TaGW2L, a GW2-like RING finger E3 ligase, positively regulates heading date in common wheat (Triticum aestivum L.)
Dijing Zhng, Xioxu Zhng,, Wu Xu, Tingting Hu, Jinhui M, Youfu Zhng, Jin Hou,Chenyng Ho, Xueyong Zhng,, Tin Li,
a College of Life Sciences, Henan Normal University, Xinxiang 453007, Henan, China
b Key Laboratory of Crop Gene Resources and Germplasm Enhancement, Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, Beijing 100081, China
Keywords:TaGW2L RING finger domain E3 ligase Heading date Triticum aestivum
A B S T R A C T RING finger E3 ligases play an important role in regulating plant growth and development by mediating substrate degradation. In this study,we identified TaGW2L,encoding a Grain width and weight2(GW2)-like RING finger E3 ligase, as a novel positive regulator of heading date in wheat (Triticum aestivum L.).TaGW2L exhibited high amino acid sequence similarities with TaGW2 homoeologs, particularly in the conserved RING finger domain. Expression analysis indicated that TaGW2L was constitutively expressed in various wheat tissues. TaGW2L showed transactivation activity in yeast and could interact with the ubiquitin-conjugating enzymes E2s. An in vitro ubiquitination assay verified that TaGW2L possessed a similar E3 ligase activity to TaGW2. Overexpression of the TaGW2L-7A homoeolog in wheat led to a significantly earlier heading date under both natural conditions and long-day conditions. Transcriptome analysis revealed that multiple known genes positively regulating wheat heading were significantly upregulated in the TaGW2L-7A-overexpression plants compared with the wild-type control. Together,our findings shed light on the role of TaGW2L in wheat heading date and provide potential applications of TaGW2L for the adaptation improvement of crops.
1. Introduction
Ubiquitination of proteins is a key mechanism regulating numerous cellular and organismal processes, which is achieved through an enzymatic cascade involving ubiquitin-activating(E1), ubiquitin-conjugating (E2), and ubiquitin-ligating (E3)enzymes [1]. The E3 ligase binds to both the substrate and the E2 thioesterified with ubiquitin (E2-Ub), catalyzing transfer of the ubiquitin from the E2 to the substrate and determining the specificity in ubiquitination [1,2]. As a major type of E3 ligases in eukaryotes,the really interesting new gene(RING)family contains a characteristic RING finger domain with a C-X2-C-X(9-39)-C-X(1-3)-H-X(2-3)-C-X2-C-X(4-48)-C-X2-C motif, which is responsible for both binding to the E2 and stimulating ubiquitin transfer [1,2]. RING E3-mediated ubiquitination can have diverse effects on the substrate, ranging from proteasome-dependent protein degradation to modulation of protein activity, stability, and localization [1,3].
In plants, increasing evidence indicates that RING E3 ubiquitin ligases are important in growth, development, and responses to various environmental stresses [4-7]. For example,COP1, the first RING finger gene identified inArabidopsis thaliana, functions as a central repressor in photomorphogenesis by targeting key lightsignaling regulators for degradation [4]. Another RING-type E3 ubiquitin ligase, salt and drought-induced RING finger1 (SDIR1),is involved in stress responses as well as development in both dicot and monocot plants [5,6].GRAIN WIDTH AND WEIGHT2(GW2), a major quantitative trait locus (QTL) in rice (Oryza sativa), encodes a previously unknown RING-type E3 ubiquitin ligase and negatively regulates grain width and weight [7]. GW2 shares high sequence similarity with DA2, and both can interact with DA1, a negative regulator of grain size, indicating a conserved function of the ubiquitin-proteasome pathway in governing organ size and grain weight [8,9]. Although RING finger ligases comprise the largest class of single-subunit E3 ligases in plants, only very few have been characterized and their biological functions are far from being elucidated.
Heading date is a critical trait for crop regional adaptation and has a substantial impact on crop yield. The important staple crop common wheat(Triticum aestivumL.)can adapt to various environments, an ability that is mainly attributed to complex genetic factors influencing the regulation of heading date[10].Wheat heading date or flowering time is to a great extent determined by gene groups that regulate the vernalization requirement (VRNgenes)and photoperiod sensitivity (PPDgenes) [11].VRN1promotes the transition from the vegetative to the reproductive phase, whereasVRN2acts as a suppressor of heading in winter wheat plants that have not been exposed to vernalization [12,13].PPD1encodes a core component of the photoperiod response pathway, and the dominant alleles promote earlier time to heading under long-day conditions[14].VRN3(TaFT1),an ortholog of ArabidopsisFLOWERING LOCUS T(FT), acts as the main integrator of the photoperiod and vernalization pathways [14,15]. Recently, numerous wheat genes have been identified as involved in the regulation of heading date; most are transcription factors, including the CCT-domain transcription factorsCO1,CO2, andTOC1[16], the MADS-box transcription factorsTaVRT2andTaSEP3[17,18], the GATA transcription factorTaZIM1[10], and the SBP-domain transcription factorTaGW8[19].Therefore,many genes are involved in the fine adjustment of heading date,and the underlying complex regulatory network still needs to be fully explored.
