Antibacterial and antibiofilm activity of peptide PvGBP2 against pathogenic bacteria that contaminate Auricularia auricular culture bags
Shen Yng, Zijin Yun, Jude Juventus Awey, Shnggui Deng,Wuyin Weng, Yueling Zhng,*, Gungming Liu,*
a College of Food and Biological Engineering, Xiamen Key Laboratory of Marine Functional Food, Jimei University, Xiamen 361021, China
b Department of Biology, Guangdong Provincial Key Laboratory of Marine Biotechnology, Shantou University, Shantou 515063, China
c College of Food and Pharmacy, Zhejiang Ocean University, Zhoushan 316000, China
ABSTRACT
Bacteria contamination in Auricularia auricular culture bags reduces yield and increases the risk of food safety.In this study, 5 species of bacteria, mainly gram-positive bacteria including three species of Bacillus spp.,Arthrobacter arilaitensis and Staphylococcus warneri, were isolated and identified from bacteria-contaminated A. auricular culture bags. An in silico predicted antimicrobial peptide from the β-1,3-glucan-binding protein sequence of Penaeus vannamei, designated PvGBP2 (FLKLGRKSRYGMLKL), was screened and its antibacterial effect and mechanism of action on the isolated Bacillus spp. explored. The minimal inhibitory concentrations (MIC) of PvGBP2 on Bacillus spp. were 15.6–31.25 μg/mL. Peptide PvGBP2 could inhibit Bacillus subtilis in A. auricular culture bags to maintain growth and yield of A. auricular. Transmission electron microscopy (TEM) revealed that PvGBP2 kills bacteria by perforating the cell wall, destroying membrane integrity and resulting in the leakage of intracellular solutes. In addition, PvGBP2 inhibits biofilm formation by B. subtilis by 90.6% at 1 × MIC. Thus, peptide PvGBP2 could be potentially applied as an antibacterial agent to control bacterial infection of A. auricular cultivation and the spread of foodborne pathogens.
Keywords:
Auricularia auricular
Antimicrobial peptide
Penaeus vannamei
β-1,3-Glucan-binding protein
Bacillus spp.e
1. Introduction
Auricularia auricular are saprophytes mostly found on angiosperm trees and fallen logs, and are consumed as food in China and Southeast Asia [1,2]. They are the fourth largest edible mushroom after Agaricus bisporus, Pleurotus ostreatus and Lentinula edodes [3].Apart from their nutritional value, A. auricular are found to possess many bioactivities including antioxidant, modulation of intestinal microbiota and anti-in flammatory [4-6]. The culture of A. auricular is a mature technology that uses bagsfilled with wood chips, wheat bran, corncob granules, lime and gypsum, which are sterilized at 100 °C for 12 h, before being inoculated with A. auricular [7]However, due to incomplete sterilization, some A. auricular culture bags become contaminated with bacteria (Fig. 1), thereby reducing yield and posing increase risk of food poisoning or safety.
