Facile and diverse logic circuits based on dumbbell DNA-templated fluorescent copper nanoclusters and S1 nuclease detection
Zefeng Gu, Anchen Fu, Ru Qiu, Ru Sun, Zhijuan Cao
Shanghai Key Laboratory of Bioactive Small Molecules & Department of Pharmaceutical Analysis, School of Pharmacy, Fudan University, Shanghai 201203,China
Keywords:Logic gates Dumbbell DNA templates Poly-thymine loops Copper nanoclusters Nucleic acid enzymes
ABSTRACT DNA-based logic gates promote the development of molecular computing and show enormous potential in the fields of nanotechnology and biotechnology.Dumbbell oligonucleotides (DNA) with poly-thymine(poly-T) loops and a nicked random double strand have been demonstrated to be an efficient template for the formation of fluorescent copper nanoclusters (CuNCs) in our previous work.Herein, a new platform technology is presented with which to construct molecular logic gates by employing CuNCs probe as a basic output generator, coupling of functional nucleases as the inputs.Two dumbbell DNAs are used with the difference in stem length (8 bp and 16 bp, respectively).The degradation of DNA templates can be tuned by various nucleic acid enzymes, single-stranded nuclease (S1), double-stranded specific nuclease(DSN), E.coli DNA ligase, exonucleases I and III.Briefly, S1 can digest both DNA templates, while the cleavage ability of DSN will be resistant by the short stem of SS-DNA (short-stem DNA).Exonuclease I and III can degrade these two nicked DNA templates, which are inhibited due to the ligation of E.coli DNA ligase.With this novel strategy, a set of logic gates is successfully constructed at the molecular level,including “YES”, “PASS 0”, “OR”, “INHIBIT”, which take the advantages of no label, easy operation, fast speed, high efficiency and low cost.Furthermore, S1 nuclease, as the biomarker of numerous carcinogens,is selectively detected in the range of 0.05–50 U/mL with the detection limit of 0.005 U/mL (1 × 10-6 U)based on this platform.
Conventionally, a logic gate is an elementary building block of a digital circuit, which are capable of performing all types of Boolean logic by receiving electronic inputs representing true (“1”, high voltage) or false (“0”, low voltage) and then generating the appropriate electronic outputs [1,2].This method is limited in siliconbased electronic devices, top-down lithographic processes, the necessity of the minimum feature size.Since the 1980s, de Silvaet al.have greatly improved the emergence of bottom-up molecular logic gates based on fluorescence response [3].In 1993 [4], de Silva developed the “AND” gate in solution by molecular logic gates which were capable of performing various Boolean logic, mostly by receiving one or more input values and generating a single logical output.In 1994, Adleman encoded a small graph in molecules of oligonucleotides to perform DNA-based logical operations [5].Along this line, molecular logic gates have stimulated the dramatic development of molecular-scale computers and shown enormous potential in nanotechnology, biotechnology, and medicine [6].Various materials, such as nucleic acids [3], organic molecules [7],proteins [8], enzymes [9], and supramolecular hydrogels [10] have been used for the development of molecular-scale logic gates.Among them, DNA-based logic gates attracted considerable research efforts owing to predominant properties, including wellordered structure, easy modification and highly specific recognition [11,12].Plentiful achievements have been made, but there are still some significant issues which should be taken seriously.For example, lots of logic devices with multi-output signals are developed by using of covalently labeled reporters (e.g., dye, redox or fluorescent quencher), which is of time and cost-consumption[13-18].Nanomaterials, such as graphene, carbon dots, need to be introduced to aid the establishment of logic gates [19], increasing the complexity of a logic system and are not conducive to the repeatability and assembly of different logic systems.In recent years, some label-free probes and straightforward strategies were also promoted to simplify the logic systems.For example, Yanget al.reviewed some homogeneous signal actuators [20] and proposed a versatile DNA-supramolecular logic platform [21].Huanget al.also made some achievements on the label-free logic devices [22].In order to avoid the covalent linking of probes or the addition of nanomaterials, Lvet al.[23] reported the logic gates based onin-situformation of fluorescent silver nanoclusters.Unfortunately, it took more than 5 h to generate the fluorescent products (AgNCs), and usually complicated oligonucleotides design was involved.Therefore, to explore a simple logic platform in a timeand cost-effective way is urgently demanded.
