Single-cell level point mutation analysis of circulating tumor cells through droplet microfluidics
Shihui Qiu, Chunjie Shen, Xioyu Jin, Yunxing Lu, Zhoduo Tong,Zhenhu Wu,*, Hongju Mo,*, Jinlong Zho
a State Key State Laboratory of Transducer Technology, Shanghai Institute of Microsystem and Information Technology, Chinese Academy of Sciences,Shanghai 200050, China
b Center of Materials Science and Optoelectronics Engineering, University of Chinese Academy of Sciences, Beijing 100049, China
Keywords:Single cell CTCs Point mutation EGFR Microfluidics
ABSTRACT Tumor heterogeneity plays a critical role in the determination of appropriate anticancer therapy.As circulating tumor cells (CTCs) contain all tumor-related information, the genetic changes on CTCs could help us choose the appropriate treatments for different patients.Single-base mutations are very common in tumor genetic changes which may result in drug resistance.Here, we introduce a single-cell mutation detection platform based on droplet microfluidics.This platform integrates cell capsulation, cell lysis, polymerase chain reaction (PCR) and the observation process.The droplets’generation speed is over 6000 per minute and more than 600 cells could be encapsulated in one second.To verify the performance of our platform in practical use, we performed the mutation analysis of 4 kinds of cells with our platform and noted that the genetic status of each single cell was clearly discriminated.Moreover, these results agreed with those from direct sequencing.Compared with other forms of single-cell mutation detection techniques, our platform has high throughput, short experimental time and less experimental operations.
Circulating tumor cells (CTCs) are rare tumor cells that shed into the peripheral blood during metastasis.CTCs contain complete tumor genetic information so that genetic analysis of circulating tumor cells can provide some guidance for early diagnosis, anticancer treatment and prognosis [1–3].Point mutation analysis of the cancer cells could guide the selection of anticancer treatment plans.For example, T790M could result in the drug resistance of non-small cell lung cancer (NSCLC) patients, thus CTC analysis for T790M mutation is of great importance [4–6].Many studies have found that epidermal growth factor receptor (EGFR) protein is abnormally expressed in NSCLC, which promotes the proliferation, invasion and metastasis of cancer cells.Therefore, the point mutation of EGFR gene in CTCs is of great significance for diagnosis, treatment and prognosis of NSCLC patients [5,7].
However, the low abundance of CTCs is the biggest problem for further studies.The large background noise from white blood cells (WBC) and circulating nucleic acid molecules would affect the CTCs’signal [8,9].Thus, identifying the genetic status of CTCs on single-cell level poses a great challenge.
Microfluidic chips are widely used in the field of single cell’s research due to their comparable scale, highly integrated design and high throughput [10–13].Most microfluidic platforms for single cell genomic analysis are based on microarray or micro-chambers[14].Shenet al.[9] introduced a micro-valve based system.This system could realize cell caption, wash, cell lysis and reverse transcription-polymerase chain reaction (RT-PCR).However, it is difficult to use in clinical applications due to its complex operations.Parket al.[15] designed a device which is integrated with MagSifter [16] for tumor cells’enrichment and a single-cell nanowell array for single-cell genetic analysis.Duet al.[17] reported a physically structured microholes for rapid generation of picoliter scale droplets.This droplet array was applied in single cell analysis but the fabrication of the microholes is complicated.Novaket al.[18] developed a single cell analysis platform based on agarose emulsion.Although all these systems are highly integrated,the preparation of magnetic beads and hydrogel beads is complex.Droplet microfluidics is now a new trend in single cell analysis for its high throughput and easy generation process [19–22].The present genome tests based on droplets suffer from low yield and efficiency which are not suitable for most kinds of cell samples[23,24].On the other hand, it is difficult to perform multi-step liquid handling.Although the gold standard method of cell genetic analysis is direct sequencing, only cell samples with a mutant proportion higher than 25% are suitable for direct sequencing [25].
Here, we report a single-cell mutation detection platform which is based on droplet microfluidic technique and single cell PCR.This platform is integrated with cell encapsulation, cell lysis, PCR reaction andin situobservation.Within just one minute, more than 600 cells could be encapsulated in droplets and stored in the glass chamber for downstream detection.The platform was employed to analyze EGFR L858R mutation [26] of 4 kinds of cells on single-cell level.The genetic statuses are successfully detected and consistent with that of direct sequencing.
