecular weights(m1A, m6A and m1I; Am and Im; Cm and Um; m3C, m5C, m3U and m5U; m3U, m5U and s2C; s2C, ho5U, s2U and s4U; hm5C, mo5U and m5s2U; m5Cm and m5Um).Under the LC-ESI-MS/MS analysis with multiple reaction monitoring (MRM) detection mode, RNA modifications with same or similar MRM mass transitions need to be efficiently separated for their identification.Thus, we optimized the chromatographic separation conditions for these 48 RNA modifications.Under the optimal conditions, all these 48 RNA modifications, especially those with same or similar molecular weights,were well distinguished from each other (Fig.2 and Table S2 in Supporting information), which allowed the simultaneous detection and quantification of 48 RNA modifications.
The optimized LC-ESI-MS/MS method was then applied to detect and quantify RNA modifications from different RNA species of thyroid tissues, including mRNA, 18S rRNA, 28S rRNA and small RNA (<200 nt).mRNA, small RNA (<200 nt) and large RNA(>200 nt) were extracted using the commercial kit according to previous report [24,25] and the extraction procedure of mRNA could be found in Supporting information (Fig.S2 in Supporting information).The 18S rRNA and 28S rRNA were isolated from the large RNA (>200 nt) by size-exclusion chromatography according to the previous reports [26,27] (Fig.S3 in Supporting information).The LC-ESI-MS/MS results showed that 10, 15, 14, and 25 kinds of RNA modifications could be detected in mRNA (Fig.3), 18S rRNA(Figs.S4 and S5 in Supporting information), 28S rRNA (Figs.S6 and S7 in Supporting information) and small RNA (<200 nt) (Figs.S8–S11 in Supporting information), respectively.The retention times of these detected RNA modifications from different RNA species were identical to those of their standards (Fig.3 and Figs.S4–S11).It is remarkable that all these detected modifications were not detectable in the enzyme control samples (Figs.3 and Figs.S4–S11),validating that these detected RNA modifications were from RNA samples.
We next evaluated the abundance of RNA modifications in thyroid carcinoma and matched tumor-adjacent normal tissues.Thus, the calibration curves were first constructed to quantify RNA modifications.A series of mixed solutions that contain different amounts of RNA modifications and a certain amount of isotopic internal standard (rC-13C5) were prepared.We constructed calibration curves by plotting peak area ratios of nucleosides/rC-13C5against the amounts of nucleosides.The good linearities were achieved with the coefficients of determination (R2) being greater than 0.99 (Table S3 in Supporting information).The limits of detection (LODs) and the limits of quantification (LOQs), which are defined as the amounts of analytes at signal-to-noise ratio of 3 and 10, respectively, were used to evaluate the detection sensitivities for RNA modifications by the established method [28].LODs of RNA modifications ranged from 0.05 fmol to 486.3 fmol and LOQs were between 0.18 fmol and 1621.01 fmol (Table S3).The precision and accuracy for RNA modifications quantification were evaluated through calculating intra- and inter-day relative standard deviations (RSDs) and relative errors (REs), which were less than 17.4%and 17.6%, respectively (Table S4 in Supporting information).These results manifested the acceptable precision and accuracy for quantification of RNA modifications.
With the validated LC-ESI-MS/MS method, we then quantified RNA modifications in mRNA, 18S rRNA, 28S rRNA and small RNA(<200 nt) of thyroid carcinoma and the matched tumor-adjacent normal tissues (Table S5 in Supporting information).The results showed that the measured contents of various RNA modifications were from 0.00010% to 11.26% in different RNA species, indicating the large content ranges of endogenous RNA modifications.Nevertheless, the current developed method showed good capability in the simultaneous quantification of these RNA modifications.In addition, the measured levels of these modifications were comparable to the previously reported levels in RNA of mammals (Tables S6–S9 in Supporting information).As for the detectable 10 RNA modifications (m1A, m6A, m6Am, Am, m7G, Gm, m3C, m5C, Cm and I) from mRNA, the level of m5C showed significant increase in thyroid carcinoma compared to the matched tumor-adjacent normal tissues (Fig.4a).In our previous study, we also observed the increased levels of m5C in RNA in circulating tumor cells of patients with lung cancer [29].These results indicated that m5C in mRNA may have important role on thyroid carcinoma.