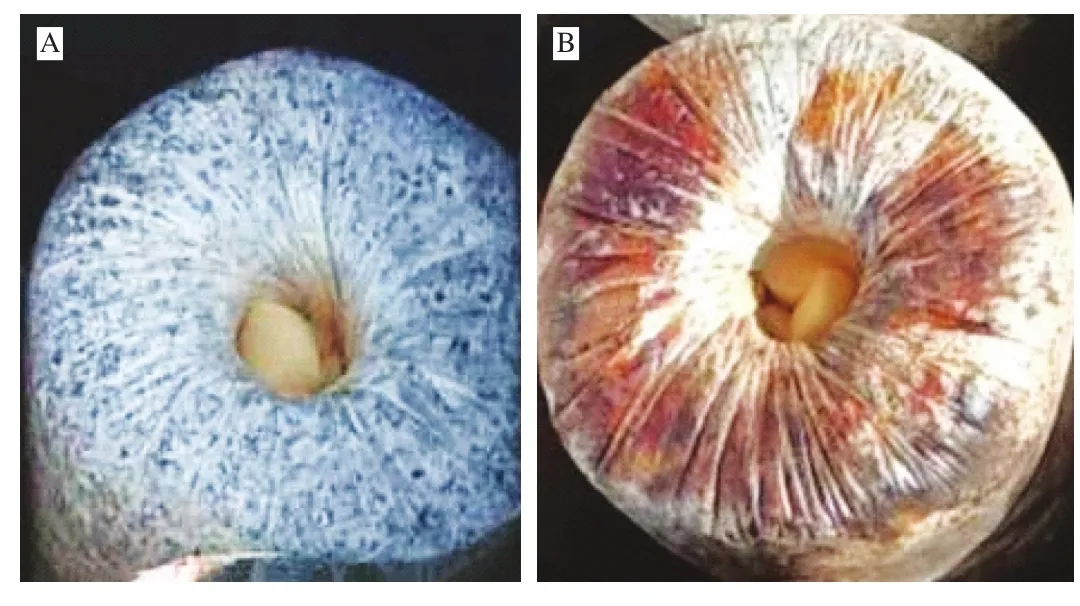
Fig. 1 A. auricular culture bags. (A) Uncontaminated A. auricular culture bag; (B) contaminated A. auricular culture bag.
Antimicrobial peptides (AMPs) are widely used as food preservatives to control food-borne pathogens in the food industry [8].Most AMPs act by destroying bacterial cell membranes, as they bind to negatively charged phospholipids on gram-positive and gram-negative bacteria [9]. Given that phospholipids constitute a very small part of fungal membranes, some cationic AMPs could be used in A. auricular culture bags to prevent bacterial contamination [10].
In invertebrates, β-1,3-glucan-binding protein (β-GBP) is found in diverse species and plays crucial roles including antibacterial activity against pathogenic gram-positive bacteria, such as Bacillus subtilis and Bacillus cereus etc. [11-13]. Despite these observations,the role played by AMPs derived from β-GBP has not been reported.With the vast improvements in the use of in silico tools for AMPs prediction [14-16], it is now feasible to leverage these tools to predict and screen AMPs derived from sequence of β-GBP against bacteria such as Bacillus spp.
In this study, an AMP designated PvGBP2, derived from the β-GBP sequence of P. vannamei, was shown to possess strong antibacterial activity against Bacillus spp. isolated from bacteria contaminated A. auricular culture bags. The antibacterial mechanism of peptide PvGBP2 was also elucidated.
2. Materials and methods
2.1 Strains and culture conditions
The strain of A. auricular and bacteria-contaminated A. auricular culture bags were obtained from Dongning Edible Mushroom Research and Development Center (Heilongjiang, China).Samples were cultured in potato dextrose agar (PDA) slants (20% potato, 2% glucose, 0.2% yeast extract, 0.4% KH2PO4, 0.2% MgSO4·7H2O, and 16% agar, pH (5.5 ± 0.1)) at 21 °C protected from light for 10 days [17]. Strains Arthrobacter arilaitensis (JN885458.1),B. subtilis (AY867792.1), B. cereus (KX890470.1), Bacillus nealsonii(JQ579625.1), and Staphylococcus warneri (KF057949.1) were isolated from the bacteria-contaminated A. auricular bags cultured at 37 °C for 24 h in nutrient broth (NB) medium.
2.2 Isolation of bacteria from A. auricular bags
Dregs (5 g) were taken from contaminated A. auricular bags and ground in a sterile mortar and pestle using 5 mL of 0.85% sterile saline before being spread on yeast extract (1%), sucrose (1%), agar (2%)plates treated with anti-fungal cyclohexamide. Next, plates were incubated at 37 °C and the bacteria colonies were sub-cultured to obtain pure cultures [18].
