Detection of Biomarker Protein PDGF-BB in Esophageal Squamous Cell Carcinoma Using a DNA Biosensor Based on Enzyme Cycle Amplification
LIU Xiao-fan,QI Chen,WANG Wei-cai,LI Xue-mei
(Shandong Province Key Laboratory of Detection Technology for Tumor Makers,School of Chemistry and Chemical Engineering,Linyi University,Linyi 276005,China)
Abstract:The condition of patients with esophageal squamous cell carcinoma(ESCC)is difficult to be found and effectively treated in the early stage of the disease due to the few types and low levels of tumor markers in the body.Therefore,it is the most urgent problem for researchers to find a suitable marker for rapid and efficient detection.Platelet derived growth factor-BB(PDGF-BB)is extremely important for the early diagnosis of malignancy.A DNA biosensor based on enzyme cycle amplification fluorescence spectroscopy was designed for PDGF-BB detection in this work,using a shearing enzyme that can cut the specified site to achieve the signal amplification,reducing the limit of detection from 1 fmol/L to 1 amol/L,greatly improving the sensitivity of detection.This method is of great value in biochemical and medical applications for its simple,rapid and efficient diagnosis and detection on esophageal squamous cell carcinoma.
Key words:tumor markers;DNA biosensors;enzyme cycle amplification;fluorescence detection
Esophageal squamous cell carcinoma(ESCC)is a global malignant tumor,especially common in eastern countries,accounting for about 90%of all esophageal cancers in the world.Due to its strong invasion,the ability of metastasis and recurrence,and lack of effective biomarkers for early detection,it has high mortality and poor prognosis[1-3].Nowadays,various tumor markers have been gradually discovered by scientists and used as diagnostic tools for cancer in biomedical research[4-5].Protein is considered one of the timely,effective,and accurate biomarkers for cancer detection[6-8].
Platelet derived growth factor(PDGF)is a basic protein that regulates the growth and proliferation of human cells[9-10].PDGF and its receptor have constructed a valuable diagnostic system for disease through specific binding[11].Among them,PDGF-BB is directly related to a variety of biological behaviors such as proliferation and metastasis of tumor cells and is not expressed in normal cells or its expression level is very low.Therefore,PDGF-BB as a target protein marker for the detection of esophageal squamous cell carcinoma plays a key role in the diagnosis and treatment of ESCC[12-15].
At present,researchers have developed a variety of detection methods for PDGF-BB,including affinity chromatography,enzyme-linked immunosorbent assay(ELISA),electrochemical analysis,etc.[11,13,16-19].These detection methods limited their application in practical clinical practice due to their price,complex operation,and long analysis time.Therefore,finding a new detection method that is efficient,simple and inexpensive is of great significance to the prevention,diagnosis,treatment,and monitoring of ESCC.It is also urgent to improve the detection sensitivity on the basis of original detection method[2,20-21].DNA,as the carrier of genetic information of organisms,is ubiquitous in organisms and has good biocompatibility.Moreover,due to its controllable size and shape,DNA biosensors based on base complementary pairs have become a hot topic for scholars of multiple disciplines[22-25].Several reports of DNA biosensors detecting proteins with similar concepts have been reported in the past.For example,Zhang et al.[26]designed a AuNPs colorimetric sensor for detecting PDGF-BB.Its limit of detection is 1.1 nmol/L,and when the concentration is beyond 4.0 nmol/L we can use the naked eye for simple and direct visualization detection.But this method needed a long time to synthesize and detect.Also,it is not enough sensitive.Gan et al.[13]developed a simple proximity hybridizationinduced on particle DNA walker for ultrasensitive detection of proteins,such as PDGF-BB.Its limit of detection is higher than the previous method.But it uses a large number of DNAs,so not cheaper.Compared with the two methods reported above,the established method uses less DNA and has an obvious quenching effect.In other words,this method is simple and takes less time in synthesis and detection.
