LAMP for the rapid diagnosis of iridovirus in aquaculture
Yepin Yu, Zituo Yng, Le Wng, Fei Sun, My Lee, Ynfei Wen, Qiwei Qin,***,Gen Hu Yue,c,*
aTemasek Life Sciences Laboratory, National University of Singapore, 1 Research Link, 117604, Singapore
bJoint Laboratory of Guangdong Province and Hong Kong Region on Marine Bioresource Conservation and Exploitation, College of Marine Sciences, South China Agricultural University, Guangzhou, 510642, China
cDepartment of Biological Sciences, National University of Singapore, 14 Science Drive, 117543, Singapore
Keywords:
Fish
Disease
Iridovirus
Diagnosis
PCR
Seabass
A B S T R A C T
Iridoviruses are DNA virus and have caused huge economic losses in the aquaculture industry. The aim of this study was to establish a colorimetric loop-mediated isothermal amplification (LAMP) protocol for the on-site detection of Singapore grouper iridovirus (SGIV). The SGIV-VP61 gene was chosen as the target gene to develop a colorimetric LAMP assay. The optimized condition of the colorimetric LAMP assay was incubation at 63 ℃ for 1 h. Samples infected with SGIV could be detected with the color change from yellow into pink. The sensitivity of the developed assay is 5.66 copies/μL of the viral DNA template. This sensitivity was about 1000 times higher than that of conventional PCR while it was slightly lower than the one-step semi-nested PCR assay.A total of 60 DNA samples extracted from the fin tissue of the SGIV-infected Asian seabass were examined for SGIV by colorimetric LAMP, semi-nested PCR and conventional PCR. The results of the colorimetric LAMP assay showed 94.87 % agreement with the semi-nested PCR. In addition, the DNA extraction method using NaOH showed a better performance in the colorimetric LAMP assay. Taken together, the colorimetric LAMP established was a sensitive, rapid and specific method for the detection of SGIV. SGIV was not detected in samples randomly taken from a genetically improved line of the Asian seabass. However, some seabass obtained from the local markets were found to contain SGIV. Thus, the LAMP assay has the potential application in the diagnosis of iridovirus diseases in the aquaculture industry.
1.Introduction
Iridoviruses are nucleocytoplasmic viruses with a large doublestranded DNA genome (Williams et al., 2005), and have caused serious systemic diseases in fish, amphibians and reptiles (Hyatt et al.,2000; Wang et al., 2007). Thus, iridoviruses not only cause major economic losses in the aquaculture industry, but also pose a huge threat to global ecological diversity (Chinchar et al., 2011; Rothermel et al.,2013). Singapore grouper iridovirus (SGIV), which is isolated from diseased grouper (Epinephelus tauvina), belongs to the genusRanavirusand familyIridoviridae(Qin et al., 2002). This virus results in a mortality rate of 50%–90% at the fingerling stage of several aquatic fish species(Qin et al., 2003; Wang et al., 2017). Developing the rapid and early detection of SGIV is essential to prevent SGIV spread and economic losses in aquaculture industry.
To date, many approaches have been developed to monitor SGIV infection and used to diagnose it in laboratories. These approaches include electron microscopy (Qin et al., 2001), the immuno fluorescence antibody test (Qin et al., 2002), some nucleic acid tests, (e.g. the polymerase chain reaction (PCR) (Qin et al., 2003), loop-mediated isothermal amplification (LAMP) (Mao et al., 2008)) and aptamer-based enzyme-linked apta-sorbent assay (ELASA) (Li et al.,2016). Although these methods work well in laboratories, they have many disadvantages and restrictions, including the requirements for expensive and sophisticated instruments, which limit their applications in the aquaculture industry, especially when required on-site. Thus, a simple and fast detection method, which can be used on-site, is urgently needed.