2.3 Identification and phylogenetic analysis of isolated bacteria
Bacteria in the isolated cultures were identified through 16S rDNA sequencing [19]. Briefly, bacteria genomic DNA was extracted and purified by DNA extraction kit (The Beijing Genomics Institute, China) followed by polymerase chain reaction (PCR)amplification of the 16S rDNA sequence using two universal primers 27F (AGAGTTTGATCCTGGCTCAG) and 1492R(TACGGCTACCTTGTTACGACT) with a Biometra thermocycler.The amplification program employed an initial denaturation at 95 °C for 4 min and then 35 cycles of 94 °C for 1 min, 1 min at 57 °C and 90 s at 72 °C. Final extension was at 72 °C for 10 min. The amplified products were analyzed by 1% agarose gel electrophoresis.The PCR products were sequenced by a commercial company Borui Biotechnology Inc. (Xiamen, China). DNA sequence homology was BLAST searched on the NCBI GenBank website (https://blast.ncbi.nlm.nih.gov/Blast.cgi). A phylogenetic tree of bacteria was constructed with the 16S rDNA sequences using the neighbor-joining method in Mega 7.0.
2.4 In silico prediction of AMPs derived from P. vannamei β-GBP
Two online in silico tools, i.e., Antibacterial peptides (AntiBP2)and the collection of antimicrobial peptides (CAMP) server were used to predict AMPs derived from the β-GBP sequence of P. vannamei (UniProtKB/Swiss-Prot: P81182) as previously reported with some modifications [20]. AntiBP2 and CAMP were used to screen and evaluate the reliability of the predicted AMPs, while the charge and hydrophobicity of the peptides were calculated by using the APD3 software.
2.5 Synthesis of predicted peptides
Peptides were synthesized by using the solid-phase procedure,as previously described by Yang et al. [21], with the N-terminal acetylated and the C-terminal amidated. High performance liquid chromatography (HPLC) was used to purify the peptides so as to attain 99% peptide purity with an Agela C18column. The molecular mass of the synthetic peptides was determined by liquid chromatography coupled to mass spectrometry (LC-MS/ESI).
2.6 Evaluation of antibacterial activity
2.6.1 Minimum inhibitory concentration (MICs) of peptides PvGBP2 and PvGBP5
B. subtilis (AY867792.1), B. cereus (KX890470.1) and B. nealsonii (JQ579625.1) were grown in nutrient broth (NB) medium at 37 °C and diluted to 106–107CFU/mL in 0.01 mol/L pH 7.2 phosphate buffer saline (PBS), while peptides PvGBP2 and PvGBP5 were diluted in sterile PBS. Next, the bacteria and diluted peptides were mixed in equal-volumes, followed by incubation for 12 h at 37 °C.The MIC was defined as the lowest concentration of peptide with no visible growth of bacteria from the microtiter plates.
2.6.2 Growth curve and time-kill kinetics
The growth curve and time-kill kinetics of peptide PvGBP2 was assessed as described by Yi et al. [22] with some modifications. First,B. subtilis (AY867792.1) was cultured for 12 h to logarithmic growth phase (OD600nm= 0.6-0.7) at 37 °C, followed by addition of PvGBP2 to the bacterial suspensions to final concentrations corresponding to 1/2, 1, and 2 × MIC, respectively. Bacterial numbers were measured each hour for 12 h at 600 nm by using an UV-5200 spectrophotometer(METASH, China). PBS buffer (0.01 mol/L, pH 7.2) was used as control.
For the time-kill kinetics analysis, culturedB. subtilis(AY867792.1) (106–107CFU/mL) was exposed to 1/2, 1, and 2 × MIC of PvGBP2. Next, bacterial suspensions were incubated at 37 °C, and at different time points (i.e., 0, 0.5, 1, 1.5, 2 and 2.5 h),0.5 mL of the bacterial suspensions were obtained, and the number of colonies counted after culturing on NB plates at 37 °C for 12 h.Control samples were treated with PBS buffer (0.01 mol/L, pH 7.2) in the same way.
2.6.3 Effect of B. subtilis on the growth of A. auricular
StrainB. subtilis(AY867792.1) was cultured in NB at 37 °C for 12 h, while PvGBP2 (1 × MIC) was added as positive control during the fermentation. Fermentation broths were centrifuged at 2 700 ×gfor 10 min at room temperature and passed through 0.22 μm pore size membranefilters to obtain culturefiltrate. Next, culturefiltrates were incubated in PDA and precooled to 50 °C. The PDA plates without culturefiltrates were used as negative control.A. auricularwas placed at the center of the culture medium and incubated at 25 °C. Thefinal growth rate was calculated as the radial extension of each mycelial colony after 7 days [23].