In this study, we identified a GW2-like RING finger geneTaGW2Las a previously unidentified positive regulator of heading date in wheat. TaGW2L shows relatively high protein sequence similarity with TaGW2 homoeologs,and they have similar expression patterns,subcellular localization,and E3 ligase activity.WhileTaGW2has a reported function in controlling grain weight,overexpressingTaGW2Lin wheat led to a significantly earlier heading date both in the field and greenhouse environments,implying thatGW2-likegenes might have divergent functions fromGW2in plant development. Our findings provide new insight into the functions ofGW2-likegenes in general and highlight the potential use ofTaGW2Lfor wheat adaptation improvement in particular.
2. Materials and methods
2.1. Plant materials and transformation
Wheat (Triticum aestivumL.) cultivar Chinese Spring (CS) was used forTaGW2Lisolation and spatiotemporal expression analysis.To construct theTaGW2L-overexpression vector for wheat transformation, the full-length coding sequence (CDS) ofTaGW2L-7Awas amplified from CS cDNA and cloned into a modified pCAMBIA3301 vector under the control of the maize(Zea mays)ubiquitin(Ubi)promoter as described[9].The resulting construct was introduced intoAgrobacterium tumefaciensstrain EHA105 and then transformed into immature embryos of wheat cultivar Fielder via a modified Agrobacterium-mediated transformation method [20].Phenotypic traits of T3generation transgenic lines were measured in two environments: greenhouses under long-day conditions(16 h light/8 h darkness)at 23°C and the Chinese Academy of Agricultural Sciences experimental field in Beijing under natural conditions.
2.2. RNA extraction and quantitative reverse-transcription PCR (RTqPCR)
Total RNA was extracted from wheat tissues using the RNA Easy Fast Plant Tissue Kit (Tiangen, Beijing, China). The cDNA was synthesized using the FastKing RT Kit (Tiangen, China). RT-qPCR was carried out on a Roche LightCycler 96 Real-time PCR system using SYBR Premix Ex Taq (Takara, Beijing, China), and the wheatActingene was used as the internal reference [9]. All RT-qPCR assays were performed three times, and the relative gene expression levels (presented as fold-change values) were calculated using the comparative CT method [21]. Primers used in this study are listed in Table S1.
2.3. Subcellular localization
The full-length CDSs ofTaGW2L-7AandTaGW2-6Awere amplified and separately subcloned into the pJIT163-GFP vector as described previously [9]. The verified fusion constructs and the control vector were introduced into wheat mesophyll protoplasts via the PEG-mediated method [22]. After a 16 h incubation, GFP fluorescence was observed using a LSM880 laser scanning confocal microscope (Zeiss, Oberkochen, Germany).
2.4. Transactivation activity assay
To explore the transactivation activity of TaGW2L in yeast, the full-lengthTaGW2L-7ACDS and its truncated variants were amplified and separately cloned into the pGBKT7(BD)vector.The resulting constructs were transformed into yeast strain AH109. After transformation, the transactivation activity was determined by the growth of transformants on a selection medium (SD/-Trp/-His/-Ade) as described [19].
2.5. Yeast two-hybrid assay
TheTaGW2L-7AcDNA encoding the N-terminus (residues 1-221), showing no transactivation activity, was used as the bait in yeast two-hybrid screening.In brief,yeast strain AH109 was transformed with BD-TaGW2L1-221, and yeast strain Y187 was transformed with an activating domain (AD) fusion library [23]. After the BD-TaGW2L1-221mating with the AD library, positive clones were selected on SD/-Trp/-Leu/-His/-Ade medium supplemented with X-α-gal, and then sequenced and annotated according to Yeast Protocols Handbook (Clontech, Palo Alto, CA, USA).