DNA-scaffolded fluorescent copper nanoclusters attract much more attention in fluorescent bio-sensing by virtue of fast synthesis (within several minutes, much faster than AgNCs), low cost and excellent photophysical properties of mega-Stokes shifting [24,25].Different types of DNA templates were reported to form copper nanoclusters (CuNCs) for various applications [26,27].Our group reported that hairpin or dumbbell DNA with a poly-T loop could be a promising template for the formation of CuNCs [28].The DNA structure was composed of two poly-T loops regions and a random dsDNA stem.By comparison of previous templates, dumbbell DNA-templated CuNCs showed enhanced fluorescent properties for biochemical sensing.Meanwhile, nucleases, such as exonucleases, endonucleases, are capable to hydrolyze phosphodiester bonds in DNA or RNA into mono- or oligonucleotide fragments [29,30], while the ligase can link two ends of the DNA template.Therefore, those nucleases, especially single-stranded nuclease (S1), play an important role in various biological procession in living systems, such as DNA replication, repair and recombination,genotyping, mapping, as well as molecular cloning.The abnormal levels of nucleases were related to the disease states [30].
In this study, regarding the distinct features and great potential in many fields of fluorescent CuNCs, we report diverse logic gates, including “YES”, “PASS 0”, “OR”, and “INHIBIT”.The logic circuit signal reporter, CuNCs can be formed within twenty minutes,alleviating the time-consuming sequence design, and also reducing the costly labeling and covalent binding procedures.In addition,these logic gates are operated in homogeneous solution to avoid the immobilization procession.Thus, it shows superior features of high speed, simplicity and integration.
The sequences of dumbbell DNA template used in this work are shown in Table S1 (Supporting information), which are the same at two poly-T loops and different in stem length.The LS-DNA (longstem DNA) template has a 16-base pair (bp) double-strand stem,while the SS-DNA (short-stem DNA) has an 8-bp one.The strongly fluorescent CuNCs were reported with an emission peak at 630 nm and an average diameter of about 2.6 nm (Fig.S1 in Supporting information) under the condition of CuSO4and ascorbic acid (Fig.S2 in Supporting information) [31].When DNA templates were destroyed by nucleases, the fluorescence would be quenched due to the absence of CuNCs.As depicted in Scheme 1, the fluorescent signal of LS-DNA templated CuNCs can be quenched by either S1 nuclease or double-stranded specific nuclease (DSN), while that of SS-DNA templated CuNCs can only be quenched by S1 nuclease,but not DSN.That is because the dumbbell DNA with a short stem(usually less than 10 bp) is resistant to the cleavage of DSN [32].Both DNA templates can be degraded by exonucleases (Exo) I and III thoroughly, but inhibited byE.coliligase which can link the 3′and 5′ terminus of the dumbbell DNA templates to be a closed loop without a nick.Based on these principles, we can simply establish several single-input and dual-input logic circuits.
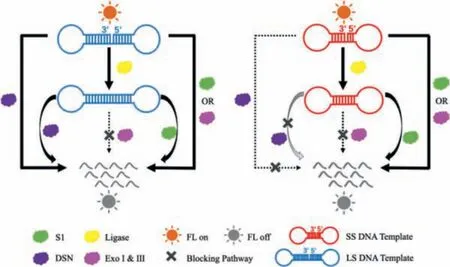
Scheme 1.Schematic principle of nucleic acid enzymes switched fluorescent signal of DNA-templated CuNCs.