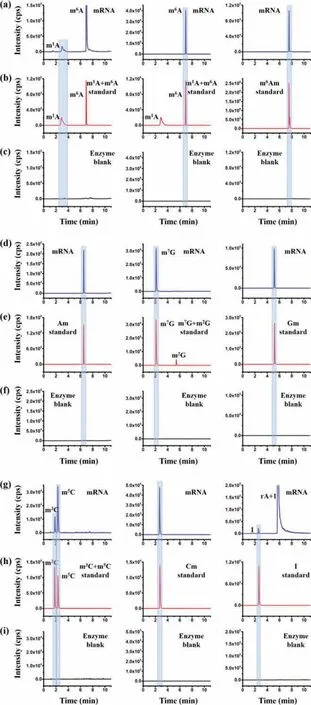
Fig.3.Extraction-ion chromatograms of m1A, m6A, m6Am, Am, m7G, Gm, m3C,m5C, Cm and I.(a, d, g) Detected RNA modifications in mRNA from thyroid carcinoma tissues.(b,e,h) Nucleoside standards.(c,f,i) Enzyme control samples for the detection of these RNA modifications.Enzyme blank samples represent the samples only containing enzymes used for digesting RNA.
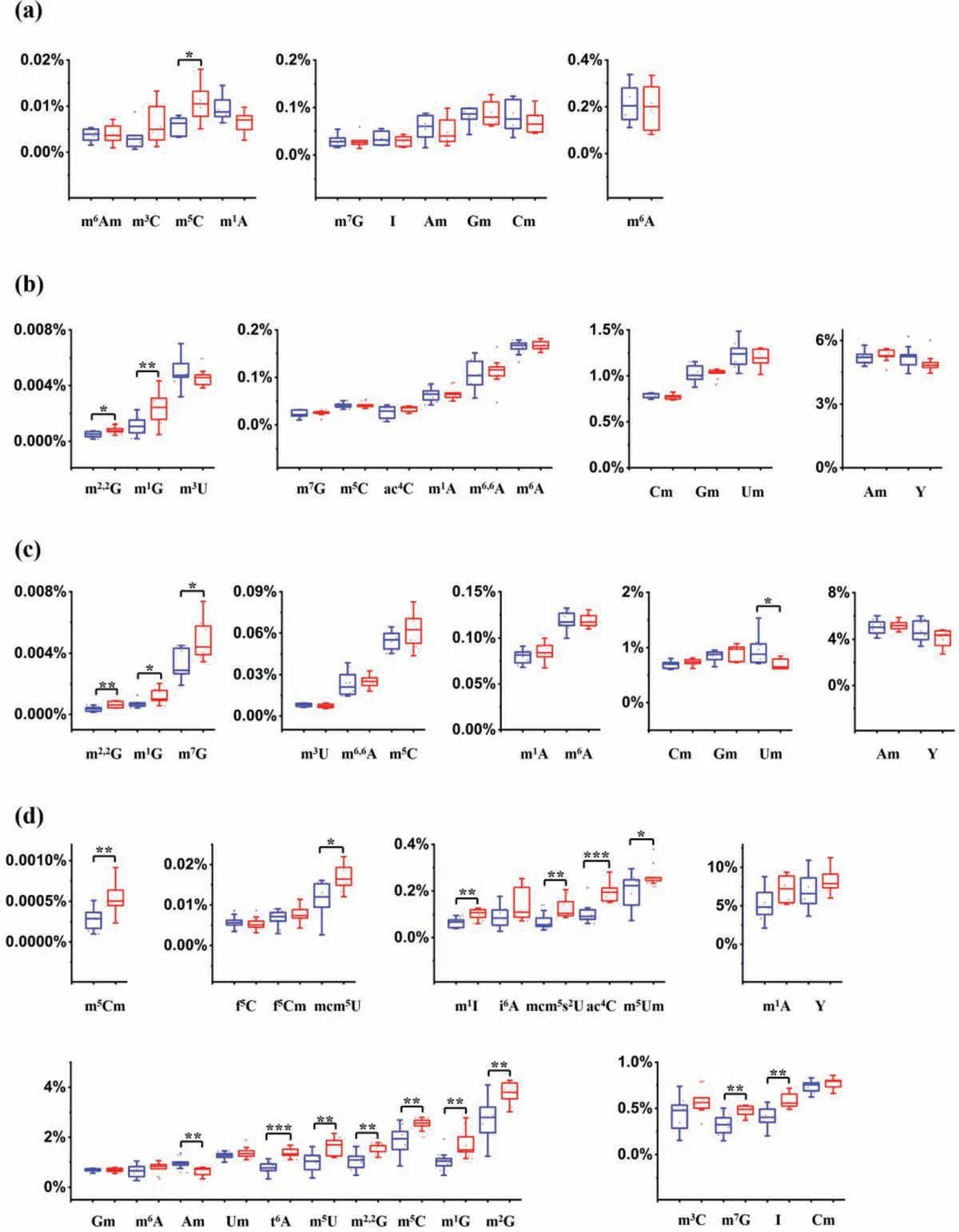
Fig.4.The measured levels of RNA modifications in different RNA species from thyroid carcinoma and the matched tumor-adjacent normal tissues.(a) mRNA.(b) 18S rRNA.(c) 28S rRNA.(d) Small RNA (<200 nt).Shown in blue and red represent the modifications from tumor-adjacent normal tissues and thyroid carcinoma tissues, respectively.The y-axis represents the levels of RNA modifications over their corresponding normal nucleosides.∗,P < 0.05; ∗∗,P < 0.01; ∗∗∗,P < 0.001.