2.6. Protein expression and in vitro ubiquitination assay
The full-lengthTaGW2L-7AandTaGW2-6ACDSs were inserted into the pET32a vector (Novagen, Madison, WI, USA) and the recombinant plasmids were transformed intoEscherichia colistrain BL21(DE3).The transformed cells were induced with 0.8 mmol L-1isopropyl β-D-thiogalactopyranoside (IPTG) for 6 h at 25 °C. For protein purification, the cultured cells were lysed by sonication,and recombinant His-TaGW2L-7A and His-TaGW2-6A fusion proteins were purified using Ni-NTA resin (Qiagen, Dusseldorf, Germany) according to the manufacturer’s instructions.
Thein vitroubiquitination assay was performed as described previously with some modifications [7]. Briefly, 0.15 μg E1,0.25 μg E2, 9 μg ubiquitin (Ub), and 1 μg purified His-TaGW2L-7A, purified His-TaGW2-6A, or purified His-tag alone were incubated in a 30 μL reaction buffer containing 40 mmol L-1Tris-HCl(pH 7.5), 5 mmol L-1MgCl2, 2 mmol L-1ATP, and 2 mmol L-1DTT.The reaction was performed at 30°C for 3 h and then stopped with 1×SDS-PAGE loading buffer(100°C,5 min).Samples(15 μL)were separated by 12%SDS-PAGE,and ubiquitinated proteins were detected by immunoblotting with an anti-ubiquitin antibody or an anti-His tag antibody(ABclonal,Wuhan,China).Human E1(UBE1),E2 (UbcH5a), and Ub were purchased from R&D Systems (Emeryville, CA, USA).
2.7. Transcriptome analysis
Total RNA was extracted from the third leaves of three-weekold wild-type (WT) andTaGW2L-7A-overexpression plants using Trizol reagent (Invitrogen, Carlsbad, CA, USA). For each genotype,a 1-g leaf sample was collected for RNA extraction and pooled for each of the three biological replicates. Library construction and sequencing were carried out on the Illumina Hiseq Novaseq platform as described [24]. Clean sequence reads were mapped to the CS reference genome v.1.0 using HISAT2 program [25]. The expression level of each gene was normalized by fragments per kilobase of transcript per million fragments(FKPM)using StringTie[25], and differential expression analyses between the samples were performed using DESeq2[26]. Differentially expressed genes between sample libraries were identified based on the threshold with fold change >2 or <0.5,P-value <0.05, and FDR <0.05.
3. Results
3.1. Isolation and characterization of TaGW2L homoeologs in wheat
To isolate potentialGW2-likegenes in wheat, we performed a bioinformatics search ofTriticum aestivumcDNA sequences in Ensembl Plants [27]. BesidesTaGW2homoeologs, aGW2-likegene was identified as a close homolog withTaGW2and designatedTaGW2L.TaGW2Lhomoeologs were located on chromosomes 7A,7B, and 7D (Table S2). Primers TaGW2L-F/R were designed to amplify theTaGW2Lgenomic and coding sequences from genomic DNA and cDNA, respectively, from CS (Table S1). The coding regions ofTaGW2L-7A, -7B, and -7Dare 3854, 3692, and 3917 bp in length, respectively, and each comprise 8 exons and 7 introns(Fig. S1). TaGW2L homoeologs (-7A, -7B, and -7D) show high amino acid sequence similarities(~50%)with TaGW2 homoeologs(-6A, -6B, and -6D), especially in the N-terminal region, including the RING finger domain (Fig. 1). LikeGW2,GW2Lgenes were also conserved in various plant species, e.g., rice, maize,Brassica rapa,and Arabidopsis (Fig. S1). The ubiquitous distribution ofGW2Lhomologs across plant species implies the potential importance of their roles in plants.
3.2. Subcellular localization and expression analysis of TaGW2L and TaGW2
To investigate the subcellular localization of TaGW2L and TaGW2, we fused the CDSs ofTaGW2L-7AandTaGW2-6Ainframe with the gene encoding green fluorescent protein (GFP)and introduced the resulting constructs into wheat protoplasts.The GFP fluorescence of the fused TaGW2L-7A-GFP and TaGW2-6A-GFP proteins were both distributed in the cytoplasm and nucleus, showing similar subcellular localization patterns(Fig. 2A). Next, we analyzed the spatial and temporal expression patterns ofTaGW2LandTaGW2in CS by RT-qPCR. Similarly, bothTaGW2LandTaGW2were constitutively expressed in various wheat tissues at different developmental stages (Fig. 2B).TaGW2showed higher expression levels in developing spikes and grains thanTaGW2L,consistent with its important function in the regulation of yield traits[28].The slightly different expression patterns ofTaGW2LandTaGW2implied that they might play divergent roles in wheat development.