According to nuclease-induced cleavage of DNA templates, the logic gates were developed by assigning the fluorescent signal as the output and nucleic acid enzymes as inputs.The normalized fluorescent signal (0.5) was set as a threshold value.The strong fluorescent signal generated from well-templated CuNCs (higher than 0.5) was defined as the output of 0, and the weak fluorescent signal from fragmented DNA templates (lower than 0.5) was defined as the output of 1 after introducing various enzymes.
The simplest “YES” and “PASS 0” logic circuits were first achieved based on the input-dependent signal variation.In this logic system, S1 and DSN with the concentration of 0 U or 10 U were defined as the input 0 or 1, respectively.As shown in Fig.1a,SS-DNA-templated CuNCs were first considered as the initial state,showing strong fluorescence and giving an output of 0.With the addition of 10 U S1 nuclease as the input 1, the fluorescent intensity obviously decreased due to the degradation of SS-DNA template, resulting in output 1.The logic circuit was defined to be“YES”.Correspondingly, with the addition of 10 U DSN as input 1,the fluorescent intensity has no obvious change (output 0) because that the short-stem of SS-DNA (less than 10 bp) was resistant to the cleavage of DSN effectively.Thus, the logic circuit was “PASS 0”.The columns in Fig.1c represented the normalized fluorescence signal corresponding to output signals of “YES” and “PASS 0” gates under different inputs, respectively, which was consistent with the fluorescence images by Camara (Canon E450, Fig.1b).
Furthermore, logic gate “OR” (seen in Figs.1d-f) was also implemented successfully on the simple platform of LS-DNA templated CuNCs.In this logic circuit platform, LS-DNA was used as the template for fluorescent CuNCs.S1 and DSN were employed as the dual inputs, respectively.As shown in Fig.1d, either of two inputs (1/0, 