2.6.4 Antibacterial assays
The method used to cultivateA. auricularwas a modification of the method by Oei [24]. To assess the growth ofA. auricular,strainB. subtilis(AY867792.1) (102–103CFU/mL) was added toA. auricularculture bags, with bags containing PvGBP2 (1 × MIC) and PBS (0.01 mol/L, pH 5.5) used as positive and negative controls,respectively. The yield ofA. auricularwas measured as the total dry weight of all mature mushrooms collected from each bag.
2.7 Antibacterial mechanism of peptide PvGBP2
2.7.1 Transmission electron microscopy (TEM) analysis of B. subtilis treated with peptide PvGBP2
StrainB. subtilis(AY867792.1) at 106–107CFU/mL were treated with 2 × MIC of PvGBP2 at 37 °C for 2 h, followed by centrifugation at 2 700 ×gfor 10 min, and washed twice with PBS (0.01 mol/L,pH 7.2). Bacterial cells were thenfixed with 10 mg/mL osmic acid and dehydrated with ethanol. Next, samples were embedded by roasting at 70 °C for 24 h, followed by preparing 70–90 nm thin sections on copper grids and stained with lead citrate and uranyl acetate. The ultrastructure was observed on a H-7650 transmission electron microscope (Hitachi Co., Ltd., Japan).
2.7.2 Effect of peptide PvGBP2 on biofilm formation
StrainB. subtilis(AY867792.1) was collected by centrifugation and resuspended in NB broth at an OD600nmof 0.1. Bacteria suspensions were mixed with equal volumes of PvGBP2 (1/8 ×MIC–1 × MIC), and pipetted into sterile 96 well plates followed by incubation at 37 °C for 72 h. Plates were washed twice with PBS (0.01 mol/L, pH 7.2) to remove unattached cells followed by cell staining with 200 μL of 1 mg/mL crystal violet solution. After 5 min staining, plates were then washed with sterile PBS to remove the crystal violet stain, followed by the addition of 200 μL of 250 mg/mL glacial acetic acid and incubated at 25 °C for 30 min to dissolve the stained biofilm. The biofilm biomass was quantified by measuring absorbance at 600 nm on a Synergy H1 Multimode Plate Reader (BioTek, USA). Samples treated with Nisin and PBS were used as positive and negative controls, respectively [25].
2.8 Statistical analysis
All experiments were carried out in triplicate, and the results were analyzed with SPSS software (version 22.0 for Windows) using one-way ANOVA. Significance was considered atP< 0.05.
3. Results and discussion
3.1 Identification of bacteria strains on contaminated A. auricular bags and phylogenetic analysis
Five species of bacteria, includeA. arilaitensis(JN885458.1),B. subtilis(AY867792.1),B. cereus(KX890470.1),B. nealsonii(JQ579625.1), andS. warneri(KF057949.1) were isolated from contaminatedA. auricularbag culture based on the nucleotide blast of 16S rRNA gene sequences with the NCBI gene bank sequences (Table 1).Most of the identified bacteria were of genusBacillus(Fig. S1),which are antagonistic bacteria against fungi [26], and are capable of producing a wide range of antagonistic compounds including lipopeptides [27], cyclic peptides [28], chitinase [29], and volatile organic compounds [30], as well as show strong antifungal activities.WhenA. auricularculture bags are not well sterilized,Bacillusspp.can survive, colonize fungal hyphae, and germinate under suitable conditions [31]. Moreover,Bacillusspp. generate amylases and proteases, which results in discoloration and slime formation, typical indicators of contaminatedA. auricularculture bags (Fig. 1) [32].Therefore, the observed inhibition ofA. auriculargrowth could be due to these three species of genusBacillusisolated from the contaminated bags.

