Dynamics and biological relevance of epigenetic N6-methyladenine DNA modification in eukaryotic cells
Yuwei Sheng, Meijuan Zhou, Changjun You, Xiaoxia Dai
State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Molecular Science and Biomedicine Laboratory, Hunan Provincial Key Laboratory of Biomacromolecular Chemical Biology, Hunan University, Changsha 410082, China
Keywords:DNA modification N6-Methyladenine Epigenetic mark Methyltransferase Demethylase
ABSTRACT DNA methylation represents a major type of DNA modifications that play key roles in diverse biological processes.With the recent development of highly selective and sensitive bioanalytical techniques,N6-methyladenine (6mA) has been characterized as an important internal DNA modification dynamically occurring in multiple eukaryotes including humans.Increasing evidence has indicated that 6mA may act as a novel epigenetic modification involved in regulation of development, stress response and diseases such as cancer and neurodegenerative disorders.We review herein the recent advances in the detection and functional studies of 6mA modification, with special emphasis on its biological consequences and human health relevance as well as its dynamic regulation by various types of methyltransferases,demethylases and 6mA-binding proteins.It can be envisaged that further chemical and biological studies of 6mA modification will lead to a better understanding about its potentially important roles in normal and pathological biological processes.
1.Introduction
DNA is the carrier of genetic information, and DNA modification can be used as a signal of DNA damage or an epigenetic regulator of diverse biological processes [1].It has been well acknowledged that DNA modifications involved in epigenetic regulation mainly include 5-methylcytosine (5mC) andN4-methylcytosine(4mC).5mC is the most abundant epigenetic modification dynamically occurring in genomic DNA, which plays important roles in regulating chromatin structure and gene expression in mammals and plants [2,3].It is called the "fifth base" in addition to adenine, guanine, cytosine, and thymine [4].4mC is mainly found in prokaryotes and functions primarily in the bacterial restrictionmodification (R-M) system [5].N6-Methyladenine (6mA) was previously regarded as a DNA modification only in prokaryotes due to its low abundance in eukaryotes.With the advances in detection methods, it has been found in many eukaryotes, such as green alga,flies, worms, plants, mice and humans [6-13].6mA plays important roles in a variety of biological processes, such as DNA replication[14-16], transcription [17,18] and nucleosome positioning [6,19].It has been suggested that 6mA may function as a novel epigenetic mark in eukaryotes [1,20,21].In this review, we primarily discuss the recent advances in the detection and functional studies of 6mA in eukaryotes.
2.Detection of 6mA
6mA was initially found in the DNA ofE.coliduring the study of R-M system [22-24].It had been considered to be a DNA modification that is only present in prokaryotes for a long time.With the development of detection techniques, 6mA has been gradually found in some unicellular eukaryotes, plants and mammals[20].6mA in DNA can be detected by multiple methods (Fig.1),such as dot blots, liquid chromatography-tandem mass spectrometry (LC-MS/MS), methylated DNA immunoprecipitation sequencing (MeDIP-seq) and single molecule real-time sequencing (SMRTseq) [8,25-32].Recently, bioinformatics tools, such as 6mA predictors 6mAPred-FO and p6mA, have been developed to predict 6mA sites based on DNA sequence characteristics [33,34].In addition,6mA-specific zinc-finger proteins have also been employed for the readout of 6mA [35].Moreover, a stable isotope tracing-based approach, which used [15N5]-dA as an initiation tracer, has been developed to quantitatively and specifically evaluate the origins and metabolism of 6mA in human cells [36].
