Second-generation DNA-encoded multiple display on a constant macrocyclic scaffold enabled by an orthogonal protecting group strategy
Qigui Nie, Shuting Zhong, Yngfeng Li, Gong Zhng,*, Yizhou Li,b,*
a Chongqing Key Laboratory of Natural Product Synthesis and Drug Research, School of Pharmaceutical Sciences, Chongqing University, Chongqing 401331,China
b Key Laboratory of Biorheological Science and Technology, Ministry of Education, College of Bioengineering, Chongqing University, Chongqing 401331, China
Keywords:DNA-encoded library Macrocyclic scaffold DNA-compatible Protein–protein interaction Chemical probe
ABSTRACT DNA-encoded chemical library (DEL) represents an emerging drug discovery technology to construct compound libraries with abundant chemical combinations.While drug-like small molecule DELs facilitate the discovery of binders against targets with defined pockets, macrocyclic DELs harboring extended scaffolds enable targeting of the protein–protein interaction (PPI) interface.We previously demonstrated the design of the first-generation DNA-encoded multiple display based on a constant macrocyclic scaffold, which harvested binders against difficult targets such as tumor necrosis factor-α (TNF-α).Here, we developed a novel strategy which utilized four orthogonal amine-protecting groups on DNA, to explore larger chemical combinations on the same constant macrocyclic scaffold, following the parallel paradigm to mimic the versatile antibody-like multivalent epitope recognition patterns.We successfully integrated these orthogonal protecting groups with acylation and made a mock second-generation DNA-encoded display combination.This work illustrates a strategy to produce larger encoded multiple display on a constant macrocyclic scaffold, which could facilitate potential binder discovery with enhanced affinity to clinically significant PPI targets.
DNA-encoded chemical library (DEL) has emerged as a widely used ligand discovery technology for the development of drug candidates and bioactive chemical probes [1–5].Recording synthetic structures with DNA barcodes provides abundant chemical combinations and enables facile hit identification, which has turned into reality with the development of DNA-compatible synthetic methodologies [6–11].While DEL has facilitated the discovery of several clinical compounds from drug-like small molecule libraries[12–15], this technology is also capable of constructing macrocycle libraries [16–23].Macrocyclic compounds, conventionally derived from natural products or discoveredviamolecular evolution tools such as phage display/mRNA display approaches, possess large interaction surfaces suitable for targeting difficult protein–protein interactions (PPIs) [24–28].Compared with phage display/mRNA display where proteinogenic amino acids are mainly accommodated as diversity elements, DEL encompasses an extended range of synthetic building blocks (BBs) into macrocycle libraries to achieve superior scaffolds or sidechain modifications [16,18,19].So far, bioactive macrocyclic compounds have been discovered through DEL to target a panel of clinically significant proteins with larger interaction surface areas, such as Src kinase [29,30], insulin-degrading enzyme (IDE) [17,31],α-1-acid glycoprotein (AGP) [18], tumor necrosis factor-α(TNF-α) [19], respiratory syncytial virus (RSV) N protein [20], S-phase kinase-associated protein 2 (Skp2) [21], p300[11] and so on.
Group Neri and our group previously reported the firstgeneration DNA-encoded multiple display on a constant macrocyclic scaffold named ETH-YL library [19] (Scheme 1A).With a structure-defined constant decapeptide scaffold and three spatially oriented sidechains, this library contains 107-chemical combinations, mimicking the versatile antibody-like recognition patterns.Specific binders against a variety of proteins including TNF-α(a target involved in PPIs) have been isolated from the encoded library, despite with single-digit micromolar range affinities.TNFαis an important inflammation-related extracellular cytokine targeted by the antibody drugs [32].We envision that a secondgeneration library design featuring larger chemical combinations may further expand the interface area to enhance binders’affinity(e.g., nanomolar affinity), by exploiting the fourth spatially oriented sidechain on the same scaffold.As carboxylic acids are the most abundant, diversified and easily accessible BBs, we plan to generate the second-generation encoded display by acylation based on our previous systematical optimization of on-DNA amide bond formation [33].Nevertheless, the essential task here is the development of orthogonal amine-protecting groups on DNA for sequential sidechain acylation.In this work, we developed a novel DNAcompatible strategy with four amine-protected sidechains, providing an avenue to produce the second-generation DNA-encoded multiple display on a constant macrocyclic scaffold, with sidechain diversity facilitating further binder discovery.