Here,we designed a new DNA biosensor,as shown in Fig.1.Based on the characteristic that PDGF-BB is specifically bound with DNA aptamer(DNA1)to form a stable complex,we modified the amino on DNA1,which was a part of double-stranded DNA(DNA3)combined with carboxyl magnetic beads to form magnetic probes through amide reaction.In the presence of PDGF-BB,PDGF-BB formed a stable complex with DNA1 with magnetic beads,and a free complementary sequence of DNA1(DNA2)was obtained after magnetic separation.Hairpin DNA was then added,which was modified with the fluorescence group(FAM)at one end and the fluorescence quenching group(BHQ1)at the other end.DNA2 could turn on the hairpin DNA and then cut specific sites of the double-stranded DNA by adding a shearing enzyme(Nb.BbvCI)to restore and amplify the fluorescence signal.On the contrary,in the absence of PDGF-BB,DNA2 could not bind to hairpin DNA,Nb.BbvCI could not perform enzyme digestion,and fluorescence signal might not be recovered,so it could not be detected.This method detected PDGF-BB simply,sensitively and efficiently,and the detection limit was reduced to 1 amol/L,which had important research significance for the prevention,diagnosis,treatment,and monitoring of ESCC.
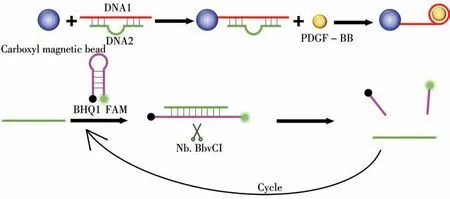
Fig.1 Schematic diagram of PDGF-BB detection by enzyme cycle amplification fluorescence spectroscopy
1 Experimental
1.1 Reagents and apparatus
The DNA sequences used in the experiment are shown in Table 1 were purchased from genscript Biotechnology(Nanjing,China).Carboxyl magnetic beads(10 mg/mL)were purchased from Shanghai Carboxylphenyls Biomedical Company(Shanghai,China).PDGF-BB was purchased from Pepro Tech Company(Cranbury,NJ,USA).Nb.BbvCI shearing enzyme was purchased from NEW ENGLAND Bio Labs(NEB,England).NHydroxysuccinimide(NHS)and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride(EDC)were all purchased from Aladdin(Shanghai,China).Tris(Hydroxymethyl)aminomethane(Tris)and chemical reagents such as magnesium chloride(MgCl2)were analytical reagents(AR).The solution and centrifuge tubes used in the experiment were treated with 0.1%DEPC ultrapure water and sterilized by high temperature and high-pressure steam.

Table 1 DNA sequence used in the experiment
F-7000 fluorescence spectrophotometer(Hitachi,Japan).Gas bath water bath constant temperature oscillator(Lichen,China).Ultrasonic cleaning instrument(Shanghai Kedao Ultrasonic Instrument Co.,LTD.),precision electronic balance(Shanghai,China).pH meter(PHS-3C)(Shanghai,China).Highspeed centrifuge(Hangzhou,China).PCR amplification instrument(Hangzhou,China).Electrophoresis apparatus and electrophoresis tank(Beijing,China).Zeta potential determination and particle size analyzer(Malvern,UK).
1.2 Activation of carboxyl magnetic beads
25μL carboxyl-modified magnetic beads(5 mg/mL)and 50μL PBS buffer solution(pH 7.4)were mixed,centrifuged at 11 000 r/min for 5 min,removed supernatant,then added another 50μL PBS buffer solution(pH 7.4),repeated 3 times.50μL EDC solution(0.2 mol/L)was added and activated for 30 min in a 37℃incubator.Next,50μL NHS solution(0.8 mol/L)was added,mixed evenly,and activated for 1 h at room temperature.Finally,the activated beads were washed with PBS buffer solution 3 times and dispersed in 50μL PBS buffer solution.
1.3 Preparation of DNA immunomagnetic probe
12.5 μL DNA1(5.0×10-8mol/L)was added to 12.5μL TM buffer solution(10 mmol/L Tris-HCl and 30 mmol/L MgCl2).The mixture was evenly mixed and the reaction lasted for 5 min at 65℃,and the temperature was slowly reduced to room temperature.After 12.5μL DNA2(5.0×10-8mol/L)and 12.5μL TM buffer were added,the mixture was homogenized and shaken at 37℃for 12 h to obtain DNA3.
The DNA magnetic probe was obtained by mixing the double stranded DNA with the activated immunomagnetic beads and shaking the DNA magnetic probe at 37℃for 30 min.The DNA magnetic probes were washed 3 times with PBS buffer solution and dispersed in 50μL PBS buffer solution.