3.3. Transactivation activity analysis of TaGW2L in yeast
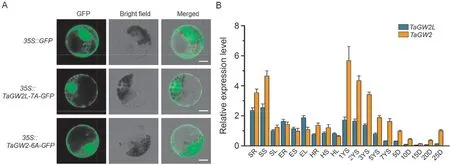
Fig.2. Subcellular localization and spatiotemporal expression of TaGW2L and TaGW2 in wheat.(A)Subcellular localization of TaGW2L and TaGW2 in wheat protoplasts.Scale bars,10 μm.(B)Expression pattern analysis of TaGW2L and TaGW2 in wheat.SR,seedling roots;SS,seedling stems;SL,seedling leaves;ER,roots at elongation stage;ES,stems at elongation stage; EL,leaves at elongation stage; HR,roots at heading stage; HS,stems at heading stage; HL,leaves at heading stage; 1YS-7YS, 1-7 cm young spikes; 5D-25D, grains at 5-25 days after pollination. Values are presented as mean ± SD.
Because GW2 could autoactivate the reporter gene when fused to the GAL4 DNA binding domain(BD)in yeast[29],we first examined whether TaGW2L also possessed similar transcriptional activation activity. The yeast harboring the full-lengthTaGW2L-7Awere able to grow and turn blue on SD/-Trp/-His/-Ade medium containing X-α-gal (Fig. 3A), confirming that TaGW2L-7A had transactivation activity and activated reporter gene expression in yeast. To locate the TaGW2L-7A activation domain, we generated a series of deletion constructs and tested their activation ability.The TaGW2L-7A N-terminal regions, including N1 (residues 1-104) and N2 (residues 1-221), exhibited no transactivation activity, while the C-terminal regions, especially C2 (residues 222-417), were required for its transactivation activity (Fig. 3A).
3.4. TaGW2L interacts with E2s
To circumvent the autoactivation activity of the C-terminus,we used the TaGW2L-7A N2 region (residues 1-221) as the bait in a yeast two-hybrid screening assay to detect its interacting proteins.After screening, we selected 37 clones showing strong interaction with TaGW2L-7A for sequencing. More than half of the clones(28/37) corresponded to E2 ubiquitin-conjugating enzymes(Table S3). The remainder mostly corresponded to ubiquitin-pro teasome-related proteins, including polyubiquitin and SINA-like E3 ligase (Table S3). To confirm the interaction between TaGW2L-7A and E2s, we performed a yeast two-hybrid assay by co-transformationTaGW2L-7AandTaE2(TraesCS3D02G271400).As expected, TaGW2L-7A strongly interacted with TaE2 (Fig. 3B).These results confirmed that TaGW2L-7A interacted with E2s,consistent with its potential E3 ligase function.
3.5. TaGW2L is a functional E3 ubiquitin ligase
Those interactions, as well as the presence of the highly conserved RING finger domain indicated that TaGW2L might be a functional E3 ubiquitin ligase,like TaGW2[30].To test this hypothesis, we performed anin vitroubiquitination assay using purified His-tagged TaGW2L-7A and TaGW2-6A recombinant proteins.Ubiquitination activity was observed in the presence of Ub, E1,E2, and His-tagged TaGW2L-7A protein by immunoblotting using an anti-Ub antibody (Fig. 4). An anti-His tag blot analysis also showed self-ubiquitination of TaGW2L-7A. The positive control His-tagged TaGW2-6A protein showed similar ubiquitination activity, whereas the negative control, His-tag alone, showed no ubiquitination activity. Absence of Ub, E1, or E2 abolished the self-ubiquitination ability of TaGW2L-7A (Fig. 4). Our results confirmed that TaGW2L is a functional RING-type E3 ubiquitin ligase.
3.6. Overexpression of TaGW2L promotes heading in wheat
To explore the biological function ofTaGW2L, we generatedTaGW2L-7A-overexpression (OE) transgenic lines in the common wheat cultivar Fielder. The transcript levels ofTaGW2L-7Awere all significantly increased in three representative transgenic OE lines(OE1-OE3), to >10-fold higher than in the wild-type (WT) control(Fig. S2). Phenotypic assessment in two environments (field and greenhouse) indicated thatTaGW2L-7Aoverexpression could lead to a much earlier heading time.Under natural conditions in the field,the heading date of OE lines was 6.5-8.5 days earlier than that of the WT (Fig. 5A, B). Similarly, under long-day conditions in the greenhouse,the heading date of OE lines was 6.8-9.1 days earlier than that of the WT (Fig. 5C, D). In addition, the plant height of OE lines was slightly higher than that of the WT (Fig. S2). We also investigated yield-related traits including tiller number, spikelet number per spike, grain number per spike, and thousand-grain weight (TGW).As shown in Fig. S2, there was no significant difference in these yield-related traits between the WT and the OE lines. Therefore,TaGW2Lshowed a major effect in promoting earlier heading.