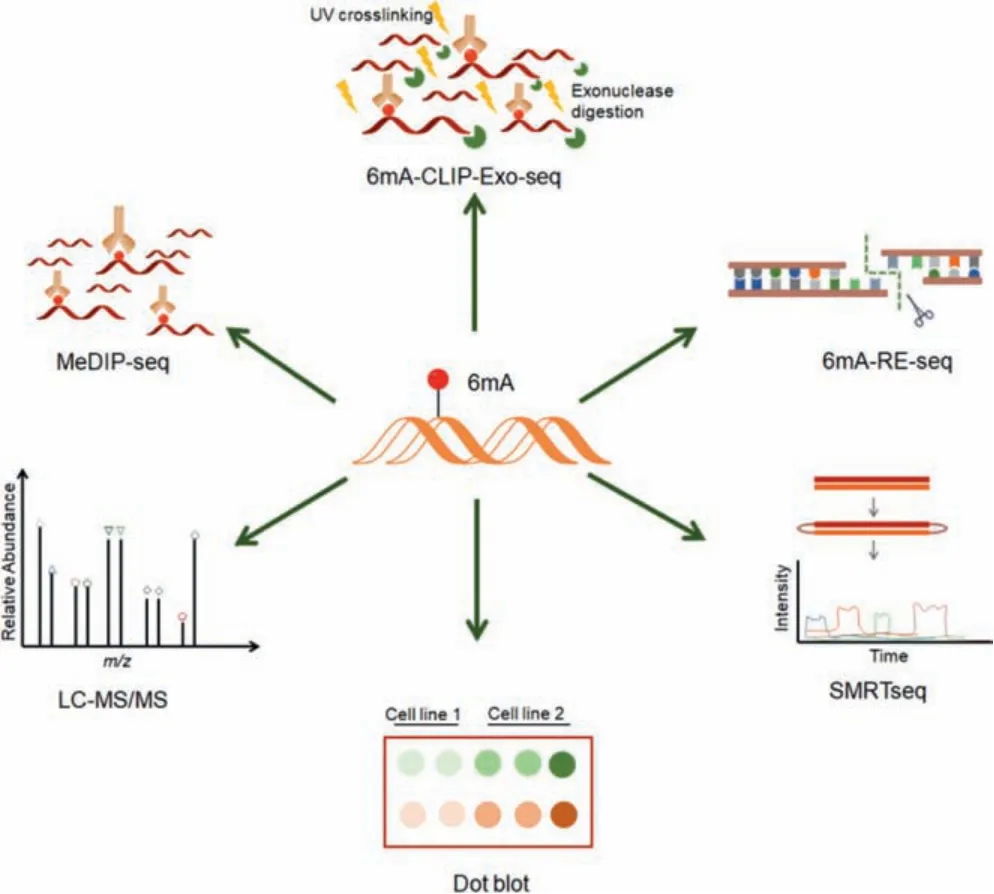
Fig.1.Various 6mA detecting methods.Dot blot and LC-MS/MS are usually used to measure the abundance of 6mA.MeDIP-seq and 6mA-CLIP-exo-seq are antibodybased methods for mapping 6mA sites, and the latter can improve resolution by using Ultravoilet (UV) cross-linking and exonuclease digestion.6mA-RE-seq can recognize 6mA sites at single-nucleotide resolution by employing methylation-sensitive restriction enzymes, whereas SMRTseq is able to distinguish different DNA modifications at single-base level by measuring the kinetic signatures of each modification.
The 6mA level is very low in eukaryotes.A previous study found that 6mA accounts for ~0.4% of total adenine in the genomic DNA ofChlamydomonasand is enriched at the transcription start site [6].The levels of 6mA inDrosophila melanogasterandCaenorhabditis elegansare about 0.001%–0.07% and 0.01%–0.4%of adenine, respectively [7,8].Many eukaryotes have low levels of 6mA, such asTetrahymena[19],Xenopus laevis[37], rice [11], zebrafish [38], pigs [38] and mice [39].6mA was identified in mouse embryonic stem cells (mESCs) by using high performance liquid chromatography (HPLC) and a SMRTseq-based strategy [9].However, some groups did not detect the existence of 6mA in mESCs by using an ultra-high performance liquid chromatography coupled with the mass spectrometry (UHPLC-MS/MS) method [40].These studies have caused controversy as to whether 6mA modification is present in mouse embryos and human genome [40].A recent study has combined multiple 6mA detection methods, including SMRTseq, MeDIP-seq, methylated DNA immunoprecipitation-quantitative PCR (MeDIP-qPCR) and LC-MS/MS, to identify the presence of 6mA in human genomic DNA.It was found that the level of 6mA in human genome is ~0.051% (6mA/A) and a higher level of 6mA was observed in subcellular organelles, especially in the mitochondria(0.184%, 6mA/A) [10].