The fluorophore-molecules and quencher-labeled Taqman probes together with unmodified PCR primers were synthesized and purified by high-performance liquid chromatography (HPLC)by Sangon biotech (Shanghai, China).The primers and probes were resuspended at a concentration of 100 μmol/L in diethyl pyrocarbonate (DEPC) treated water and stored at -20 °C.The sequences are listed in Table S1 (Supporting information).
H1975 cell line, carrying EGFR L858R point mutation gene, was used in this experiment.A549 and H446 that were reported with no EGFR L858R mutation gene were used as a wild type control group.All these 3 cell lines were purchased from National Collection of Authenticated Cell Culture.Human peripheral blood was obtained from healthy people at Changzhou Ruikang Zhi’an Medical Examination Center with informed consent.White blood cells(WBC) were extracted from human peripheral blood samples by Ficoll-PaqueTMPlus (GE Healthcare) density gradient centrifugation kit according to the manufacturer’s instructions.The genomic types of the cells are listed in Table S2 (Supporting information).
Genomic DNA was extracted from the four kinds of cells mentioned above using ArcturusTMPicoPureTMDNA Extraction Kit(Thermo Fisher Scientific) according to the manufacturer’s instructions.Genomic DNA was used as a template for the subsequent PCR process.The PCR process included a denaturation step at 95 °C for 5 min, followed by 45 cycles of heating at 95 °C for 10 s, 58 °C for 40 s, and 72 °C for 15 s sequentially.Then, direct sequencing of the PCR products was performed by BGI (Beijing, China).
To fabricate the chip, the pattern of the channel was fabricated on a wafer with SU8 negative photoresist through lithography,exposure and development.Polydimethylsiloxane (PDMS) mixture was prepared by mixing the pre-polymer and cross-linker at the ratio of 10:1 (w/w).After that, PDMS mixture was poured onto the SU8 mold.Baked at 95 °C for 2 h, the cured PDMS layer was peeled off carefully and cut into PDMS chips.Holes were punched at the corresponding place to make the inlet and outlet.Finally,the PDMS chips were bound onto clean slide glasses and baked at 105 ˚C overnight.Droplet storage chamber was made of two slide glasses to avoid the evaporation of oil and the fusion of droplets.
Two dispersed phase channels were designed for the single cell encapsulation (Fig.S1a in Supporting information).A serpentine channel was designed after two dispersion phases’inlet channels (Fig.S1b in Supporting information).Two dispersed phases would join and mixed in the serpentine channel (Fig.S1c in Supporting information).Then the generated droplets were storage in glass chamber (Fig.S1d in Supporting information).The continuous phase is prepared with mineral oil (Thermo Fisher Scientific,USA), with 3% w/w Abil EM 90 (Degussa/Goldschmidt, Germany)and 0.1% w/w Triton X-100 (Sigma-Aldrich, USA).Afterwards, 12.5 μL dispersion phase I and 12.5 μL dispersion phase II were added to the two dispersion phases inlet ports.Meanwhile, 50 μL oil mixture was added to continuous phase inlet port and then the three inlet ports were pressurized with Fluidiclab Pressure Controller (Prinzen Biomedical, China).The pressure of three phase were tuned to ensure the steady generation of droplets with a diameter of 50 μm.The volume ratio of dispersion phase I to II was 1:1.The component of two dispersion phases are listed in Table S3(Supporting information).
Thermocycling of the single cell RT-PCR was performed on an Eppendorf Gradient Mastercycler.The PCR process included a reverse transcription step at 50 °C thermal for 30 min, denaturation step at 95 °C for 5 min, followed by 45 cycles of heating at 95 °C for 10 s, 58 °C for 40 s, and 72 °C for 15 s sequentially.The chip was preserved at 10 °C afterward.Finally, the droplets signals were observed with a fluorescence microscope (BX51, OLYMPUS, Japan).The images were captured by a Charge-coupled Device (CCD) camera.
The schematic diagram is shown in Fig.1a.Cells, PCR mix and cell lysis solution were encapsulated in droplets simultaneously.Then the droplets were stored in the glass chamber for PCR reaction and fluorescent observation.Taqman probe with carboxyfluorescein (FAM) dye [27] is for EGFR mRNA detection.Another probe with hexachloro fluorescein (HEX) dye [28] is used for reporting EGFR L858R mutation (Fig.1b).Thus, positive FAM fluorescence reports the presence of EGFR mRNA and the positive HEX signal indicates L858R mutant alleles were carried in the cell.