LAMP is a one-step DNA synthesis that is performed withBstDNA polymerase and 4–6 primers recognizing 6–8 distinct regions of target DNA (Caipang et al., 2012). It has been developed and used as a rapid diagnostic method in many disease diagnosis (Gill & Ghaemi, 2008;Tomlinson, 2013). As the assay can be completed within 1 h, it shortens the time for the pathogen detection. In the aquaculture industry, the LAMP assay has been applied in detecting IHHNV (infectious hypodermal and hematopoietic necrosis virus) (Sun et al., 2006), WSSV (white spot syndrome virus) (Tomoya et al., 2004), RSIV (red sea bream iridovirus) (Christopher et al., 2004), KHV (koi herpes virus) (Soliman &El-Matbouli, 2010) and LMBV (largemouth bass ranavirus) (Zhu et al.,2020). The visual detection of LAMP using pH-sensitive dyes, including neutral red and phenol red needs no specialized or expensive instrumentation (Tanner et al., 2015). Therefore, with the advantage of being easy to perform, rapid and highly sensitive in the detection of viruses(Tanner et al., 2015), the colorimetric LAMP method is ideal for on the spot diagnosis of aquatic pathogens.
The Asian seabass,Lates calcarifer, is one of the most popular food fish in South-East Asia and Australia (Jerry, 2013; Yue et al., 2001).Although a breeding program for genetic improvement for growth,disease resistance and meat quality has been started since 2004 and has improved these traits (Shen et al., 2020; Yue et al., 2017). The SGIV still causes diseases and great economic losses in the Asian seabass aquaculture industry in Southeast Asia (Gibson-Kueh et al., 2011; Wang et al., 2017; Yu et al., 2021). The rapid and early diagnosis of SGIV in cultured Asian seabass could help prevent the spread of the virus and reduce the economic losses.
The purposes of this study were (1) to develop a sensitive and rapid LAMP assay for the detection of SGIV on the spot for aquaculture, (2) to check whether there are any SGIV-infected individuals in a genetically improved Asian seabass broodstock, and (3) to find a better choice among the various DNA extraction methods, which can be applied to the established colorimetric LAMP assay. Our results show that developed LAMP method is sensitive, specific, cost-effective and is suitable for onthe-spot detection of SGIV in the aquaculture industry. In a genetically improved broodstock, LAMP of random samples detected no SGIV infection, while in samples obtained from local fish markets, SGIV was detected (5/16). Therefore, it is meaningful to inspect the aquatic pathogen infection. And the developed colorimetric LAMP is a useful and reliable method to support the quality assurance of fishery products.
2.Materials and methods
2.1.Ethics statement
The handling of Asian seabass used in this study strictly followed the guidelines of the IACUC of our Institute. The IACUC approval number for this study is TLL (F)-17-001.
2.2.Cells and virus
An SB epithelial-like cell line from Asian seabass (Lates calcarifer)fries was established and cultured in L15 medium (Life Technologies,Carlsbad, USA), which contains 10 % heat-inactivated FBS (fetal bovine serum) (Life Technologies, Carlsbad, USA) at 28 ℃ as previously described (Liu et al., 2016; Yu et al., 2021). SGIV (Singapore grouper iridovirus, strain A3/12/98 PPD) was propagated in grouper embryonic cell line (GP) (Qin et al., 2001) and was stored at −80 ℃. The same conditions of SB cell culturing were employed in GP cell generation.
2.3.PCR amplification of SGIV-VP61 gene and reconstruction of the pGEM®-T Vector
Primers SGIV-VP61-PF/PR (Table 1) were used to amplify the SGIVVP61 gene and clone into the pGEM®-T Vector (Promega, SG,Singapore) to construct pT-VP61 vector. The PCR for the amplification of SGIV-VP61 open reading frame (ORF) was performed in a volume of 25 μL reaction mixture consisting of 1 μL (2.25 ng) of template (SGIV-infected SB cell lysate), 1 ×PCR buffer (containing 1.5 mM MgCl2), 0.5 μL of dNTPs (10 mM), 0.2 μL of each primer (10 μM), and 1 unit ofTaqDNA Polymerase (Finnzymes, Espoo, Finland). The ORF of SGIV-VP61 was cloned into pGEM®-T Vector (Promega, SG, Singapore), and the reconstructed vector was used as the DNA templates in the following experiments.