Due to the low abundance of 6mA in eukaryotes, different detection methods, technical error and environmental contamination have a great impact on 6mA level.It is controversial whether the low abundance of 6mA detected in some species is real.To convincingly identify 6mA, some researchers have optimized the pretreatment method and redetermined the 6mA contents of 16 eukaryotic genomes, includingChlamydomonas, C.elegans, mouse and human genomes.They found that the levels of 6mA in these species were significantly different from the previous reports [41].These studies suggested that, compared with UHPLC-MS/MS, SMRTseq can identify 6mA at single-base resolution, but has a higher false positive rate [41].Therefore, it is critical to combine multiple detection methods and prevent the external contamination when measuring 6mA level in eukaryotes.
3.6mA regulating proteins
Nucleic acid modifications can be dynamically regulated according to their roles in growth, development and reproduction,including the generation or elimination of modifications.For instance, RNAN6-methyladenosine modification (m6A) affects multiple cellular processes through dynamic regulation by its writers(methyltransferases), readers (m6A-binding proteins), and erasers(demethylases) [42].Methyltransferases and demethylases are particularly important in this process.A critical step in the confirmation of 6mA as a regulatory biological mark has been the identification of proteins that install and remove it.
3.1.Methyltransferases
6mA methyltransferases mainly belong to methyltransferase MT-A70 family (Fig.2).These enzymes have a conservedα-helical domain and a 7-β-chain methyltransferase domain at the N- and C-terminus, respectively, which utilizeS-adenosyl-L-methionine(SAM) as the methyl donor [2].Methyltransferases-like 3 and 14(METTL3 and METTL14) in MT-A70 family belong to RNA methyltransferases [43], whereas METTL4 may function as a 6mA methyltransferase in eukaryotes [44].
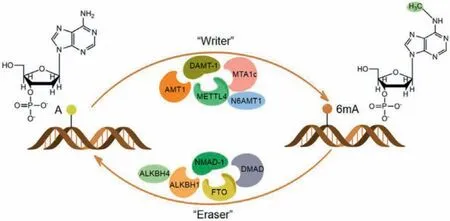
Fig.2.Regulating proteins of 6mA.6mA can be dynamically regulated by methyltransferase (writer) and demethylase (eraser).6mA methyltransferases mainly belong to MT-A70 family, whereas 6mA demethylases are primarily ALKB family proteins.
3.1.1.METTL4
The silence of METTL4 reduced the levels of 6mA in several eukaryotes, such as silkworm [45], mice [46,47] and humans [44].DAMT-1, a homolog of METTL4 inC.elegans, also acts as a 6mA methyltransferase which affects the fertility of worms by regulating 6mA level [8].In humans, 6mA is mainly distributed in mitochondria.METTL4 has been recognized as a mitochondrial protein which can catalyze 6mA modification of mitochondrial DNA and modulate gene expression in human hepatocellular carcinoma(HepG2) cells [44].Together, these findings support that METTL4 is a conserved 6mA methyltransferase in eukaryotes.However, some studies found that METTL4 is a nuclear protein which is involved in the methylation of U2 small nuclear RNA (snRNA) in human HEK293T cells [48,49], indicating that the localization and function of METTL4 is cell line-dependent.
3.1.2.N6AMT1
N6adenine-specific DNA methyltransferase 1 (N6AMT1) has the characteristic Asn-Pro-Pro-Tyr (NPPY) motifs, which may be involved in the regulation of 6mA level in human genome [50,51].The overexpression of N6AMT1 increases the level of 6mA [10], but the absence of N6AMT1 in glioblastoma stem cells (GSCs) has no effect on 6mA level [52].The crystal structure of human N6AMT1-TRM112 in complex with cofactor SAM demonstrates that N6AMT1 is a protein methyltransferase rather than a DNA methyltransferase[53].Therefore, whether N6AMT1 is a real 6mA methyltransferase in mammals remains controversial.