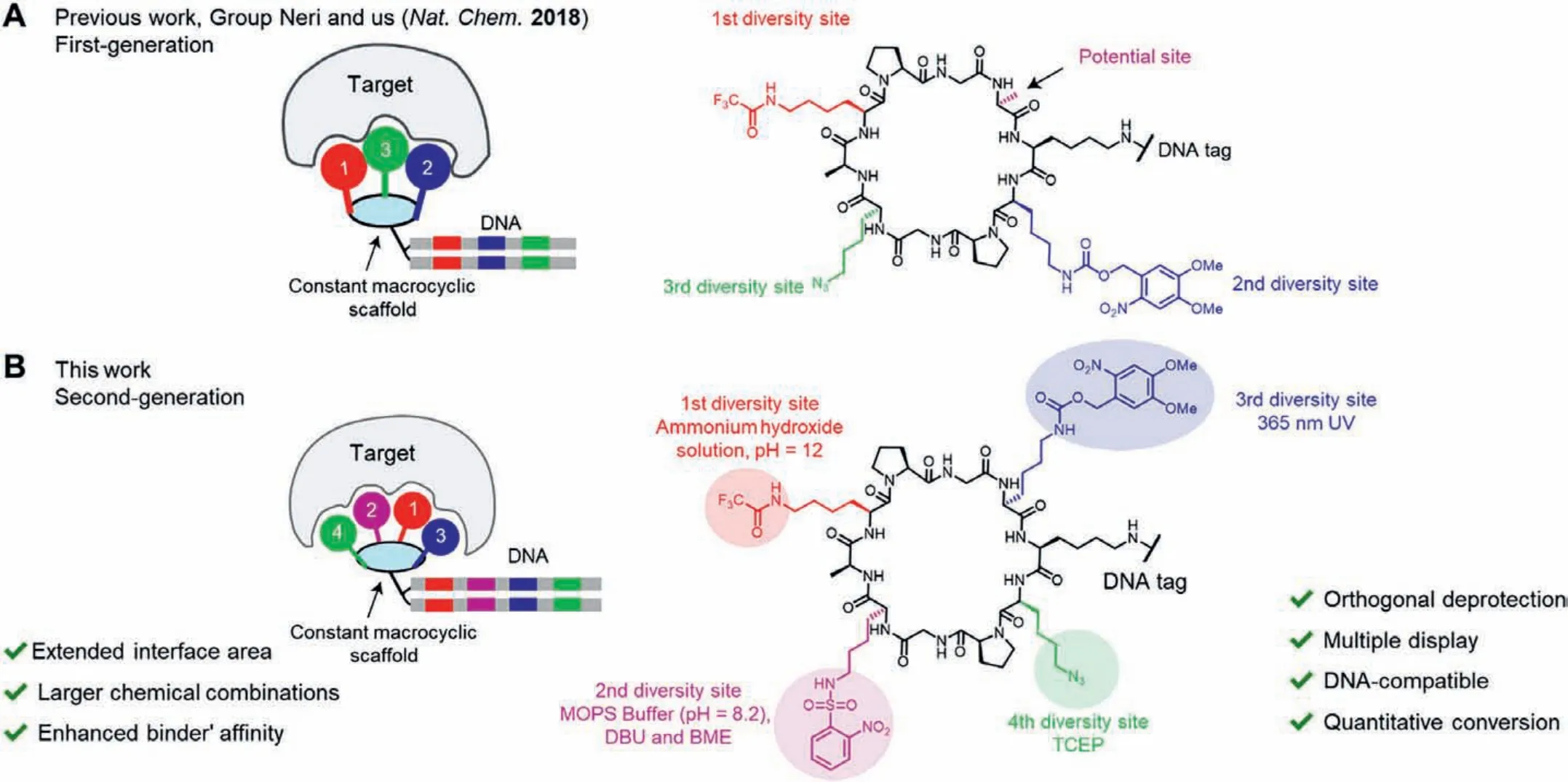
Scheme 1.Design of the second-generation DNA–encoded multiple display on a constant macrocyclic scaffold.(A) The first-generation DNA–encoded multiple display reported in our previous work.(B) Illustration of the second-generation DNA-encoded multiple display in this work, with four orthogonal protecting groups and larger chemical combinations.The numbering of protection groups indicates the deprotection order.