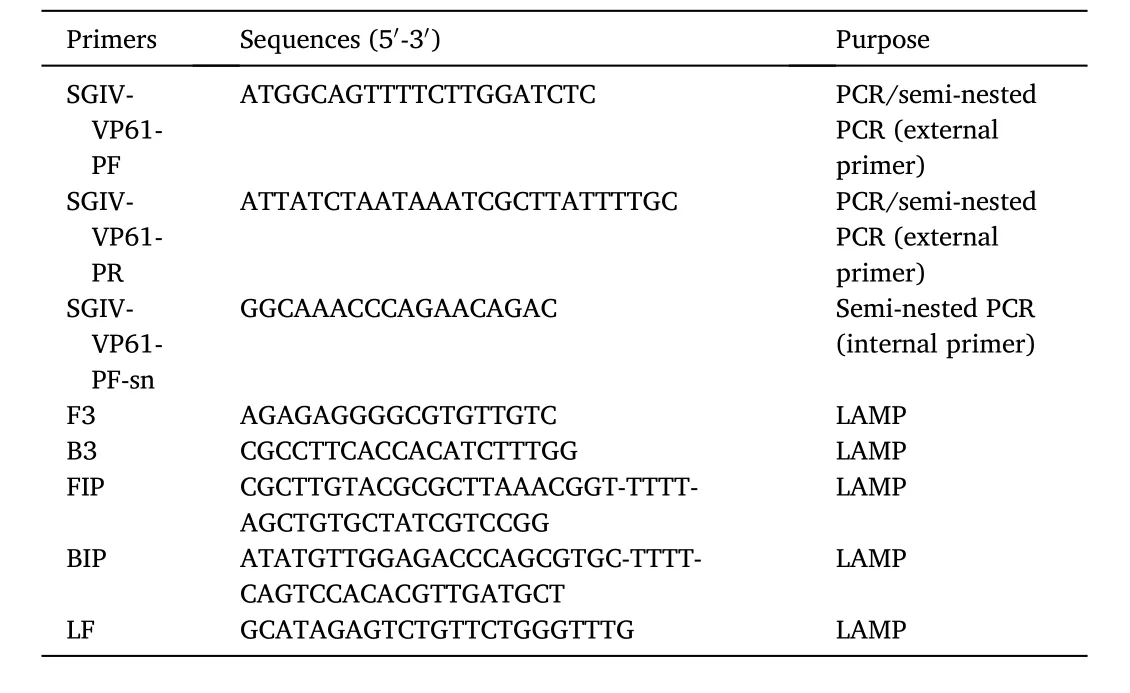
Table 1The primers used for loop-mediated isothermal amplification, semi-nested PCR and PCR amplification. The amplicon size: conventional PCR: 612 bp; seminested PCR: 612 bp and 474 bp.
2.4.LAMP primer design and LAMP assay conditions
LAMP primers (Table 1) were designed based on the ORF of SGIVVP61 gene using the Primer Explorer Version 5 (https://primerexplo rer.jp/e/). The primer sets of the LAMP reaction include two outer primers (F3/B3), two inner primers (Forward Inner Primers, FIP and Backward Inner Primers, BIP), and one loop primer (Loop Forward, LF).
The colorimetric LAMP reaction was carried out in a 25 μL system containing 2.5 μL 10 × Isothermal Amplification Buffer (NEB, MA, USA),1.5 μL MgSO4(100 mM), 3.5 μL dNTPs (10 mM), 1 μLBst2.0 DNA polymerase (8000 Unit/mL, NEB, MA, USA), 1.6 μM FIP and BIP primers, 0.2 μM F3 and B3 primers, 0.4 μM LF primer, 1 μL template (pTVP61 plasmid, SGIV-infected SB cell lysate or the DNA extracted from the SB cell line or the fin tissue), and topped up using water with neutral red (N-red, with a final concentration of 100 μM) to 25 μL. The negative control used 1 μL of nuclease-free water or mock-infected SB cell lysate as the template. The LAMP reaction was performed at different temperatures (60, 63 and 65 ℃) for different elongation times (30, 45, and 60 min). The LAMP products were also detected through electrophoresis on 2.0 % agarose gels.
2.5.Observation of LAMP products with naked eyes
The results of the colorimetric LAMP reaction were directly observed with the naked eye by adding N-red with a final concentration of 100 μM before the LAMP reaction. After the incubation at 63 ℃ for 1 h, the color change of the solution was observed. The color of the solution changed from light yellow to pink in the presence of LAMP amplicons, while it remained light yellow with no amplification.