3.1.3.6mA methyltransferases in Tetrahymena thermophile and ciliates
Adenine methyltransferase 1 (AMT1) is a distinct MT-A70 family methyltransferase, which maintains 6mA modification at transcriptionally related adenine-thymine (ApT) dinucleotides inTetrahymena thermophile[54].6mA methyltransferase complex (MTA1c) in ciliates consists of two MT-A70 proteins and two homeobox-like DNA binding proteins.The disruption of the catalytic subunit of MTA1c results in the loss of 6mA throughout the genome and affects the growth cycle of ciliates [55].The homologues of AMT1 are usually found in protists and basic fungi but lacking in animals,plants, and real fungi [54], suggesting that AMT1 homologues are prototypical 6mA methyltransferases in eukaryotes.
Noteworthy, there are some controversies as to the origin of 6mA and the presence of methyltransferase-mediated 6mA in mammalian cells [56,57].It has been recently reported that the 6mA may be formed by free m6A in RNA through nucleotidesalvage pathway and misincorporated into DNA by DNA polymerases.DNA polymeraseλwas identified as one of major polymerases responsible for 6mA accumulation in late G1 phase [56].Therefore, the origin and regulatory mechanisms of 6mA remain to be further elucidated.
3.2.Demethylases
Most of the 6mA demethylases belong toα-ketoglutaratedependent dioxygenase ALKB family (Fig.2) [4].The ALKB family contains nine homologues in humans, including ALKBH1-ALKBH8 and FTO (fat mass and obesity-associated protein).The catalytic core region of the ALKB family is conserved in humans and bacteria, but the adjacent substrate recognition subdomain has variability and plays a key role in its interaction with the substrates[58,59].
3.2.1.ALKBH1
Previous studies have identified ALKBH1 as a 6mA demethylase in human cells, mice and rice [10,11,52,60-62].In mESCs, the young LINE-1 transposons were significantly enriched with 6mA when knocking out ALKBH1, thus acting as a silencing center for neighboring genes [9].However, another group did not observe noticeable changes at 6mA level after ALKBH1 knockout in mESCs[63].This divergence may be attributed to different 6mA detection methods or external contamination.A recent study reported that ALKBH1 may function as a nuclear eraser of 6mA in unpaired DNA,such as "bubble" DNA and circular DNA (R-loop,D-loop, stem-loop,etc.) [64], providing new directions for revealing the real substrate of ALKBH1.
3.2.2.FTO and ALKBH4
ALKBH5 and FTO have been extensively studied as m6A demethylases in RNA [65].However, the demethylation activity of ALKBH5 on 6mA is very weak or even completely inactive.FTO can catalyze the removal of 6mAin vitroand has a negative correlation with 6mA level [26].In addition, FTO can promote the maintenance of lipid content in mature adipocytes by regulating 6mA level in the promoter of enhancer binding proteinδ(CEBPD) [66].In vitroexperiments have proved that FTO mainly catalyzes 6mA on single-stranded DNA [66], but whether FTO plays a similar rolein vivoremains to be further elucidated.Moreover, murine ALKBH4 can catalyze the demethylation of 6mAin vitro[46], indicating that ALKBH4 might play a complementary role with other demethylases to control the 6mA level in mammalian genomes.
3.2.3.Other 6mA demethylases
InDrosophila, 6mA removal is catalyzed by a Tet-like protein DMAD (DNA 6mA demethylase) which can cause severe developmental defects by disturbing the level of 6mA [7,67].InC.elegans,ALKB family member NMAD-1 (N6-methyladenine demethylase 1)has been recognized as a 6mA demethylase which can regulate 6mA level and affect the fertility of worms [8].The homolog of NAMD-1 in lepidopteran silkworm is named NMAD which can affect the overall level of 6mA and induce cell cycle arrest [45].Although ALKBH1 has been recognized as m6A demethylase in rice[11,68], whether other ALKB proteins are also involved in 6mA demethylation in plants remains to be further investigated.