To reserve the constant scaffold design which benefited structure-defined spatial orientation, we exploited the macrocyclic scaffold previously developed by Mutteret al.[34,35].The scaffold contains two proline-glycine turns and spontaneously folds into a constant anti-parallelβ-sheet plane [36,37].Four sidechain-protected lysine derivatives for sequential acylation are spatially oriented and scattered on the same side of the planar scaffold, while another lysine handle is ready for the stepwise DNA barcode conjugation on the contrary side of the macrocyclic scaffold (Scheme 1B).The development of the four orthogonal on-DNA amine-protection for lysine sidechains is critical for the production of the second-generation encoded multiple display.We previously demonstrated three mutually orthogonal amine-protecting groups with DNA-compatibility:a trifluoroacetyl (Tfa) group removed under basic conditions (final pH 12), ano-nitroveratryloxycarbonyl (Nvoc) group cleaved by 365 nm UV irradiation [38], and an azido group reduced by tris(2-carboxyethyl)phosphine (TCEP)viaStaudinger reduction in solution.According to structural analysis, there is a fourth potential site on the scaffold [19].To turn this site into an additional encoded display site, here we sought a fourth orthogonal amineprotecting group.The 2-nitrobenzenesulfonamide (Ns) protecting group could be removed under reductive (like 2-mercaptoethanol,BME) and mild basic conditions (e.g., MOPS buffer, pH 8.2, with 1,8-diazabicyclo[5.4.0]undec-7-ene, DBU), which was previously reported in DNA synthesis [18,39].We envisioned that the Ns group could be orthogonal to the other three amine-protecting groups and applicable for the DNA-compatible sequential acylation here.
In the practice, we first set out to build the constant macrocyclic scaffold with four sidechain protections in the adjusted sequence.The linear decapeptide 1 was synthesizedviasolidphase peptide synthesis (SPPS), followed by an intramolecular ring-closure of the pre-organized linear configuration to gain cyclized decapeptide 2 [35].After the de-Boc with trifluoroacetic acid, a carboxylic acid handle for DNA conjugation was introduced with intermediate 3, yielding compound 4 (Fig.S1 in Supporting information).Later, we produced the DNA–scaffold conjugate I through amide bond formation between cyclopeptide 4 and a commercially available amino-modified oligonucleotide headpiece–primer (HP–P) (Fig.S2 in Supporting information).This purified DNA–scaffold conjugate I served as the starting point for verification of the protecting group strategy as well as library synthesis.
We then examined the orthogonality (whether compatible with each other) and DNA-compatibility (whether deprotection conditions cause DNA damage) of the four amine-protecting groups, respectively.(1) Tfa-deprotection was quantitatively completed with>95% conversion in 25% ammonium hydroxide solution (final pH 12) at 25 °C in 2 h.(2) Ns-deprotection yielded the target product with>90% conversion in MOPS buffer (pH 8.2)/DBU/BME (final pH 11) at 25 °C in 10 h, while the byproduct with additional Tfa-deprotection was observed (<10%).Therefore, Ns-deprotection was not fully orthogonal with Tfa but was compatible to be performed after Tfa-deprotection.(3) Nvoc-deprotection was triggered by 365 nm UV irradiation on ice for 1 h with>90% conversion.(4) Reduction of azide to amine was induced by TCEP at 25 °C for 12 h with>85% conversion (Fig.1).The conversion and purity of these products were examined by UPLC–MS, indicating no detectable DNA damage occurred (Figs.S3–S6 in Supporting information).