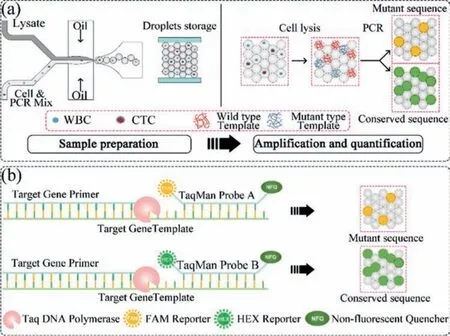
Fig.1.(a) The flow of point mutation analysis on single-cell level.(b) The schematic of PCR reactions in droplets.
The direct mixing of lysate and PCR reaction solution could not generate droplets successfully (Fig.S2 in Supporting information).In order to keep the steadiness of droplet generation, the concentration of cell lysis solution should be considered.The problem here is the balance between the cell lysis efficiency and the steadiness of droplet generation.We diluted the lysis solution in dispersion phase II by 2×, 4×, 6× and 8× (Table S4 in Supporting information).6× dilution of lysis solution is considered suitable for this experiment (Fig.2b).We also observed the cell lysis in droplets.As shown in Fig.2a, cells were encapsulated in droplets with 6× lysis solution and RT-PCR mix.In 5 min, there was vesica occurred from the cell membrane.Within 20 min, all cells were lysed.The downstream PCR results also proved that cell lysis succeeded (Fig.2c).
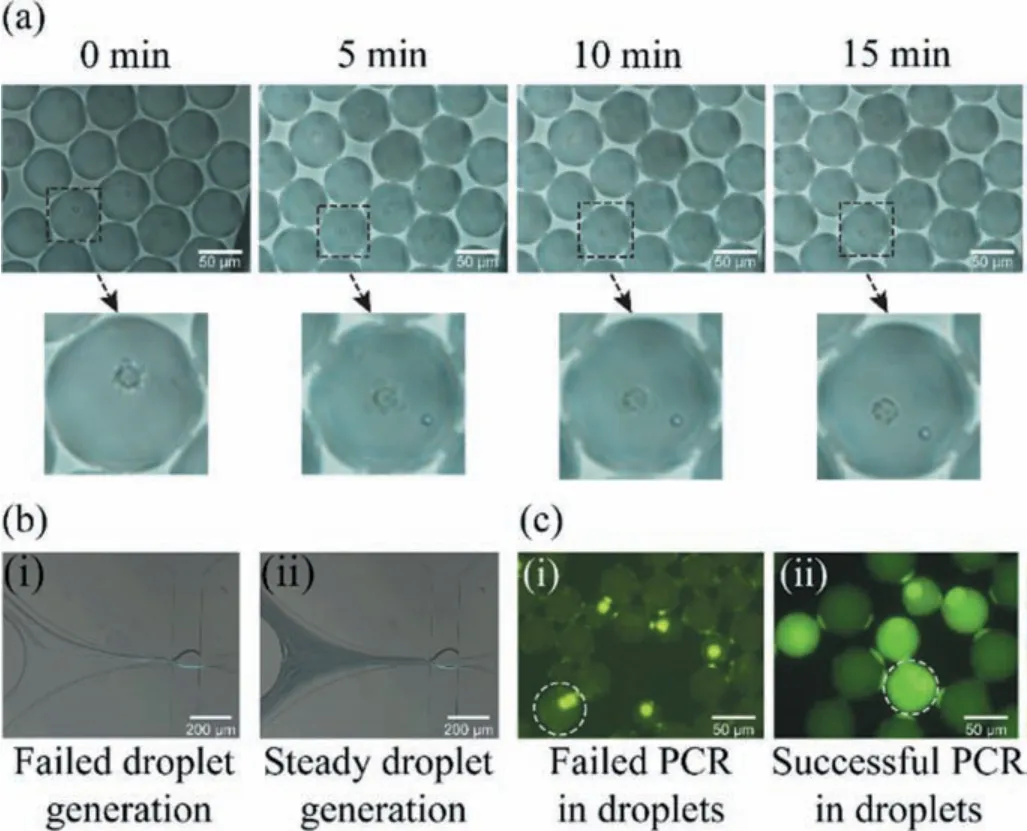
Fig.2.(a) The cell lysis process in droplet.(b) i) Unstable droplet generation with 4× lysis solution; ii) Stable droplet generation with 6× lysis solution.(c) i) Failed PCR results with 8× lysis solution; ii) Successful PCR results with 6× lysis solution.(scale bar ~100 μm).