4.6mA reading proteins
In addition to the enzymes adding or removing 6mA, 6mA also functions as a regulatory signal to modulate multiple biological processes.6mA can be recognized by specific reader proteins which can directly or indirectly recruit other proteins to change the chromatin structure or transcriptional state [69].The readers mainly refer to those proteins that can bind to the modification sites and recruit other molecules, which can act as an intermediate bridge for signal transmission pathways.YTH (YT521-B homology) family proteins (YTHDF1-YTHDF3, YTHDC1, and YTHDC2) are the readers of m6A modification in RNA, which can regulate the localization, splicing, stability, and translational efficiency of mRNA[70,71].Our previous studies have also identified YTH family proteins as the readers forN1-methyladenine and 5mC in human RNA[72,73].However, so far, fewer 6mA readers have been found in eukaryotes.
InE.coli, SeqA can recognize and bind to hemimethylated adenine sites to prevent DNA replication in this region [74].InDrosophila, Fox family protein Jumu has been recognized as a 6mAbinding protein, which is involved in maternal-to-zygotic transition[75].In addition, quantitative proteomics methods have been employed to identify a series of potential 6mA-binding proteins in human cells, such as single-stranded DNA-binding protein 1 (SSBP1)and YTH family proteins [61].In this vein, YTH domain of YTHDC1 preferentially binds to single-stranded 6mA-containing DNA, while YTH domains of YTHDF1 and YTHDF2 bind to m6A-carrying RNA more easily [76].
Moreover, 6mA also functions by preventing the interaction between proteins and DNA.For instance, 6mA in AAC (adenineadenine-cytosine) motif can inhibit the interaction between R2R3-MYB protein WEREWOLF (WER) and its target DNA to regulate the growth and development ofArabidopsis[77].A recent study found that the 6mA level in the stress-induced DNA double helix destabilization (SSID) regions was upregulated during the development of mouse trophoblast stem cells and 6mA can regulate chromatin structure by hindering the interaction between regulatory protein SATB1 (special AT-rich sequence binding protein 1) and SSID to modulate early embryoic development [78].
5.Biological functions of 6mA
DNA is the carrier of genetic information and DNA modifications play important roles in the transmission of genetic information.In prokaryotes, 6mA is mainly used to protect bacterial genomes against the invasion of foreign DNA [79]; in eukaryotes,the biological functions of 6mA are far beyond the defense of foreign DNA [80,81].
5.1.Effects of 6mA on transcription, DNA replication and repair
As a part of the antiviral R-M system, 6mA can regulate bacterial gene expression and is closely related to transcription [82].The absence of 6mA methyltransferase in pathogenicE.coliled to changes in global transcription, indicating that 6mA has important regulatory functions beyond the markers of host genomes[83,84].Among the genes transcribed by three RNA polymerases(POL I, II, III), 6mA is specifically associated to POL II-transcribed genes [85].The presence of 6mA on DNA template will cause the site-specific transcription arrest of the POL II-transcribed genes inSaccharomyces cerevisiae[86].In addition, 6mA is an indispensable component in chromatin group, which is involved in nucleosome positioning and chromatin remodeling [6,9,19,85].In mice,6mA accumulation in genic elements is related with transcriptional silencing [9,46].In humans, a correlation between 6mA abundance and the transcription levels of genes has also been reported[44,52,60].In this respect, 6mA may affect transcription indirectly by modulating the binding of transcription factor to its target sequence or changing the chromatin structure in the promoter regions [9,44,78].