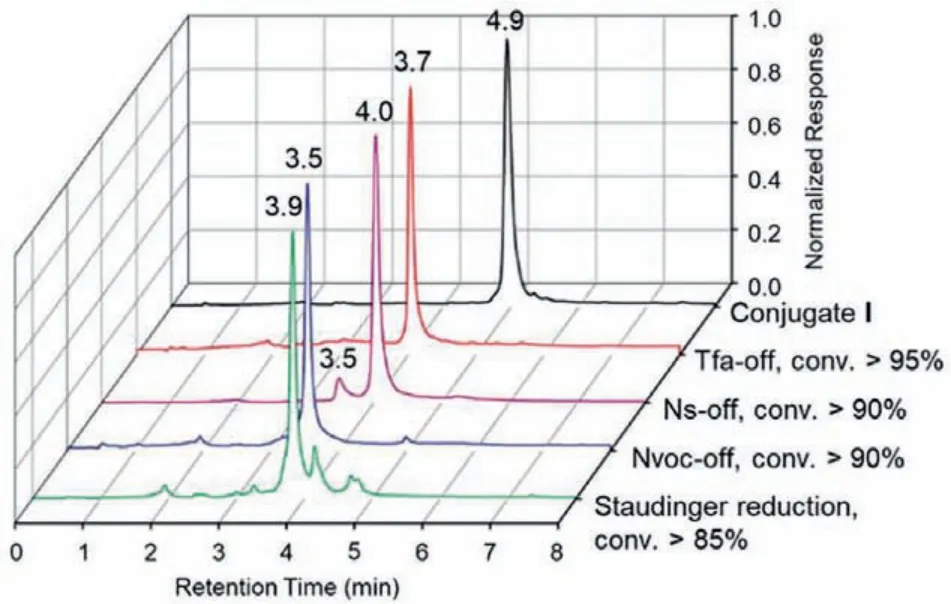
Fig.1.Verification of orthogonal deprotection on the DNA–scaffold conjugate I.Different products are shown in different colors:Tfa-off (red); Ns-off (magenta); Nvocoff (blue); azide reduction (green).Conversion characterized by UPLC–MS.
Since Ns-deprotection should proceed after Tfa-deprotection while the other reactions were mutually orthogonal, we decided a synthetic progress as shown in Fig.2A, and then examined the products’molecular weight during sequential amine-deprotection and acylation with four carboxylic acids (II to IX) (Fig.2B and Figs.S7–S14 in Supporting information).Altogether, the results presented the feasibility of multiple display of chemical combinations on the constant macrocyclic scaffold sequentially with the orthogonal strategy.
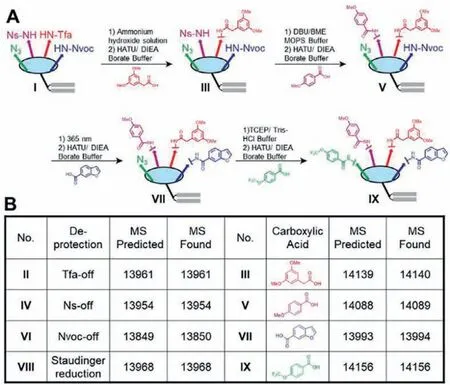
Fig.2.Sequential display of chemical combinations on the DNA–scaffold conjugate.(A) Progress of the four sequential amine-deprotection and acylation rounds.(B) UPLC–MS determination of the sequential amine-deprotection and acylation products.
Furthermore, we aimed to construct a second-generation DNAencoded multiple display with>109(~200 × 200 × 300 × 300)chemical combinations to increase binders’ affinity and isolate novel binders against different targets.For phage display selections, the multivalent epitope display on the antibody could not only increase library combination diversity but also increase candidate binding affinity exponentially [40,41].By analogy with phage display, such a second-generation DNA-encoded multiple display could promisingly facilitate binders’affinity maturation, for example, isolation of nanomolar affinity binders to PPI targets such as TNF-α, from the larger chemical combinations.Prior to synthesizing a highly cost large encoded library (>109), we decided to validate the synthetic route and selection process with a much smaller mock library with four-dimensional combinations of 24(2 × 2 × 2 × 3) using the same design.