2.6.Conventional PCR for SGIV
The reaction mixtures of the conventional PCR contained 1 ×PCR buffer (1.5 mM MgCl2contained), dNTPs (50 μM each), 1 unit of DNA polymerase (Finnzymes, Espoo, Finland), 1 μL template (pT-VP61 plasmid or the DNA extracted from fin tissue of Asian seabass), the forward and reverse primers (SGIV-VP61-PF/PR listed in Table 1, 200 nM each), and nuclease-free water in a final volume of 25 μL. The conventional PCR was conducted on the thermal cycler (Bio-rad, SG,Singapore) with a program of a denaturation step at 94 ℃ for 5 min,followed by 34 cycles of 94 ℃ for 30 s, 55 ℃ for 30 s, 72 ℃ for 60 s with an extension at 72 ℃ for 5 min. PCR products were checked through electrophoresis on 2.0 % agarose gels.
2.7.Semi-nested PCR to detect SGIV
Primers designed for semi-nested PCR are listed in Table 1. The onestep semi-nested PCR mixtures contained 1 ×PCR buffer (1.5 mM MgCl2contained), dNTPs (50 μM each), 1 unit of DNA polymerase (Finnzymes,Espoo, Finland), 1 μL DNA template (pT-VP61 plasmid or the DNA extracted from fin tissue of Asian seabass), two external primers (40 nM SGIV-VP61-PF and 200 nM SGIV-VP61-PR), one internal primer (160 nM SGIV-VP61-PF-sn), and nuclease-free water in a final volume of 25 μL. The semi-nested PCR program consisted of a denaturation step at 94 ℃ for 5 min, followed by 9 cycles of 94 ℃ for 30 s, 55 ℃ for 30 s,72 ℃ for 60 s with an extension at 72 ℃ for 5 min. After denaturation at 94 ℃ for another 5 min, the reaction continued with 25 cycles of 94 ℃ for 30 s, 58 ℃ for 30 s, 72 ℃ for 30 s with another extension at 72 ℃ for 5 min. The PCR was performed on the thermal cycler (Bio-rad, SG,Singapore). PCR products were also examined through electrophoresis on 2.0 % agarose gels.
2.8.Analytical sensitivity of the different diagnostic methods
The analytical sensitivity comparison of the different diagnostic methods was evaluated using 10-fold serial dilutions of pT-VP61 plasmid (at the original concentration of 5.66 × 1010copies/μL). After that, 1 μL of each dilution was used as the templates for the conventional PCR, semi-nested PCR and colorimetric LAMP reaction. The colorimetric LAMP reaction was performed at 63 ℃ for 60 min and compared with the conventional PCR and semi-nested PCR assay.
2.9.Application of LAMP to detect virus infection in SB cells and in fish tissues
SB cells were seeded into the 24-well plate for 18 h before infected with SGIV (at a multiplicity of infection (MOI) of 0.2). The infected cells were collected at the indicated time points (0, 1, 3, 6, 12, 24, 36 and 48 h post infection (p.i.)) followed by DNA extraction from TRIzol reagent(Life Technologies, USA) preparations. In brief, 200 μL isopropanol was added for every 1 mL of TRIzol used to harvest the cells/tissue before centrifuging at 12,000 ×gfor 10 min. The upper phase was removed,while the interphase and organic phase were mixed with 500 μL Back Extraction Buffer (BEB, containing 4 M Guanidine Thiocyanate, 50 mM Sodium Citrate NaCl and 1 M Tris base) for every 1 mL TRIzol used initially. The tubes were centrifuged at 12,000 ×gfor 30 min at room temperature. Then, the upper phase was transferred to a clean tube. 400 μL of ice-cold isopropanol was added to precipitate the DNA. After centrifuging the tubes at 12,000 ×gfor 15 min at 4 ℃, the supernatant was removed and 500 μL of 70 % ethanol was added to wash the DNA pellet. The tubes were centrifuged at 12,000 ×gfor 15 min at 4 ℃ and the washing step was repeated. The DNA pellet was dissolved in 50 μL of nuclease-free water, and the DNA were used as the templates in the virus detection.