As for 18S rRNA, 14 RNA modifications (m1A, m6A, m6,6A, Am,m7G, m1G, m2,2G, Gm, m5C, ac4C, Cm, m3U, Um and Y) could be quantified except m2G (Fig.4b).Although m2G was detectable in both 18S rRNA and 28S rRNA (Figs.S4 and S6 in Supporting information), it was not quantified in all the analyzed tissues due to its relatively low abundance.Thus, m2G was not included in the quantification data for 18S rRNA and 28S rRNA.The quantification results illustrated that m2,2G and m1G in 18S rRNA exhibited significant increase in thyroid carcinoma compared to the matched tumor-adjacent normal tissues.As for 28S rRNA, 13 RNA modifications (m1A, m6A, m6,6A, Am, m7G, m1G, m2,2G, Gm, m5C, Cm,m3U, Um and Y) could be quantified (Fig.4c).Among these modifications, m2,2G, m1G, and m7G showed significantly higher levels in thyroid carcinoma compared to the matched tumor-adjacent normal tissues.In addition, we also observed the significant decline of Um in 28S rRNA of thyroid carcinoma.It is worth noting that m2,2G and m1G showed significant increase in both 18S rRNA and 28S rRNA, indicating that these two RNA modifications in 18S rRNA and 28S rRNA should have important impact on thyroid carcinoma development.
Since the greatest number of modifications (25 RNA modifications) were detected in small RNA (<200 nt) (Fig.4d), we found 14 modifications (m5Cm, mcm5U, m1I, mcm5s2U, ac4C, m5Um,m7G, I, t6A, m5U, m2,2G, m5C, m1G and m2G) exhibited significant increase in thyroid carcinoma compared to the normal tissues (Fig.4d).Moreover, we observed the level of Am was significantly decreased in small RNA (<200 nt) of thyroid carcinoma tissues (Fig.4d).
The quantification results demonstrated an overall increased trend of RNA modifications in thyroid carcinoma compared to the matched tumor-adjacent normal tissues.As previous studies also demonstrated the increased levels of some RNA modifications in the tissues of cancer patients.For example, there are studies reporting that m6A in total RNA of gastric cancer tissues and m6A in mRNA of human hepatocellular carcinoma tissues was increased compared to the controls [30,31].The levels of m5Cm in total RNA of human lung carcinoma tissues and m5C in mRNA of urothelial carcinoma tissues showed increasing trends compared to the matched normal tissues [32–34].Our results, together with these previous studies, revealed that aberrant levels of RNA modifications were closely correlated to certain human diseases, which lays a foundation for diagnosis, monitoring disease evolution, and predicting response to treatment through specific RNA modification patterns.
We revealed that totally 16 RNA modifications exhibited significant difference (14 modifications increase and 2 modifications decrease) between thyroid carcinoma and matched tumor-adjacent normal tissues.Although the precise underlying mechanisms that explain how RNA modifications link to tumorigenesis remain unknown, we reason that the differentiated levels of RNA modifications may derive from the aberrant expression of RNA modifying enzymes.The contributions of these modifying enzymes and respective modification to tumor initiation, growth, and metastasis need to be further investigated in future study.
In summary, here we established the LC-ESI-MS/MS method for the simultaneous detection of 48 different types of RNA modifications.With the developed method, we carried out the comprehensive profiling and evaluation of the alteration of RNA modifications in thyroid carcinoma.We detected 10, 15, 14, and 25 modifications in mRNA, 18S rRNA, 28S rRNA and small RNA (<200 nt),respectively.Compared to the normal tissues, the quantification data showed a total of 14 RNA modifications exhibited significant increase and 2 RNA modifications showed significant decrease in thyroid carcinoma tissues.The specific pattern RNA modifications offer the new strategy for the diagnosis of thyroid cancer.Future investigation on the underlying mechanisms of the alteration of RNA modifications may also provide new opportunity on the thyroid carcinoma treatment through targeting RNA modifications and RNA modifying enzymes.
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgments
The work is supported by the National Natural Science Foundation of China (No.22074110), and the Fundamental Research Funds for the Central Universities (No.2042021kf0212).
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2021.12.008.
 Chinese Chemical Letters2022年8期
Chinese Chemical Letters2022年8期
- Chinese Chemical Letters的其它文章
- Adsorptive removal of PPCPs from aqueous solution using carbon-based composites: A review
- A review on hollow fiber membrane module towards high separation efficiency: Process modeling in fouling perspective
- Recent advances in DNA glycosylase assays
- Chiral pillar[n]arenes: Conformation inversion, material preparation and applications
- Recent progress in carbon-based materials boosting electrochemical water splitting
- Working principle and application of photocatalytic optical fibers for the degradation and conversion of gaseous pollutants