In this demo study, we initially performed the mock library synthesis using the “split-and-pool” strategy which could be applied for a larger second-generation encoded multiple display construction [11,42].Generally, four rounds of DNA barcode encoding,deprotection, and acylation were employed (Fig.3A).In the first step, the DNA–scaffold conjugate I was separately ligated with the code 1 sequences, then the Tfa group was removed, and the corresponding carboxylic acid BBs were conjugatedviaamide bond formation.The products were then pooled and split again for the next three rounds similarly (Figs.S15–S18 and Tables S1–S4 in Supporting information).After four rounds, the mock library was successfully constructed with high efficiency and good quality, monitored by the polyacrylamide gel electrophoresis (PAGE) (Fig.3B) and MS data (Figs.S19–S22 in Supporting information).
We later conducted a proof-of-concept target-based affinity selection of the mock library.Biotin, a positive hit, was installed as a 4th-dimensional BB encoded by code 4c and pooled into the final mock library with a 1% ratio (a:b:c = 49.5%:49.5%:1%) [43].Then we incubated the mock library with immobilized streptavidin beads and washed away the non-binders.The enriched binders with DNA barcodes were amplified with polymerase chain reaction(PCR) and subjected to Sanger sequencing (Fig.3C).The sequencing result of the library before selection presented a mixed peak majorly featuring codes a and b.In contrast, the sequencing result of code 4 region after selection showed significant enrichment of code c, indicating the biotin-containing positive hits were successfully fished out from the pool, albeit with only 1% initial portion(Fig.3D).
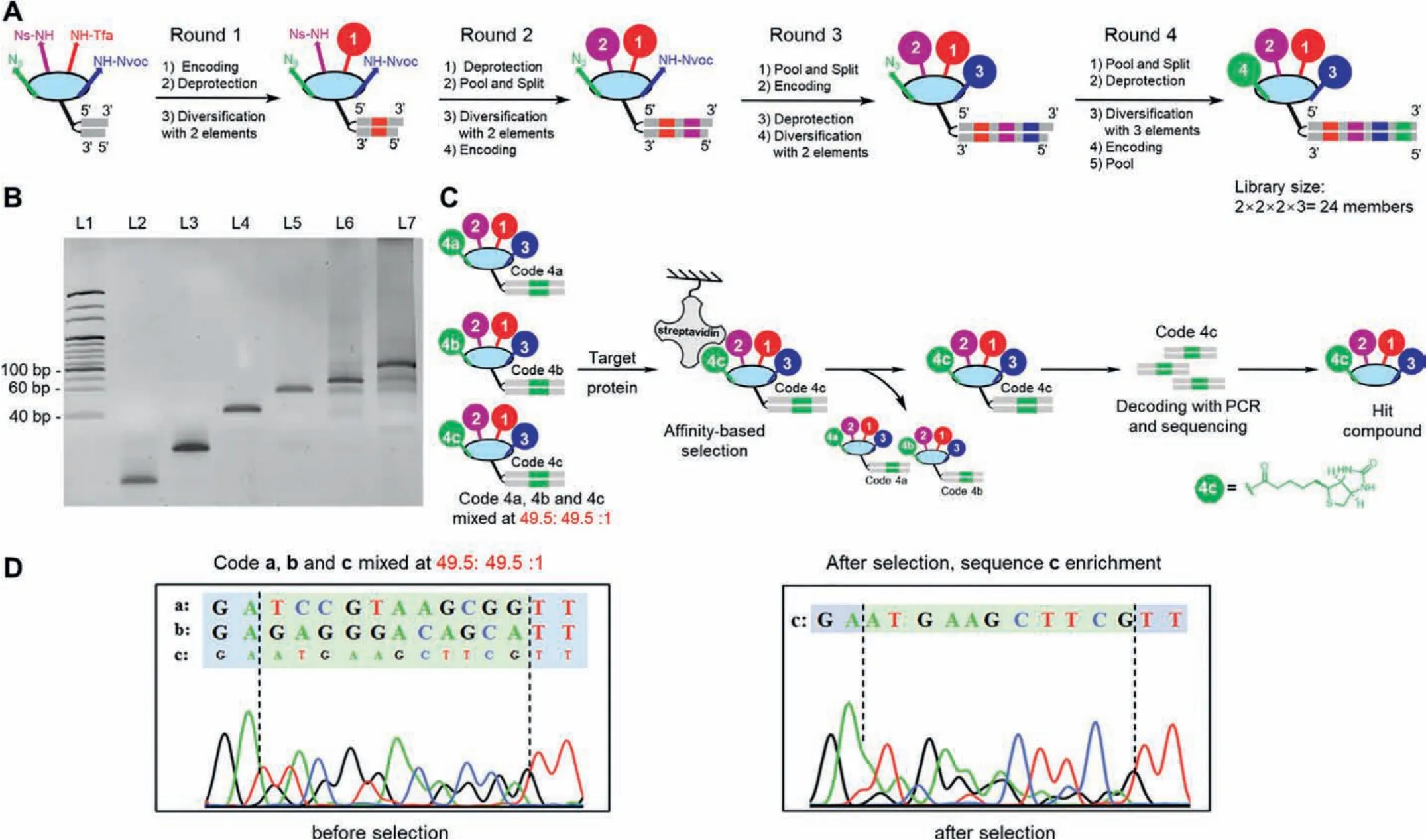
Fig.3.Synthesis and affinity selection of the mock library.(A) Workflow of mock library synthesis by the “split-and-pool” strategy employing four rounds of encoding,amine-deprotection, and acylation.(B) PAGE analysis of the pooled mock library in each round.L1, marker; L2, HP–P; L3, HP–P–scaffold conjugate; L4, 1st pool; L5, 2nd pool;L6, 3rd pool; L7, 4th pool.(C) Affinity selection workflow of the mock library against immobilized streptavidin beads.(D) Sanger sequencing of code 4 region before and after selection.