Fin clips (~ 3 mm ×3 mm in size) from 16 fish purchased from the market and the fin clips of 16 fish randomly picked from a genetically improved line of Asian seabass from the Marine Aquaculture Center(MAC), Singapore, were collected and stored in 75 % ethanol. The DNA from fin clips were extracted using a method developed previously(Wang et al., 2017). DNA quality and quantity were checked with Nanodrop (Thermo, SG, Singapore). DNA was used for the detection of SGIV using the developed LAMP assay as described above.
2.10.DNA extraction methods
SB cells were seeded into the 24-well plates for 18 h following by SGIV infection at different concentrations (at MOI of 0.5, 1, 1.5 and 2).After 24 h p.i., cells were harvested and centrifuged to discard the supernatant. DNA was extracted from the SGIV-infected cells using the following methods: (1) the TE buffer method, samples homogenized with 50 μL of TE buffer and boiled for 10 min following by centrifuged(500 ×g, 5 min) as described in a previous study (Mao et al., 2008); (2)the Triton X-100 method, samples incubated in 50 μL of 1 % Triton X-100 were boiled for 10 min following by centrifuged at 500 ×gfor 5 min (Sun et al., 2014); (3) the NaOH method, 50 μL of 0.02 N NaOH solution was added into the samples and incubated at room temperature or in the boiled water for 10 min before centrifuged at 500 ×gfor 5 min(Sun et al., 2014). The DNA extraction by TRIzol method (used as the control method) was described in the Section 2.8. The extracted DNA templates were 10-time diluted and used to conduct the colorimetric LAMP assay. The results were summarized in the Table 3.
3.Results
3.1.Optimized LAMP reaction to detect SGIV
The primer sets of LAMP assay for SGIV-VP61 amplification were listed in Table 1, and the positions of the primers were shown in Fig. 1B.The LAMP reaction was performed using SGIV-infected cell lysis as template to determine the optimal temperature and time of reaction.The amplicon was formed at 60, 63 and 65 ℃ and the clear products could be detected at all three temperatures. The color changes could also be observed with the naked eye. The color change from light yellow to pink suggested a positive detection (Fig. 2A). Taking the on-site conditions into consideration, the temperatures of the water bath or the heat block might not be very precise. Thus, the optimal temperature of 63 ℃ was used in LAMP assays.
On the other hand, the LAMP products could be detected at 45 and 60 min at 63 ℃ with both the N-red staining and electrophoresis(Fig. 2B). The LAMP product digested by enzymeXmaIwas also analyzed with electrophoresis and shown in Fig. 2C. The results indicate that the LAMP amplification of SGIV-VP61 was specific.
3.2.Sensitivity of LAMP assay in SGIV detection
The detection limit of the colorimetric LAMP reaction was determined under optimal conditions using the primer sets for SGIV-VP61 detection at 63 ℃ for 60 min. The reaction was tested using 1 μL 10-fold serial dilutions of plasmid (pT-VP61) DNA (at the original concentration of 5.66 × 1010copies/μL) and compared with the conventional PCR and semi-nested PCR assay. The semi-nested PCR showed the highest sensitivity among these methods (Fig. 3D). Compared with the detection limit of the conventional PCR (Fig. 3C), the colorimetric LAMP was about 1000 times more sensitive with the positive detection at 5.66 copies/μL (~2.25 ng) (Fig. 3A). And the LAMP products could be observed in 2.0 % agarose gel at 5.66 × 10−1copies/μL group (Fig. 3B).
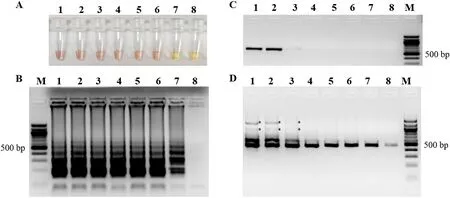
Fig. 3.Comparative sensitivities of colorimetric LAMP, conventional PCR and semi-nested PCR for the detection of plasmid pT-VP61. (A) Colorimetric LAMP products. (B) Shows a ladder-like pattern of LAMP products. (C) Conventional PCR amplification products. (D) Semi-nested PCR amplification products. M, 100 bp DNA marker; 1–8, reaction carried out using 10-fold serial dilutions of plasmid pT-VP61 DNA (at an original concentration of 5.66 × 1010 copies/μL): 5.66 × 105,5.66 × 104, 5.66 × 103, 5.66 × 102, 5.66 × 101, 5.66 × 100, 5.66 × 10−1, and 5.66 × 10−2 copies/μL, respectively. All the products were electrophoresed on a 2.0 % agarose gels and stained with ethidium bromide. *, unspecific amplicons.