Furthermore, we also generatedTaGW2-6AOE transgenic lines and surveyed their phenotypes in the field environment.As shown in Fig. S3, three representativeTaGW2-6AOE lines all demonstrated decreased grain size and grain weight compared with WT andTaGW2L-7AOE lines, consistent with its known function in negatively regulating grain size and weight [7,9]. Different withTaGW2L-7AOE lines,TaGW2-6AOE lines only showed slight change in heading data(<2 days)compared with WT(Fig.S3).These phenotypic data clearly indicated thatTaGW2Lhad a different function withTaGW2.
3.7. Identification of differentially expressed genes between the WT and TaGW2L-OE1 by RNA-seq
We performed RNA-seq transcriptome analysis of three-weekold seedling leaves of the WT and theTaGW2L-7AOE1 line. Based on a fold-change threshold of >2 or <0.5, with aP-value and FDR <0.05, we determined that a total of 1375 and 3623 genes were up- and down-regulated, respectively, in the OE1 line(Fig. 6A). Notably, many known genes promoting wheat heading or flowering were significantly upregulated in the OE1 line(Fig.6B),consistent with its earlier heading phenotype.These were mainly genes involved in the regulation of vernalization and photoperiod, such asTaVRN1,TaVRN3(TaFT1),TaPHYA,TaPPD1, andTaCO1(Fig.6B).Both CCT family and MADS-box transcription factors are important regulators of heading date or flowering time [14].Interestingly, multiple CCT and MADS-box family genes were differentially expressed between WT and OE1 plants, and most were upregulated in the OE1 line(Fig.S4),indicating that induced expression of CCT and MADS-box regulators might play a role in accelerating heading in transgenic wheat. Next, a RT-qPCR analysis was conducted to further confirm the expression patterns of three representative genes (TaVRN1,TaVRN3, andTaCO1) in the WT and theTaGW2L-7AOE lines.The expression levels of these three genes were all increased in three-and six-week-old OE lines compared with the WT (Fig. 6C-E). Therefore, our transcriptome data supported thatTaGW2Lcould induce the expression of key vernalization and photoperiod regulators includingTaVRN1,TaVRN3, andTaCO1.