Prior to the point mutation detections on the single-cell level,the PCR reagent system should be verified.For this reason, droplet digital PCR (ddPCR) was used to evaluate the performance of the PCR reagent system used in our study mentioned above.The ddPCR results (Fig.3a) agree with that of direct sequencing (Fig.3b).A549, H446 and WBC are EGFR L858R wild type while H1975 is L858R mutant type.Some droplets that contain H1975 cells are both FAM positive and HEX positive while others are only FAM positive.This means not all EGFR mRNA in H1975 cells are L858R mutant type.
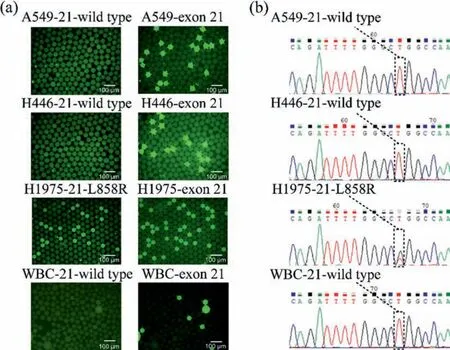
Fig.3.(a) Droplets digital PCR results of 4 kinds of cells (scale bar ~100 μm).(b) Sequencing results of 4 kinds of cells.
Here we defineDMas the number of cells that carry L858R point mutation whileDEis the number of cells that carry EGFR gene.The mutation frequencyfMis calculated as Eq.1:

After performing single cell mutation detection, we noticed that the results also agreed with that of direct sequencing.A549, H446 and WBC are EGFR exon 21 wild types while H1975 is L858R mutant type.Like ddPCR, not all H1975 cells are EGFR L858R mutant type (Figs.4a and b).Repeating more than three times of each experiment, we found that the results of both ddPCR and singlecell droplet PCR (sc-dPCR) indicated that there is a mutation ratio of L858R in H1975 cell line.Based on our sc-dPCR experimental results, the average L858R mutation proportion of H1975 cells is 53.87%.To test the performance of this platform for rare cells, 100 H1975 cells were added into 25 μL reaction volumn (4 cells/μL) to perform EGFR L858R mutation detection.As shown in Fig.4c, EGFR L858R mutation of H1975 was detected successfully and has an efficiency over 91%.Cell samples with a 2.5% mutation ratio could be detected which has a great advantage over direct sequencing (Fig.S3 in Supporting information) [25].
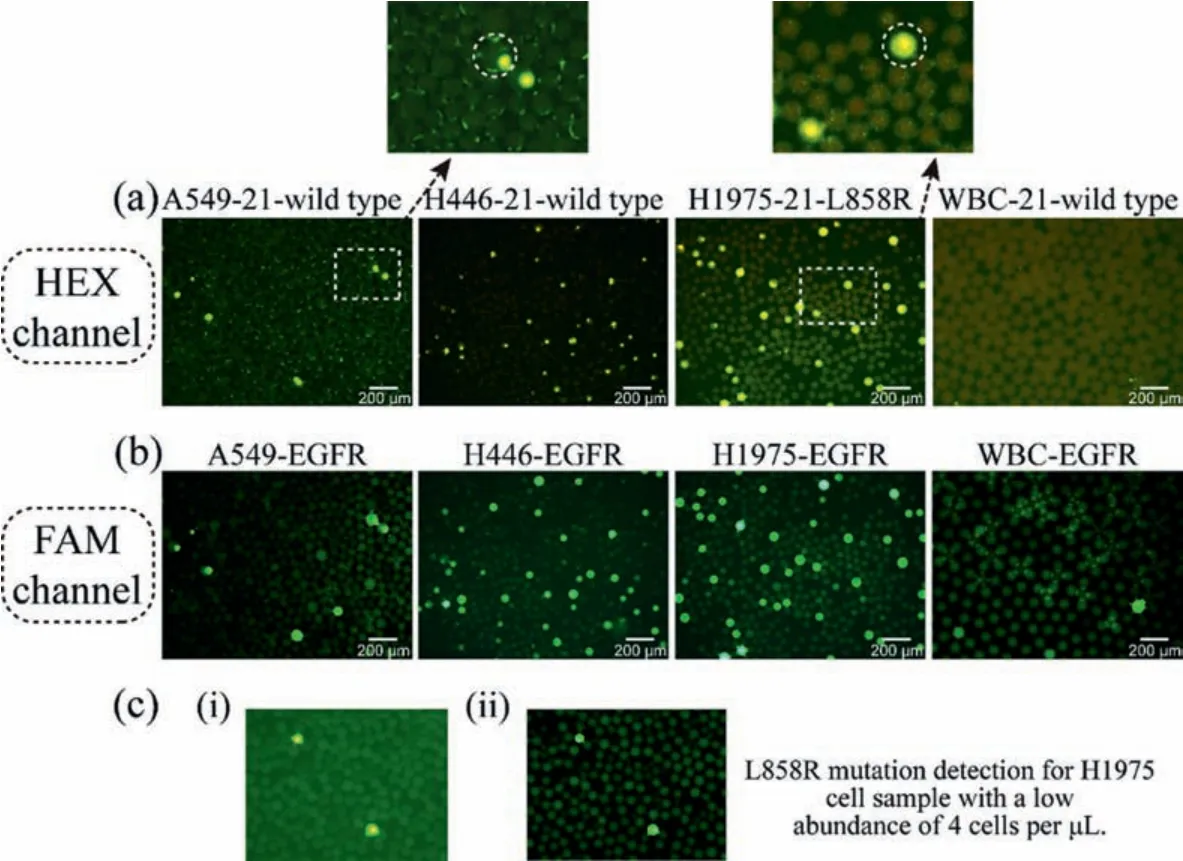
Fig.4.(a) Hex probe shows the existence of EGFR L858R mutation.(b) FAM probe shows the existence of EGFR mRNA.(c) Successful L858R mutation detection of H1975 cells under the concentration of 100 cells in 25 μL reaction volume.i) HEX positive droplets of H1975 cells.ii) FAM positive droplets of H1975 cells.Scale bar ~50 μm.