3.3.Applications of the colorimetric LAMP assay for the detection of SGIV infection in cells and in samples from Asian seabass
The colorimetric LAMP assay was carried out to detect SGIV using the DNA extracted from SGIV-infected SB cells. As shown in Fig. 4, SGIV was detected as early as 6 h p.i. in SB cells with the color change observed. In addition, colorimetric LAMP could also be used in detection of SGIV from the virus-infected cell lysate (data no shown). The detection of SGIV has also been performed in DNA templates extracted from the fin tissue of virus-infected Asian seabass. The result of the detection is shown in Fig. S1 and summarized in Table 2. Sixty- five percent (39/60) of the DNA samples were positively detected by the semi-nested PCR and 61.67 % (37/60) by colorimetric LAMP (Table 2). And the results of the colorimetric LAMP assay shared 94.87 % agreement with the seminested PCR. Thus, there was a good concordance between colorimetric LAMP and semi-nested PCR in SGIV detection both in cells and the samples from fish.
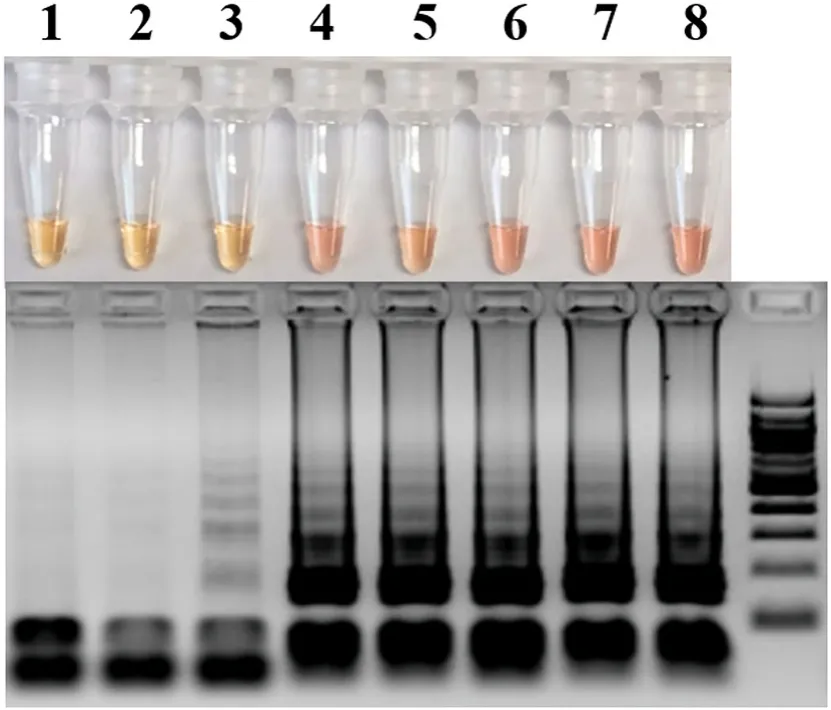
Fig. 4.Detection of SGIV in infected Asian seabass (SB) cells by colorimetric LAMP assay and electrophoretic analysis on a 2.0 % agarose gels and stained with ethidium bromide. M, 100 bp DNA marker; 1–8, detection of SGIV in infected SB cells at 0, 1, 3, 6, 12, 24, 36 and 48 h post infection, respectively.
The developed LAMP assay was employed in the detection of SGIV in a genetically improved line of the Asian seabass in MAC, Singapore and fin clips from the market. As shown in Fig. S2, all the collected samples from MAC, Singapore are free of SGIV infection (Fig. S2 A), while in 5 samples from market, SGIV was detected (Fig. S2 B).
Table 2Comparative analysis of samples for SGIV positivity by colorimetric LAMP assay,semi-nested PCR and conventional PCR.