Table 1Identification of bacterial strains from contaminated A. auricular culture bags.
3.2 Screening of in silico predicted AMPs from P. vannamei β-GBP with potential application in inhibiting bacteria contamination of A. auricular culture bags
In invertebrates, β-GBP consists of 1 454 amino acids, plays a crucial role in the innate immune response, and when activated,can mediate the recognition of invading pathogens, and agglutinateBacillusspp., includingB. subtilis,B. cereusandB. thuringiensis[33,34].In recent years,in silicotools have become one of the most important assisted technologies in the design and application of AMPs [35].For instance, recently used such tools to design and screen AMPs derived from the hemoglobin ofPseudosciaena crocea, and revealed that one AMP (designated LCH4) had strong inhibitory effects onS. aureusandVibrio parahaemolyticus, which could therefore be used as an antimicrobial agent in soy sauce [36]. Using a similar approach,eight potential AMPs were predicted from the β-GBP sequence ofP. vannameiusing the AntiBP2 and CAMP servers (Table 2).Based on the calculation of positive charge and hydrophobicity of the predicted peptides using the APD3 server, peptides PvGBP2 and PvGBP5 had synonymous features as most AMPs, including possessing net positive charges of between 3 to 6 and having hydrophobicity ranging from 40% to 60% [15]. On the basis of this,the antibacterial activities of peptides PvGBP2 and PvGBP5 were further explored.
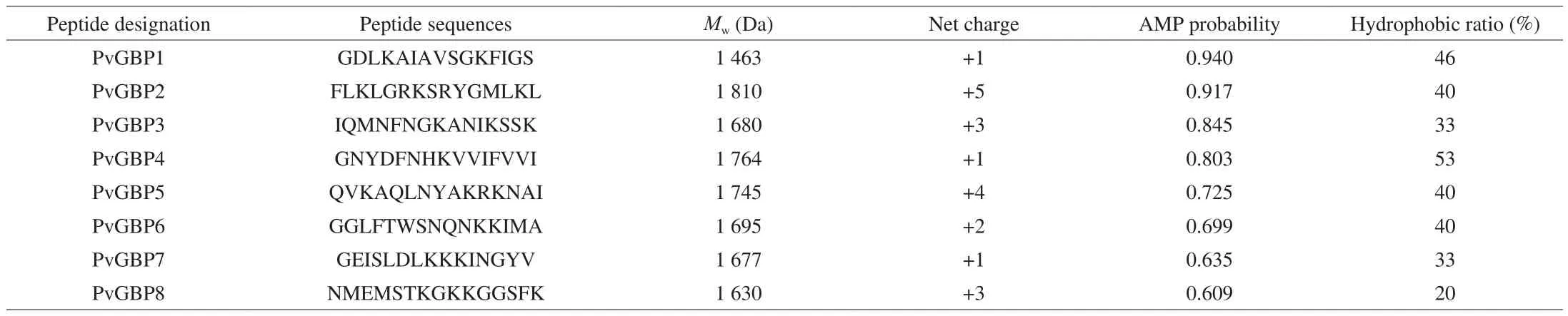
Table 2In silico predicted potential antimicrobial peptides from P. vannamei β-GBP.
3.3 Antibacterial activity of peptides PvGBP2 and PvGBP5 against isolated bacteria
To ascertain the antibacterial activity of PvGBP2 and PvGBP5,the MICs of chemically synthesized PvGBP2 and PvGBP5 were determined againstB. subtilis(AY867792.1),B. cereus(KX890470.1),B. nealsonii(JQ579625.1) isolated from bacteria contaminatedA. auricularculture bags. Peptide PvGBP2 had strong antibacterial activities against these bacteria, with the MICs being 15.6–31.25 μg/mL (Table 3), and stronger than PvGBP5. Moreover,the MICs of PvGBP2 was stronger than nisin, a cationic natural AMP produced byLactococcus lactissubsp, which has been widely used as a food preservative, due to its strong inhibitory effect on heat-resistant,spore-forming microorganisms, especially theBacillusspp. [37].