0/1) and both of them (1/1) were able to present output 1, which was the result of no formation of CuNCs, indicating that these two nucleases trigger cleavage procession on LS-DNA templates.The fluorescent images (Fig.1e) and normalized fluorescent signals (Fig.1f) confirmed the above results.Thus, a typical “OR”logic gate was achieved.
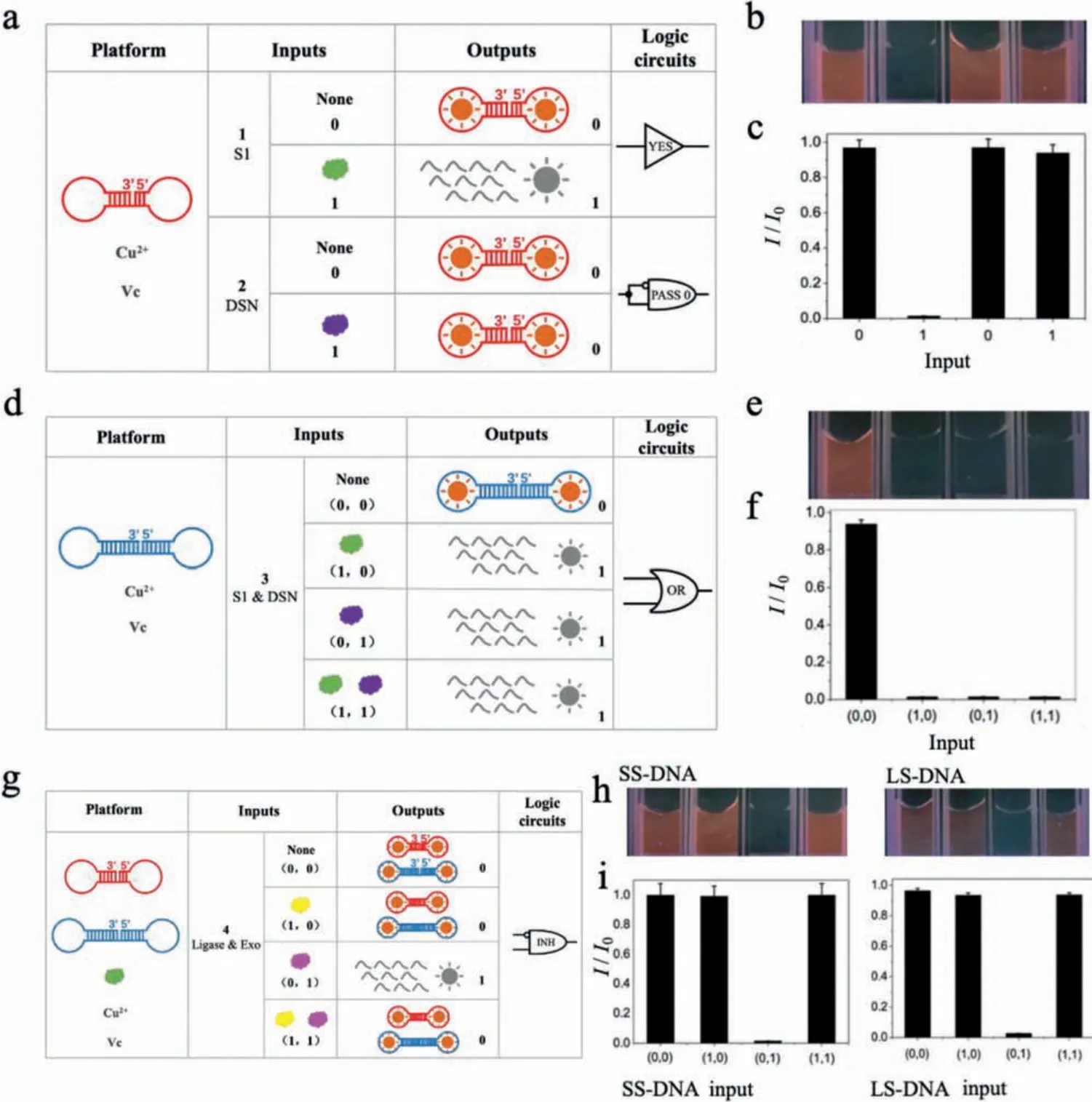
Fig.1.(a) Schematic description of SS-DNA-templated CuNCs undergoing structural and fluorescent change upon addition of enzyme inputs.(b) Fluorescence images of CuNCs with different inputs S1 (0 and 1) or DSN (0 and 1) in MOPs buffer.(c) Normalized fluorescence intensities of CuNCs in two types of single-input logic circuits (“YES”and “PASS 0”).Measurement conditions:SS-DNA (50 pmol), 50 μmol/L CuSO4, 2 mmol/L ascorbic acid, S1 0 U (input 0) and 10 U (input 1), DSN 0 U (input 0) and 10 U(input 1).(d) Schematic description of LS-DNA-templated CuNCs undergoing structural and fluorescent change upon addition of enzyme inputs.(e) Fluorescence images of CuNCs with input S1 (0 and 1) or DSN (0 and 1) in MOPs buffer.(f) Normalized fluorescence intensities of CuNCs in the two types of dual-input logic circuits (OR).Measurement conditions:LS-DNA (50 pmol), 50 μmol/L CuSO4, 2 mmol/L ascorbic acid, S1 0 U (input 0) and 10 U (input 1), DSN 0 U (input 0) and 10 U (input 1).(g)Schematic description of “INHIBIT” logic gate based on SS-DNA or LS-DNA-templated CuNCs undergoing structural and fluorescent change upon addition of enzyme inputs.(h) Fluorescence images of CuNCs with inputs of E.coli DNA ligase/Exo (0,0; 1,0; 0,1 and 1,1) in MOPs buffer.(i) Normalized fluorescence intensities of CuNCs in the logic circuit (“INHIBIT”).Measurement conditions:SS-DNA or LS-DNA (0.25 μmol/L), 50 μmol/L CuSO4, 2 mmol/L ascorbic acid, 50 μmol/L NAD+, E.coli DNA ligase 0 U (input 0)and 10 U (input 1), Exo 0 U (input 0) and 10 U (input 1).