The newly established colorimetric LAMP assay was also performed in the SGIV detection using the DNA templates extracted with different methods. The TRIzol method was used as the control method. As shown in Table 3, the DNA templates extracted from the 0.02 N NaOH (boiling)showed better quality in the colorimetric LAMP detection, and the detection result was 100 % consistent with the control method. The Triton X-100 method shared 89 % agreement with the control method,while the 0.02 N NaOH (room temperature incubation) method and the TE buffer method shared only 72 % and 61 % agreement, respectively(Table 3). Thus, among the virous DNA extraction methods, the 0.02 N NaOH (boiling) method was the best choice in the on-site SGIV detection with colorimetric LAMP assay.
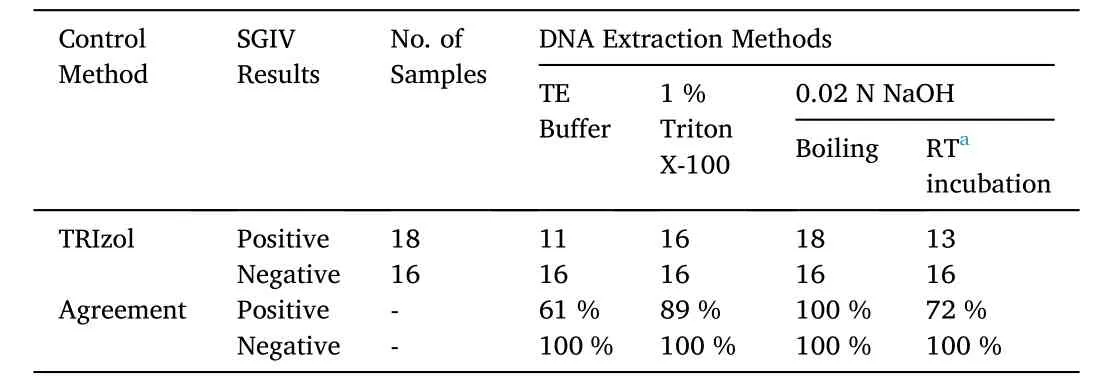
Table 3A comparison of colorimetric LAMP assay results using templates extracted by various methods for the detection of SGIV in virus-infected SB cells.
4.Discussion
Early and rapid diagnosis of the exact pathogen is a key process to reduce losses (Mzula et al., 2021). In the present study, a rapid, simple,visible and sensitive colorimetric LAMP protocol was developed for on-the-spot detection of SGIV infection for the aquaculture industry.
The whole procedure of the developed LAMP assay could be performed with a water bath or a heat block at a temperature of 63 ℃ within 1 h. In practice, the time required for diagnosis is a crucial factor(Kibenge & Godoy, 2016). Reducing the time and simplifying the processes are critical factors in the disease diagnosis (Kibenge & Godoy,2016). Our LAMP assay took 60 min, while the conventional PCR and nested PCR method took at least 2–3 h. Therefore, our LAMP assay is a more efficient method in detecting SGIV.
Post analysis of LAMP amplicons always requires nucleic acid electrophoresis staining with ethidium bromide, or with DNA-intercalating dyes added into the LAMP product (Cai et al., 2012; Hill et al., 2008;Parida et al., 2005). A combination method with LAMP and lateral flow dipstick (LFD) has been reported to advance the specificity and simplify the visual detection (Andrade & Lightner, 2009; Zhai et al., 2019).However, the LFD is expensive. Antibodies, modified probes, nano-gold particlesetc. are necessary for the construction of the LFD (Zhai et al.,2019; Zhang et al., 2019). Therefore, in this study, a simple and low-cost method was developed to detect the products of LAMP. By adding the N-red into the LAMP reaction system, the positive reactions would show a color change from light yellow to pink, which could be seen with the naked eye. With regards to the ease of detection visually, our method is comparable to previously reported methods for LAMP detection (Tanner et al., 2015).