There are few studies about the effect of 6mA on DNA replication.InE.coli, 6mA mostly appears in the 5′-GATC-3′ motif which is usually distributed in the origin of replication, suggesting that 6mA inE.colimight affect DNA replication [87].In addition, 6mA can reduce the incorporation frequency of dTTP (deoxythymidine triphosphate) during DNA replication directed byBst, Klenow and gp90 exo-DNA polymerasesin vitro[88,89].Moreover, 6mA on template strand would hinderin-vitroDNA replication mediated by Y-family DNA polymerases Polι, Polηand Dpo4, resulting in reduced incorporation efficiency of dTTP or dTMP and the inhibition of next-base extension process [14-16].
InE.coli, 6mA plays an important role in DNA mismatch repair pathways.Recent studies have shown that 6mA might also be involved in DNA repair in eukaryotes.8-Oxo-2′-deoxyguanosine(8-oxoG) is an important DNA damage resulting from oxidative stress, which can lead to gene mutation by mispairing with adenine.It has been found that 6mA:8-oxoG is significantly less stable than A:8-oxoG, suggesting that 6mA may play a role in DNA repair by minimizing incorporation of 8-oxoG opposite adenine by DNA polymerases [90,91].
5.2.Effects of 6mA on cell growth and differentiation
6mA plays important roles in cell growth and differentiation.InCaulobacter, the expression of cell-cycle controlling factors is related with 6mA level in promoter regions, suggesting that 6mA may be involved in the regulation of cell cycle [92].InDrosophila,6mA is closely associated with embryonic growth and development.6mA demethylation mediated by DMAD regulates transposon expression in ovary and differentiation of early germ stem cells[7].In mouse trophoblast stem cells, 6mA can affect early embryoic development by hindering the interaction between SATB1 and SSID regions [78].ALKBH1-mediated 6mA demethylation is also correlated with axon regeneration and the differentiation of mouse skeletal muscle [62,93].In human mesenchymal stem cells (MSCs),the accumulation of 6mA in the promoter region of osteogenic differentiation factorATF4significantly inhibits the differentiation of MSCs, resulting in abnormal bone formation [60].In addition, the level of 6mA is also closely related with plant growth and development.In rice, the increase in 6mA level can lead to an earlier heading date [11].The mutations in nuclear small plasticizer DDM1(deficient in DNA methylation 1) can decrease the level of 6mA, resulting in reduced plant height and number of grains [94].
5.3.Relationship between 6mA and human diseases
6mA can regulate the expression of disease-related proteins,thus providing a molecular basis for the treatment of human diseases.It was found that the reduction of genomic 6mA level could induce the growth of cancer cells and promote tumorigenesis in humans [10].This finding draws people’s attention to exploring the relationship between 6mA and human diseases.
5.3.1.Effects of 6mA on tumor cell proliferation
The levels of 6mA decreased in gastric and liver tumor tissues,indicating that 6mA might be correlated with the occurrence of tumors [10].In addition, the up-regulation of 6mA level has an inhibitory effect on the migration and development of tumor cells[10], suggesting that 6mA could be used as a potential therapeutic target for cancer treatment.In glioblastoma, the deletion of ALKBH1 increased the level of 6mA and induced transcriptional silencing of the oncogenic pathway, thereby inhibiting the proliferation and metastasis of tumor cells [52].Moreover, the expression levels of ALKB family demethylases significantly increased in many cancer tissues, such as bladder, prostate and pancreatic cancers, which may reduce the levels of 6mA and promote the proliferation of cancer cells and drug resistance [95–98].In triple negative breast cancer (TNBC) tissue, the level of 6mA was significantly decreased, which is closely related with drug resistance [99].Together, 6mA is the most regulated DNA modification in cancer, implying that 6mA could be used as a biological marker and therapeutic target for cancer treatment.