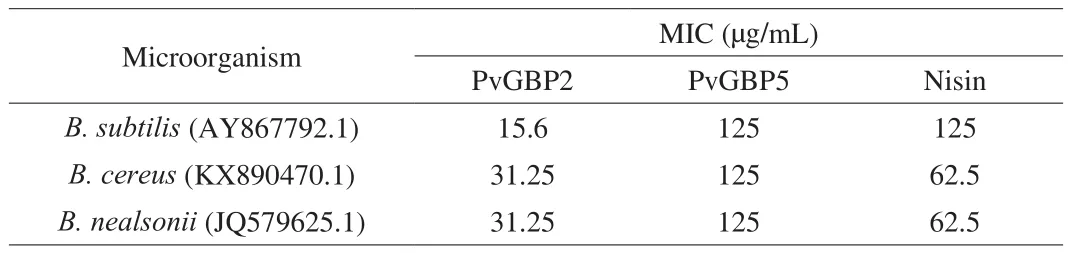
Table 3Antibacterial activity of peptides PvGBP2 and PvGBP5 against three Bacillus spp.
To further explored the antibacterial activity of PvGBP2, HPLC and LC-MS/ESI was used for purification and identification (Figs. S2A and S2B). The growth curve and time kill kinetics of isolateB. subtilis(AY867792.1) were measured as shown in Fig. 2A, the growth ofB. subtiliswas completely inhibited during 16 h incubation with 1 × MIC concentration of PvGBP2. Control group had a lag phase in the first 2 h, and kept exponential-phase growth after lag-phase. PvGBP2 (1 × MIC) was added at 4 h in control group, resulting in significantly inhibition at 6 h of incubation. Moreover, there was an increase of OD600nmfor 1 × MIC (4 h) addition group beyond 14 h of incubation, the numbers were still much lower than the control group.This trend in antibacterial activity is similar to that of monolauroylgalactosylglycerol againstB. cereus[38]. In the time-kill curve analysis (Fig. 2B), the bacterial numbers decreased with increasing PvGBP2 concentration. When the concentration of PvGBP2 was 1 × MIC, there was a 76.3% decrease in bacteria number after 1.5 h,with 96.8% decrease when 2 × MIC of PvGBP2 was used. There was 100% decrease in bacteria number upon treatment with 1 × MIC and 2 × MIC for 2.5 h.

Fig. 2 Growth curve and time-kill kinetics. (A) Growth curve of B. subtilis,(B) time-kill kinetics of B. subtilis. The MIC used was 15.6 μg/mL.
3.4 Effect of the antibacterial peptides on the growth of A. auricular
In order to leverage the use of thesein silicodesigned antibacterial peptides in controlling bacteria contamination ofA. auricularculture bags, the synthetic peptides should not affect the growth ofA. auricular. Thus, to assess the effect of peptide PvGBP2 on inhibitingB. subtilisand any consequence on the growth ofA. auricular, the growth rate of mycelium was determined by colony diameter. As shown in Fig. 3A, compared with the control group, the growth of mycelium was significantly inhibited by B. subtilis, which was reduced by 49.4% after 7 days, due to the antibacterial activity of PvGBP2 against B. subtilis, the mycelial growth of mycelium was almost the same as that of the control group. The mushroom yields in the PvGBP2 treatment group averaged 50.3 g/bag, with a 48.7% decrease in yield in the B. subtilis treatment group (Fig. 3B).