The colorimetric LAMP developed in this study for the detection of SGIV is much more sensitive compared to the conventional PCR assay.The SGIV DNA template could be detected as low as 5.66 copies using our colorimetric LAMP method. This level of sensitivity is similar to that in HBV (hepatitis B virus) (Notomi et al., 2000) and SGIV (Mao et al.,2008) LAMP detection. However, it is to note that in the present study,the sensitivity of the LAMP assay is slightly lower than the semi-nested PCR assay for DNA extracted from a cell line. Similar results have also been reported in the detection ofStreptococcus equisubsp. equi(Hobo et al., 2012). On the other hand, our colorimetric LAMP was highly sensitive and accurate in the detection of the DNA extracted from the fin tissue of SGIV-infected Asian seabass. The results of the colorimetric LAMP assay performed in 60 samples from Asian seabass fin tissue showed 94.87 % agreement with the semi-nested PCR, while the conventional PCR only shared 84.62 % agreement. Many reports reveal that the LAMP assay would be equally or even more sensitive as compared to the semi-nested PCR (Hobo et al., 2012; Ramos et al., 2017). However,in the present study, although the LAMP assay showed a lower sensitivity towards the semi-nested PCR, there is a good consistency between these two methods. Therefore, both the developed colorimetric LAMP and semi-nested PCR provide an accurate diagnosis of SGIV, while the colorimetric LAMP has the advantage of being easy to perform, rapid,and highly sensitive for the aquaculture industry. Moreover, the colorimetric LAMP assay needs no precision instruments or complex procedures, so it is convenient and reliable for the on-the-spot diagnosis.
The developed colorimetric LAMP assay was employed in SGIV detection in a broodstock of a genetically improved line of Asian seabass to ensure the health of the breeding stock. No SGIV was detected in samples from the improved broodstock while 5 out of the 16 samples from the market contained SGIV, suggesting the broodstock of the improved line in MAC Singapore is free of SGIV.
As suggested in a previous study on DNA extraction quality evaluation for hepatitis B virus (HBV) detection, simple boiling showed a slightly lower efficiency than NaOH lysis (Lu et al., 2006). However, in another study, the DNA extracted using boiling method was the best method among the three methods (boiling, boiling in 1 % Triton X-100,and treating with 0.02 M NaOH) (Sun et al., 2014). The present study suggested that among the virous DNA extraction methods, the one carried out with 0.02 N NaOH (boiling) should be the best choice for the on-site condition. It was a simple method and the extracted DNA templates were of great quality for colorimetric LAMP detection.
In summary, a colorimetric LAMP protocol was developed together with a simple DNA extraction method to detect SGIV on the spot. The colorimetric LAMP consists of a single step (i.e. application at 63 ℃ for 60 min with 100 μM of N-red added) using the DNA extracted with samples boiling in 0.02 N NaOH solution as templates. After ampli fication, the results can be seen with the naked eye. The sensitivity (5.66 copies/μL) of the colorimetric LAMP was 1000 times higher than the conventional PCR, but lower than the semi-nested PCR. And the colorimetric LAMP shared 94.87 % agreement with the semi-nested PCR in SGIV detectionin vivo. Therefore, the developed colorimetric LAMP is simple, sensitive and specific. It can be easily applied both in the laboratory and out in the field for specific detection of SGIV infection.
Declaration of competing interest
The authors declared that they have declared no conflicts of interest.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (41806161) and the internal funds of Temasek Life Sciences Lab, Singapore.
Appendix A.Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.aaf.2021.08.002.
Credit authors contribution statement
QWQ, GHY and YPY: Conceptualization, YYP, ZTY, FS, LW, ML and YFW: Methodology, QWQ and GHY: Software, YYP, ZTY, FS, LW, ML and YFW: Data curation, YPY, GHY and QWQ: Writing YYP: Original draft preparation. YYP, ZTY, FS, LW, ML and YFW: Visualization, YYP,ZTY, FS, LW, ML and YFW: investigation, GHY and QWQ: Supervision.:YYP, QWQ and GHY: Writing- Reviewing and Editing.
 Aquaculture and Fisheries2022年2期
Aquaculture and Fisheries2022年2期
- Aquaculture and Fisheries的其它文章
- An overview of disruptive technologies for aquaculture
- CRISPR-Cas9 sgRNA design and outcome assessment: Bioinformatics tools and aquaculture applications
- The integrated analyses of metabolomics and transcriptomics in gill of GIFT tilapia in response to long term salinity challenge
- Phenotyping and phenomics in aquaculture breeding
- VNN disease and status of breeding for resistance to NNV in aquaculture
- Insects as a feed ingredient for fish culture: Status and trends