Figs.1g-i depicted the mechanism of “INHIBIT” logic gate.Of note, both SS-DNA and LS-DNA templates can be employed to establish this logic gate.E.coliDNA ligase and Exo (including Exo I& III) were used as input pairs and the normalized fluorescent signals as the output.Only in the presence of the input of Exo (0/1)did the fluorescence decrease, leading to output 1.Otherwise, any other inputs including none of them (0/0),E.coliby itself (1/0) or both of them (1/1) did not change the fluorescence intensity, which yielded output 0.The reason was thatE.coliDNA ligase could link two ends of the DNA templates, which inhibited the cleavage of Exo.Thus, the DNA templates were retained to form fluorescent CuNCs.The fluorescent images and normalized fluorescent signal were shown in Figs.1h and i.
Then, we studied the cleavage ability of S1 and DSN on the SSDNA template with or without the presence ofE.coliligase or Exo I & III.As shown in Fig.2, existence of either Exo I & III or S1 decreased the fluorescence signal (columns 2 and 3), while DSN did not (column 4).E.coliDNA ligase inhibited the cleavage function of Exo I & III (column 6), but not S1 (column 7).When the presence ofE.coliDNA ligase on its own (column 5), or with DSN(column 8), showed almost the same fluorescence signal with the blank (only DNA templates without any enzymes, column 1).Those results achieved the IDENTITY logic gate.That is, the fluorescence signal was quenched due to the existence of S1 no matter that other four enzymes coexisted or not (output 1), which was further confirmed by the results of gel electrophoresis.
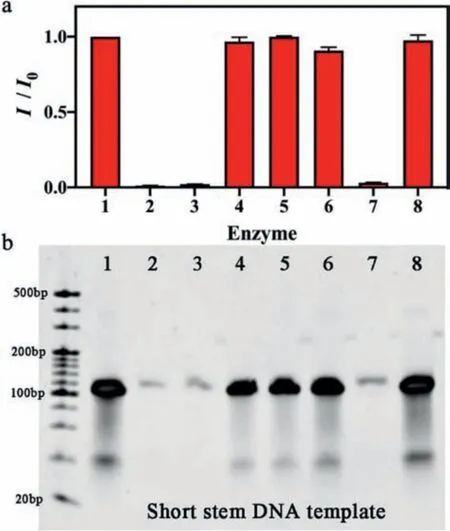
Fig.2.(a) Normalized fluorescence intensities of SS-DNA-templated CuNCs with S1,DSN, E.coli ligase or Exo I & III (Column:1.none, 2.Exo I & III, 3.S1, 4.DSN, 5. E.coli DNA ligase, 6. E.coli DNA ligase + Exo I & III, 7. E.coli DNA ligase + S1, and 8.E.coli DNA ligase + DSN).(b) Gel electrophoresis image.Measurement conditions:SS-DNA (50 pmol), 50 μmol/L CuSO4, 2 mmol/L ascorbic acid, 50 μmol/L NAD+, S1 10 U, DSN 10 U, E.coli DNA ligase 10 U and Exo I & III 10 U.
S1 nuclease protection is the easiest way to map cleavage sitesin vivoand a positive result here is definitive evidence of endonucleolytic cleavage, which has been particularly used to probe the disruption of the DNA structure by numerous carcinogens and antimitotic drugs [33,34].Here, based on this new strategy, S1 nuclease activity was detected selectively based on SS-DNA template CuNCs which would not be influenced by DSN and Exo I & III.Before the quantification, we studied and optimized various experimental conditions, including SS-DNA template in the range of 0.125–0.5 μmol/L, CuSO4in the range of 12.5–200 μmol/L, ascorbic acid in the range of 0.25–4 mmol/L, NAD+(nicotinamide adenine dinucleotide) in the range of 12.5–200 μmol/L, Exo I & III andE.coliDNA ligase in range of 1.25–20 U.Then, we obtained the optimal reaction conditions.That was 50 pmol DNA template (0.25 μmol/L),10 U ofE.coliDNA ligase, 10 U Exo I & III, 50 μmol/L NAD+, 50 μmol/L, and 2 mmol/L ascorbic acid (Figs.S2 and S3 in Supporting information).As shown in Fig.3, a good linear correlation existed between the fluorescent ratio [(I0-I)/I0] and S1 concentration in a broad range of 0.05–50 U/mL.The limit of detection (LOD) for S1 nuclease was calculated to be 0.005 U/mL (10-6U, 3σ).