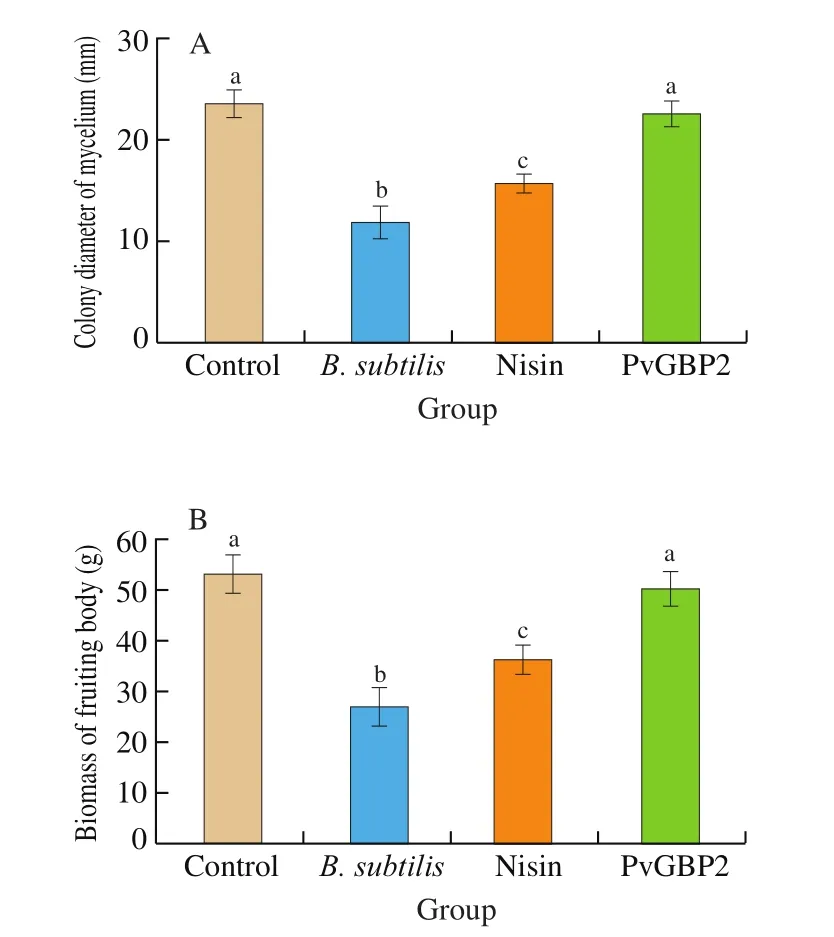
Fig. 3 Inhibition of B. subtilis in A. auricular culture bags by peptide PvGBP2. (A) Colony diameter of mycelium, (B) biomass of fruiting bodies.Significance is considered at P < 0.05 and labeled by letters. Different letters indicates significant difference.
It has been shown that Bacillus spp. inhibit growth of fungi by producing antifungal agents such as lipopeptides, cyclic peptides and chitinase etc., which results in a decline in A. auricular yields [27-29].Peptide PvGBP2 had strong inhibitory effects on Bacillus spp., but had no effect onfilamentous fungi, which is synonymous with nisin [39].The main antibacterial mechanism of nisin is through adsorption onto bacteria surface and membrane disruption, which might not be effective on fungal cell membranes [40].
3.5 Ultrastructural changes in the bacteria membranes induced by peptide PvGBP2
The ultrastructural changes in the membranes of B. subtilis(AY867792.1) upon treatment with PvGBP2 were observed by TEM.The membranes of untreated bacteria were uniform with normal cytoplasm, undeformed cells, and no cytoplasmic leakage (Fig. 4A).However, when bacteria cells were treated with peptide PvGBP2 for 1 h, the bacteria cell membranes began to become blur and thin, with inhomogeneous electron density in the cytoplasm (Fig. 4B). After 2 h post-PvGBP2 treatment, there was complete vacuolation and disruption of bacteria cell membranes (Fig. 4C). These observations indicate that peptide PvGBP2 exerts its antibacterial activity against B. subtilis by altering the membrane structure and permeability [41],which is synonymous with the action of nisin against B. subtilis,where nisin induces vacuolation and destruction of bacteria cell membranes and internal structures [42].