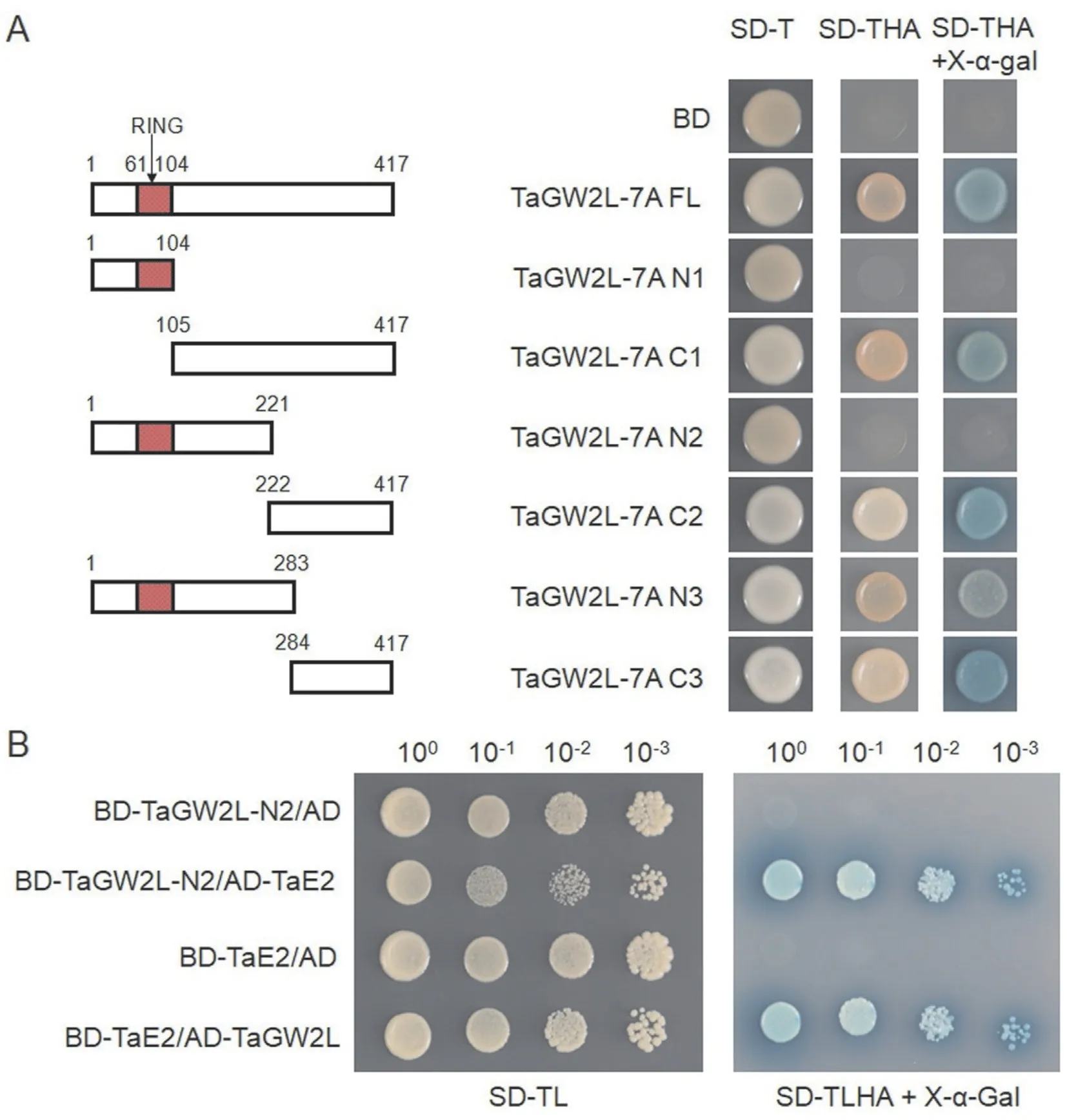
Fig.3. Transactivation activity and interaction assays of TaGW2L in yeast.(A)Transactivation activity assay of full-length and truncated TaGW2L-7A.SD-T,SD/-Trp;SD-THA,SD/-Trp/-His/-Ade.(B)Confirmation of the interaction between TaGW2L-7A and TaE2(TraesCS3D02G271400)by yeast two-hybrid assay.SD-TL,SD/-Trp/-Leu;SD-TLHA,SD/-Trp/-Leu/-His/-Ade.
4. Discussion
4.1. TaGW2L is a novel positive regulator of heading date in wheat
In plants,RING-type E3 ubiquitin ligases are the most abundant E3 enzymes, and a total of 1255 putative RING proteins have been predicted in wheat[31].Many RING finger E3 ligases participate in regulating plant development and stress responses by targeting various substrates for degradation. However, very few have been characterized and identified in the regulation of heading date or flowering time. In Arabidopsis, HOS1 is a RING-type E3 ubiquitin ligase and negatively regulates flowering by mediating CO degradation [32]. In rice, the RING E3 ligase HAF1 modulates circadian accumulation of ELF3 to control heading date under long-day conditions [33]. In the current study, we identified TaGW2L, a GW2-like RING E3 ligase, as a novel regulator of heading date in wheat.Transgenic wheat plants constitutively overexpressingTaGW2L-7Ahad a significantly earlier heading date:6.5-8.5 days earlier under natural field conditions and 6.8-9.1 days earlier under long-day conditions in the greenhouse compared to the WT(Fig. 5). Consistent with the phenotype, our transcriptome data and RT-qPCR assay indicated that some key genes positively regulating wheat heading time were significantly upregulated in theTaGW2L-7Aoverexpression lines, such asTaVRN1,TaVRN3, andTaCO1(Fig. 6).Thus,our findings provide evidence that TaGW2L is a positive regulator of wheat heading date mainly by inducing the expression of key vernalization and photoperiod regulators.