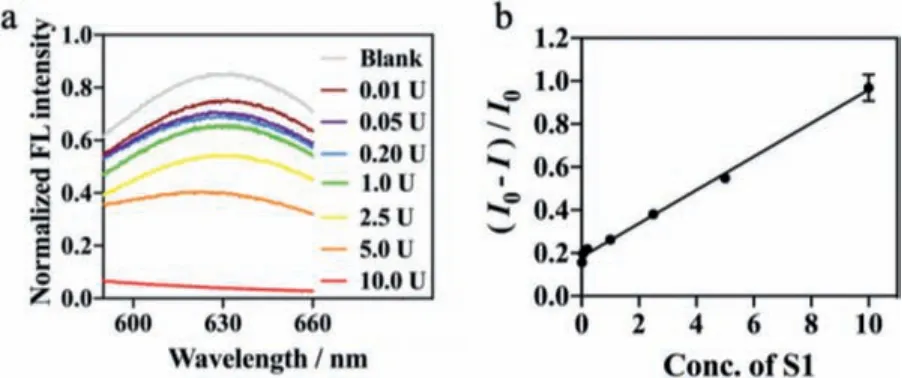
Fig.3.(a) Fluorescent signal responses with different concentrations of S1 nuclease.(b) Plot of FL intensity ratio [(I0-I)/I0] versus the concentration of S1 nuclease(y = 0.0777x + 0.182 with a correlation coefficient of 0.9963).Measurements were performed by using 0.25 μmol/L short stem DNA, 10 U E.coli DNA ligase, 50 μmol/L NAD+, 50 μmol/L CuSO4, and 2 mmol/L ascorbic acid in MOP buffer.The error bars denote the standard deviation for three independent measurements.
In summary, we have constructed a series of logic gates (“YES”,“PASS 0”, “OR” and “INHIBIT”) by using dumbbell DNA template fluorescent CuNCs as the functional component (output), and multiple nucleic acid enzymes as inputs.As compared to previous ones, the dumbbell DNAs can template the formation of CuNCs within several minutes which contributes to time- and costeffective protocol for molecular logic gates.The design also eliminates the usage of organic dyes and nanomaterial quenchers, and other components, greatly simplifying the establishment of the logic circuit platform.In further, S1 nuclease has been detected in a wide range with low sensitivity (10-6U).Together, these results introduce a new platform technology for logic gate operation that enables the simple circuits required for communication between various computational elements.We expect that this work can be an important point for research on nucleases logic systems and will be highly beneficial in future molecular computing and biochemical and electronic applications.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors are grateful to the projects of Innovative research team of high-level local universities in Shanghai and a key laboratory program of the Education Commission of Shanghai Municipality (No.ZDSYS14005), Program for high-level local universities in Shanghai (No.IDF301027/022) and Shanghai Agriculture Science and Technology Support Project (No.21N31900500), and the National Natural Science Foundation of China (No.21505023).
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2021.09.020.
 Chinese Chemical Letters2022年5期
Chinese Chemical Letters2022年5期
- Chinese Chemical Letters的其它文章
- Recent advances in enhancing reactive oxygen species based chemodynamic therapy
- An integrative review on the applications of 3D printing in the field of in vitro diagnostics
- Recent developments of droplets-based microfluidics for bacterial analysis
- Dynamics and biological relevance of epigenetic N6-methyladenine DNA modification in eukaryotic cells
- Recent progress in advanced core-shell metal-based catalysts for electrochemical carbon dioxide reduction
- Recent advances in carbon-based materials for electrochemical CO2 reduction reaction