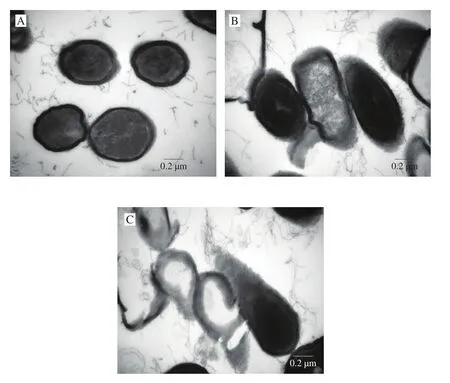
Fig. 4 TEM images of B. subtilis. (A) Untreated B. subtilis, (B) treatment of B. subtilis with PvGBP2 for 1 h, (C) treatment of B. subtilis with PvGBP2 for 2 h. The MIC used was 15.6 μg/mL, the concentration of PvGBP2 used was 2 × MIC, while the concentration of B. subtilis was 106–107 CFU/mL.
3.6 Effect of PvGBP2 on biofilm formation
To assess the effect of PvGBP2 on biofilm formation, B. subtilis(AY867792.1) was treated with PvGBP2 at concentrations lower than 1 × MIC or with an equal concentration of nisin as a positive control,to observe the effect on bacterial cell membrane formation. Nisin was used as control due to its known antibiofilm activity [43]. As shown in Fig. 5, PvGBP2 was found to show anti-biofilm activity on B. subtilis in a concentration-dependent manner, as PvGBP2 had no effect on biofilm formation at concentrations greater than 1/4 × MIC. When B. subtilis was treated with 1 × MIC of PvGBP2, biofilm formation decreased by 90.6%. Biofilms represent a protected mode of growth that allows bacteria cells to survive in hostile environments [44].Members of the Bacillus genus also possess swarming motility, which seems to facilitate microbial survival in the environment and surface colonization, leading to biofilm formation [45]. The antifungal activity of Bacillus spp. is believed to relate to biofilm formation.For this reason, the anti-biofilm activity of peptide PvGBP2 against B. subtilis indicates that it could be used as an antibacterial agent in A. auricular cultivation.
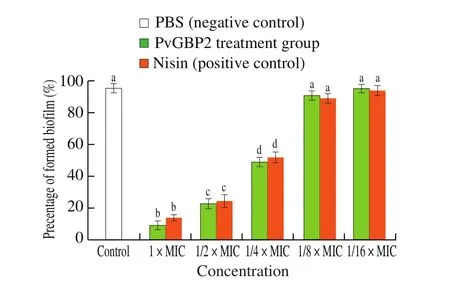
Fig. 5 Effect of peptide PvGBP2 on biofilm formation. Significance is considered at P < 0.05 and labeled by letters. Different letters indicates significant difference.
4. Conclusion
Three species of Bacillus spp., including B. subtilis (AY867792.1),B. cereus (KX890470.1), B. nealsonii (JQ579625.1), were isolated and identified from contaminated A. auricular culture bags. These bacteria seem to inhibit the growth of A. auricular. An in silico predicted AMP, i.e., PvGBP2, found on the β-GBP sequence of the P. vannamei, was screened and found to possess strong antibacterial activity against the three isolates of Bacillus spp. Peptide PvGBP2 had MICs of 15.6–31.25 μg/mL and killed bacteria by cell membrane destruction. Moreover, PvGBP2 decreased biofilm formation by B. subtilis by 90.9%, and could inhibit the growth of B. subtilis in A. auricular culture bags. Collectively, the current results showed that peptide PvGBP2 could be used as an antibacterial agent in the cultivation of A. auricular.
Declaration of competing interest
The authors declared that they have no con flicts of interest to this work.
Acknowledgement
This work was sponsored by National Key R&D Program of China (2020YFD09009).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://doi.org/10.1016/j.fshw.2022.06.019.
- 食品科学与人类健康(英文)的其它文章
- A review on mechanisms of action of bioactive peptides against glucose intolerance and insulin resistance
- Adropin as an indicator of T2DM and its complications
- Isolation and characterization of novel peptides from fermented products of Lactobacillus for ulcerative colitis prevention and treatment
- The immunity-promoting activity of porcine placenta in mice as an immunomodulator for functional foods
- Nutritional properties of Europen eel (Anguilla anguilla) bone peptide-calcium and its apoptosis effect on Caco-2 cells
- A comprehensive method to explore inhibitory kinetics and mechanisms of an anticoagulant peptide derived from Crassostrea gigas