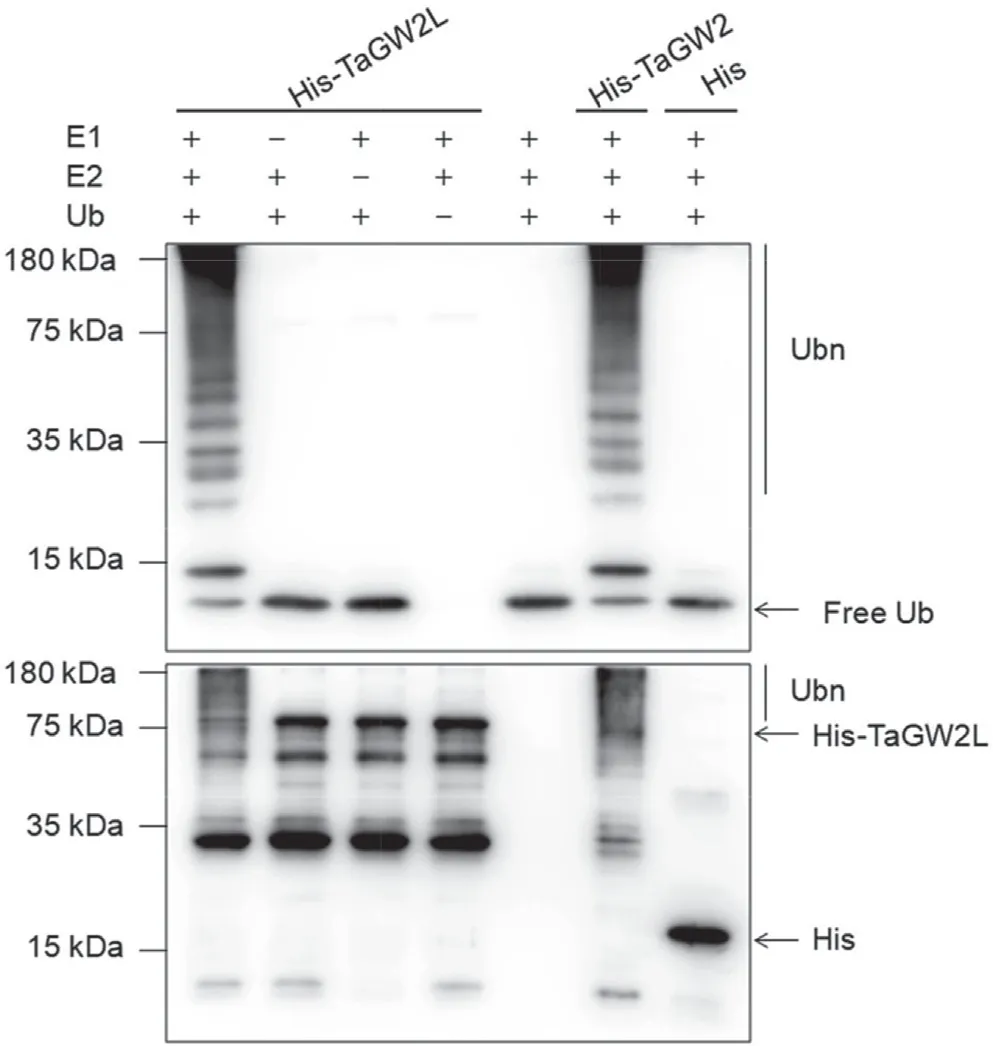
Fig. 4. In vitro ubiquitination assay of TaGW2L-7A. His-TaGW2L-7A recombinant protein was assayed for E3 ligase activity in the presence of E1, E2, and Ub. His-TaGW2-6A was used as a positive control and His-tag alone was used as a negative control.An anti-Ub antibody was used to detect free Ub and ubiquitinated proteins(Ubn), and an anti-His antibody was used to detect His-tagged recombinant proteins.
4.2. Similarities and differences between TaGW2 and TaGW2L
As close homologs, TaGW2L homoeologs share high sequence similarities with TaGW2 homoeologs, especially in the Nterminal regions including the RING finger domain (Fig. 1). Interestingly, TaGW2 and TaGW2L also showed similarities in other aspects, e.g., protein structure and biochemical function. First,TaGW2 and TaGW2L both have a conserved RING finger domain in the N-terminus, conferring similar E3 ubiquitin ligase activities(Fig.4). Second,TaGW2 and TaGW2L exhibited a similar subcellular localization pattern and spatial and temporal expression patterns in wheat (Fig. 2). Third, both TaGW2 and TaGW2L showed transactivation activities in yeast, and their C-terminus was required for the transactivation activities(Fig.3A).Previous studies have demonstrated a role ofTaGW2in the regulation of yield traits including grain width and grain weight [9,28]. Consistently, overexpression ofTaGW2-6Aresulted in significant reduction of grain size and weight (Fig. S3). However,TaGW2Lshowed a functional difference fromTaGW2in our transgenic research.Overexpression ofTaGW2L-7Acaused no significant effects on wheat yield traits,including tiller number and grain weight (Fig. S2). In contrast,TaGW2Lshowed a major effect in promoting earlier heading, andTaGW2L-7Aoverexpression led to a much earlier heading time(Fig. 5). A similar phenomenon has been observed for two barley(Hordeum vulgare)genes homologous toGW2,HvYrg1andHvYrg2:down-regulation ofHvYrg1resulted in earlier heading time and a prolonged grain-filling period,whereas down-regulation ofHvYrg2delayed heading time, showing a somewhat opposite effect [34].Taken together, these findings indicated thatTaGW2Lmight play divergent roles in wheat development and act non-redundantly withTaGW2.