1.4 Combination of PDGF-BB and immunomagnetic probe
After mixing 5μL PDGF-BB solution with DNA immunomagnetic probe,the reaction was performed on a constant temperature shaker at 37℃for 30 min,then centrifuged at 12 000 r/min for 3 min,and the supernatant was removed for later use.
1.5 Hairpin DNA reacts with supernatant
The 50μL hairpin DNA(5.0×10-8mol/L)was mixed with the supernatant and reacted at 37℃for 30 min.The fluorescence value was measured.
1.6 Nb.BbvCI cycle amplification reaction
2μL Nb.BbvCI shearing enzyme and 10μL Nb.BbvCI buffer were added to the system,and shaked at 37℃for 40 min.Then the fluorescence values of the system which through Nb.BbvCI shearing enzyme cyclic amplification were measured.
2 Results and discussion
2.1 Experimental feasibility study
2.1.1 Synthesis and characterization of DNA3According to theoretical support[27],the synthesized doublestranded DNA was more unstable than the single-stranded DNA,so the absolute value of the measured zeta value was large.We characterized the synthesized DNA3 by zeta potential measurement and agarose gel electrophoresis experiment.The characterization results were as follows.According to the data in Fig.2A,the potential of DNA3 was more negative,which was consistent with the theory,indicating the successful synthesis of DNA3.According to research,the larger the molecular weight of DNA had,the slower it swam in the electric pool under a given electric field[28].Therefore,in the electrophoretic diagram,the synthesized DNA3 should swim more slowly than the single-stranded DNA1 and DNA2.According to the data obtained in Fig.2B,the DNA3 swam the slowest,indicating that the synthesis of DNA3 was successful.Combining the above two characterization results,the successful synthesis of DNA3 laid a good foundation for the development of subsequent experiments.
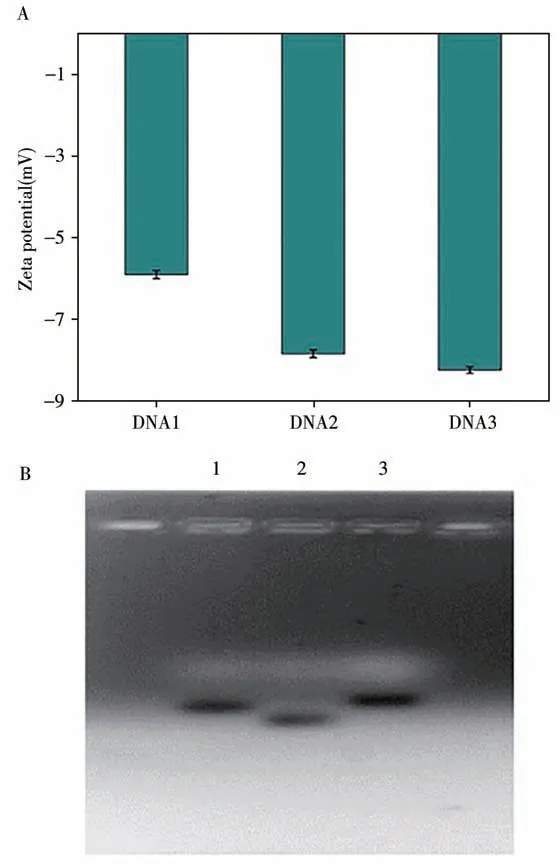
Fig.2 Synthesis characterization of DNA3
2.1.2 Feasibility study of enzyme cycle amplificationWe used the methods from step“1.5”,and determined fluorescence,then obtain the curve in Fig.3.Under the same conditions,50μL hairpin structure DNA(1.0×10-7mol/L)was used as a blank test to measure fluorescence,and a control curve was obtained.Finally,the fluorescence curve of the enzyme digestion system was obtained.As hairpin structure DNA has both FAM fluorescence groups and fluorescence quenching groups,fluorescence is very low when it is alone.The results showed that the fluorescence was increased several times after cyclic amplification under the same conditions.Therefore,this method can reduce the detection limit concentration of PDGF-BB.