In recent years, with scientists’efforts, the research of singlecell genetic analysis has made remarkable progress.Compared with sequencing techniques or even single-cell sequencing methods, PCR detection is still the most widely used genetic detection method.Especially for cancer patients in hospital, it is necessary to monitor whether the genotype of the cells has changed during anticancer treatment.PCR-based methods are faster and lower-cost alternative.For real-time monitoring, the droplet-based single-cell mutation detection method used in this article has the following advantages:(1) Compared with the microcavity method, the operation is more convenient based on its one-step operation including the cells ’change to encapsulation, cell lysis, RT-PCR reaction and signal observation; (2) more adjustable storage capacity options, if the total amount of cells is large, a larger glass chamber can be used for droplets storage and signal observation; (3) its cost is much lower, given that only a pump and fluorescent microscope are needed to complete the entire experiment; (4) compared with the single-cell sequencing method based on droplets, the droplet generation speed of this platform is much higher (~100 droplets/s)which means it is suitable for most samples.
Declaration of competing interest
The authors declare that they do not have any known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This study was supported by the National Key Research and Development Program of China (No.2017YFA0205300), National Natural Science Foundation of China (Nos.61971410, 61701171,61801464, 61801465 and 62001458), Shanghai Sailing Program (No.20YF1457100), Shanghai Engineer & Technology Research Center of Internet of Things for Respiratory Medicine (No.20DZ2254400),and the Science and Technology Commission of Shanghai Municipality (No.19511104200).
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2021.08.128.
 Chinese Chemical Letters2022年5期
Chinese Chemical Letters2022年5期
- Chinese Chemical Letters的其它文章
- Recent advances in enhancing reactive oxygen species based chemodynamic therapy
- An integrative review on the applications of 3D printing in the field of in vitro diagnostics
- Recent developments of droplets-based microfluidics for bacterial analysis
- Dynamics and biological relevance of epigenetic N6-methyladenine DNA modification in eukaryotic cells
- Recent progress in advanced core-shell metal-based catalysts for electrochemical carbon dioxide reduction
- Recent advances in carbon-based materials for electrochemical CO2 reduction reaction