In conclusion, we developed a facile, orthogonal and compatible approach for constructing the second-generation DNA-encoded multiple display on a constant macrocyclic scaffold.We sought the novel orthogonal and DNA-compatible Ns-protecting group,and proved the feasibility to integrate four rounds of aminedeprotection (Tfa, Ns, Nvoc and azide) and acylation reactions sequentially on the DNA–macrocyclic scaffold conjugate.Then we constructed a mock encoded multiple display library with high efficiency, and selected out positive hits from the library with significant enrichment.These altogether provided an avenue to produce the second-generation DNA-encoded multiple display on a constant macrocyclic scaffold with large chemical combinations(>109).
Macrocycles have grown into a vital category of bioactive compounds privileged to target PPI targets with flat and extended interfaces [24,25].Our second-generation DNA-encoded multiple display permitted the installation of diversified carboxylic acid combinations onto the constant macrocyclic scaffold.This secondgeneration library provides a larger interaction surface and the capability of multivalent interactions.Due to the ligand-binding multivalency, focused libraries could be straightforwardly produced for affinity maturation of weak micromolar affinity binders to target proteins such as TNF-α.Moreover, the topological distribution of four display sites provides the opportunity to accommodate molecular glues in the library:Multivalent macrocycles may recruit or stabilize the PPI between two targets or serve as bifunctional degraders [44–46].Further studies are underway to explore the potential of DNA-encoded multiple display on the macrocyclic scaffold in our group, in pursuit of discovering hit compounds and developing molecular probes to regulate protein–protein interactions.
Declaration of competing interest
The authors declare no competing financial interests.
Acknowledgments
This work was supported by the grants from the National Natural Science Foundation of China (No.21907011), the Fundamental Research Funds for the Central Universities (Nos.2020CQJQYZ002, 2021CDJYGRH-002, China), the Natural Science Foundation of Chongqing (No.cstc2020jcyj-jqX0009, China), and the China Postdoctoral Science Foundation (No.2020M683251).
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cclet.2021.09.041.
 Chinese Chemical Letters2022年5期
Chinese Chemical Letters2022年5期
- Chinese Chemical Letters的其它文章
- Recent advances in enhancing reactive oxygen species based chemodynamic therapy
- An integrative review on the applications of 3D printing in the field of in vitro diagnostics
- Recent developments of droplets-based microfluidics for bacterial analysis
- Dynamics and biological relevance of epigenetic N6-methyladenine DNA modification in eukaryotic cells
- Recent progress in advanced core-shell metal-based catalysts for electrochemical carbon dioxide reduction
- Recent advances in carbon-based materials for electrochemical CO2 reduction reaction