5.3.2.Impact on immune system diseases and other diseases
In addition to the regulation of tumor cell proliferation, 6mA is also related with immune system diseases and other diseases.For instance, 6mA can stimulate the production of interleukins and enhance the immune activity induced by cytosine-guanine dinucleotide (CpG) in DNA [100].A higher 6mA level was found in the patients with systemic lupus erythematosus (SLE), which is associated with the early development of SLE inflammatory process, indicating that 6mA could be a promising early diagnosis marker and treatment target of SLE [101].In addition, 6mA levels in patients with type 2 diabetes, essential hypertension, and atherosclerosis are lower than those of healthy people, suggesting the potential roles of 6mA in diagnosis and treatment of these diseases [102–104].A recent study found that ALKBH1-mediated 6mA demethylation is also related to the vascular calcification in chronic kidney disease [105].Moreover, 6mA also plays important roles in activityinduced gene expression, fear regression, memory formation and neuropsychiatric diseases in adult mice brains [39,106].Therefore,the studies on 6mA may benefit the clinical diagnosis of these diseases and the development of therapeutic drugs.
5.4.Other functions of 6mA
6mA may function as a carrier of genetic information across generations.For instance, 6mA is required to transfer mitochondrial stress adaptation to progeny inC.elegans.The level of 6mA significantly increased upon mitochondrial disorder, which promotes the transcription of 6mA-labeled mitochondrial stress response genes and reduces the mitochondrial stress of offspring[107].In addition, 6mA can extend life span of progeny by promoting the transcription of stress response genes [108].
In addition, 6mA is also required for transponson activity and cell fate transition of mammalian cells [7,9,109].6mA at special sites can affect the stem-loop structure of c-kit1 G-quadruplex(G4), thereby decreasing the thermal stability of the c-kit1 G4 structure [110].Furthermore, 6mA can also be converted intoN6-hydroxymethyladenine and hypoxanthine by ALKB proteins and deaminase, respectively [111–113].These conversion products can continue to play important roles in various biological processes(Fig.3).
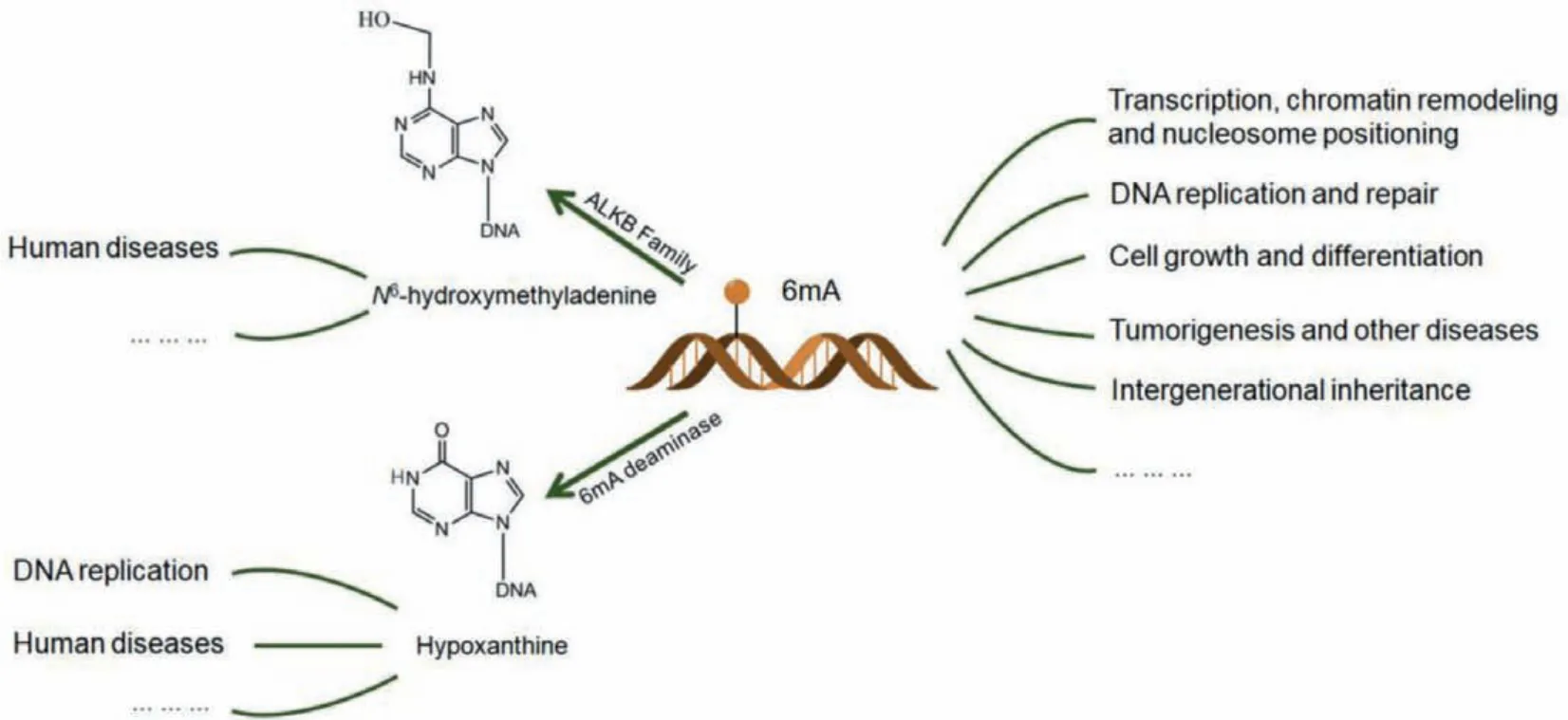
Fig.3.Biological functions of 6mA.6mA is involved in a variety of biological processes in eukaryotes, such as transcription, DNA replication, cell differentiation and intergenerational inheritance.6mA can be oxidized by ALKB family enzymes to produce N6-hydroxymethyladenine which is closely related to human diseases; 6mA can also be converted into hypoxanthine by 6mA deaminase, which can hinder DNA replication.