4.3. Possible functional mechanisms of TaGW2L in regulating heading date
Heading date is a key agronomic trait affecting crop yield and regional adaptability. Numerous genes have been found to help regulate heading date in wheat by influencing multiple signaling pathways, such as photoperiod and vernalization [14]. Therefore,there is a complex regulatory network fine-tuning wheat heading date. Herein, we identifiedTaGW2Las an important positive regulator of heading date, which, given its E3 ligase activity, might function by targeting certain substrate proteins for degradation.Further investigation will be needed to identify these target substrates. In our first attempt to identify TaGW2L-interacting proteins through yeast two-hybrid screening, most clones we identified corresponded to ubiquitin-proteasome-related proteins,including the ubiquitin-conjugating enzymes E2s, polyubiquitin,and SINA-like E3 ligase (Table S3). These results indicated that the TaGW2L N-terminus containing the RING finger domain is mainly responsible for binding to the E2 and stimulating ubiquitin transfer, while the C-terminus with autoactivation activity might interact with substrate proteins,but may be difficult to investigate through yeast two-hybrid screening.Consequently,other methods such as immunoprecipitation-mass spectrometry (IP-MS) may be applied to identify authentic substrates of TaGW2L in wheat.Moreover, the transactivation ability of TaGW2L in yeast also implies that, aside from its E3 ubiquitin ligase function, it might also be directly involved in transcriptional regulation.Future studies will need to address whether TaGW2L can act as a transcriptional co-activator or chromatin modifier.
4.4. Potential application of TaGW2L in wheat adaptation improvement
Due to climate change,water deficit and temperature extremes influence the reproductive phase of plant growth and always cause considerable yield reductions of cereal crops [35]. Breeding early maturing varieties is an important strategy to resist various stresses such as drought, heat and fungal diseases during wheat reproductive stage and grain formation. Moreover, early heading and maturity are also favorable agronomic traits in China’s major wheat production regions with multiple cropping systems.TaGW2Lis a positive regulator in heading date, supporting it as a potential target gene in wheat adaptation improvement and early heading variety breeding. Although slight increase in plant height was observed in some transgenic lines, overexpression ofTaGW2Lhad a major effect in early heading and showed no significant decrease in yield traits including tiller number and grain weight(Fig. S2). Therefore, it will be promising in breeding early heading varieties without yield punishment based on marker-assisted selection by discovering and applying functional alleles ofTaGW2Lcorresponding to high enzymatic activities or high expression levels.
CRediT authorship contribution statement
Daijing Zhang:Conceptualization, formal analysis, writing -original draft.Xiaoxu Zhang:Investigation, formal analysis.Wu Xu:Investigation.Tingting Hu:Investigation.Jianhui Ma:Data curation.Youfu Zhang:Resources.Jian Hou:Resources.Chenyang Hao:Resources.Xueyong Zhang:Funding acquisition,supervision,writing - review & editing.Tian Li:Funding acquisition, supervision, writing - review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (32172045, 31671687), the National Key Research and Development Program of China (2016YFD0100302), and the Agricultural Science and Technology Innovation Program of the CAAS.
Appendix A. Supplementary data
Supplementary data for this article can be found online at https://doi.org/10.1016/j.cj.2021.12.002.
- The Crop Journal的其它文章
- Research progress on the divergence and genetic basis of agronomic traits in xian and geng rice
- From model to alfalfa: Gene editing to obtain semidwarf and prostrate growth habits
- The chloroplast-localized protein LTA1 regulates tiller angle and yield of rice
- Genome-wide association study and transcriptome analysis reveal new QTL and candidate genes for nitrogen-deficiency tolerance in rice
- Advances in the functional study of glutamine synthetase in plant abiotic stress tolerance response
- Identification of microRNAs regulating grain filling of rice inferior spikelets in response to moderate soil drying post-anthesis