6.Conclusion and perspectives
6mA is an emerging epigenetic mark which plays important roles in a variety of cellular processes.The detection of 6mA in eukaryotes is very challenging due to its low abundance.Experimental conditions (e.g.,detection methods and external contamination)have a great impact on the measurement of 6mA in eukaryotes[41,114].In addition, it is generally believed that 6mA is generated and eliminated by 6mA methyltransferases and demethylases.However, some studies suggested that the 6mA may be formed by free m6A in RNA through nucleotide-salvage pathway and misincorporated into DNA by DNA polymerases [56,57].These findings have raised doubts regarding the entity of the examined 6mA as well as its proposed functions and the related methyltransferases(e.g.,METTL4) and demethylases (e.g.,ALKBH1 and ALKBH4).Thus,further studies are required to determine the exact origin, regulatory mechanisms and functions of 6mA in eukaryotic cells.m6A in RNA regulates multiple biological processesviaits readers, such as YTH family proteins [42].However, so far, there are few studies about the readers of 6mA and some researchers argue against 6mA as an epigenetic marker in eukaryotes [115].Further studies are required to discover some more 6mA-binding proteins and their possible biological functions in the 6mA-associated epigenetic regulation in eukaryotes.It can be envisaged that such chemical and biological studies of 6mA modification would provide a better understanding about its potentially important roles in the regulation of development and human diseases such as cancer.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Nos.21807030, 21907028), the Science and Technology Innovation Program of Hunan Province (No.2019RS2020),Natural Science Foundation of Hunan Province (No.2020JJ5046),and the Fundamental Research Funds for the Central Universities(Nos.531118010061, 531118010259).
 Chinese Chemical Letters2022年5期
Chinese Chemical Letters2022年5期
- Chinese Chemical Letters的其它文章
- Recent advances in enhancing reactive oxygen species based chemodynamic therapy
- An integrative review on the applications of 3D printing in the field of in vitro diagnostics
- Recent developments of droplets-based microfluidics for bacterial analysis
- Recent progress in advanced core-shell metal-based catalysts for electrochemical carbon dioxide reduction
- Recent advances in carbon-based materials for electrochemical CO2 reduction reaction
- Encapsulation strategies on 2D materials for field effect transistors and photodetectors