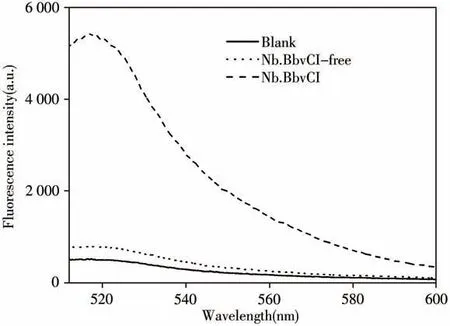
Fig.3 Fluorescence signal spectra of different samples
2.2 Selection of experimental conditions
To ensure the accuracy and sensitivity of this assay,we examined the system fluorescence value of PDGF-BB concentration of 1.0×10-7mol/L under different pH values,temperatures,and time conditions after the addition of the shearing enzyme.As shown in Fig.4,it could be determined from Fig.4A that the fluorescence signal was significantly reduced in both acidic and alkaline environments,indicating that either too high or too low pH will affect the activity of the shearing enzyme,thus weakening the fluorescence signal.In addition,the experimental results in Fig.4B showed that temperature also affected the shearing enzyme.Within a certain range,the temperature from 15℃to 45℃,the fluorescence value gradually increased with the increase of temperature,and when the temperature reached 45℃,the fluorescence signal decreased significantly,similar to the situation without the addition of shearing enzyme.The results showed that the enzyme activity was lost due to high temperature,so the purpose of cyclic amplification could not be achieved.Finally,we explored the effect of reaction time on the experiment.In Fig.4C,as the reaction time increased,the fluorescence signal gradually increased.When the time was extended to 40 min,the fluorescence signal intensity reached its maximum.When the reaction time was increased,the fluorescence value remained unchanged,indicating the high efficiency of the enzyme.We determined the optimal conditions of the experiment by detecting the fluorescence value of the 1.0×10-7mol/L PDGF-BB at different pH values,different temperatures,and different reaction times.The experimental results were satisfactory at pH 7.4,temperature of 35℃,and reaction time of 40 min.

Fig.4 Fluorescence spectra detected in different pH values(A),different reaction temperatures(B)and different reaction time(C)error bars show the standard deviation of three experiments
2.3 Specificity of PDGF-BB detected by immunomagnetic probes
To avoid false positive or false negative phenomena in the experiment and future application of the technique,we conducted specific detection for the probe.Here,bovine serum albumin(BSA),human serum albumin(HSA),and PDGF-BB were selected to evaluate the specificity of the technique.The experimental results are shown in Fig.5,in which,when PDGF-BB was co-incubated with the probe,the fluorescence intensity was higher.The fluorescence signal of other proteins was not significantly enhanced.The results preliminarily confirmed that the technique had relatively high specificity in detecting PDGF-BB.
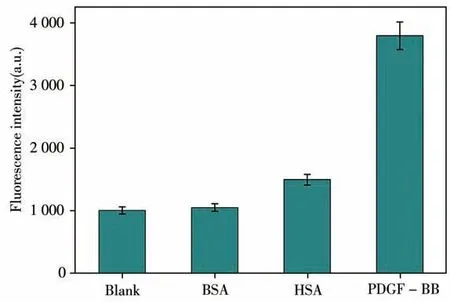
Fig.5 Selectivity of immunomagnetic probes based on enzyme cycle amplification technology to BSA,HSA and PDGF-BB all with concentrations of 1.0×10-7 mol/L;error bars show the standard deviation of three experiments
2.4 Detection of PDGF-BB by DNA biosensor
In order to explore the advantages of this method,a comparative study was conducted on the enzyme-free system and the system with the shearing enzyme cycle.It was found that the enzyme-cycle system had great advantages,with improved sensitivity and reduced detection limit.
2.4.1 Detection limit analysis of PDGF-BB in enzyme-free systemWe performed fluorescence analysis on the enzyme-free system,and the results were shown in Fig.6A and 6B.The fluorescence signal intensity increased with the increase of PDGF-BB concentration.The fluorescence signal at 518 nm and the PDGFBB concentration had a good linear relationship in the range of 1.0×10-15mol/L to 1.0×10-10mol/L.The linear regression equation of the standard curve wasY=53.662 9X+1 317.052 3(Y:fluorescence signal intensity,X:log value of PDGF-BB concentration;the same below),r2=0.999 2.The results showed that the detection limit concentration of PDGF-BB by DNA biosensor without enzyme system was 1.0×10-15mol/L.
2.4.2 Detection limit analysis of PDGF-BB by Nb.BbvCI shearing enzyme cycle systemIn order to improve the detection sensitivity,Nb.BbvCI shearing enzyme was added in the experiment,as shown in Fig.6C and 6D.When the concentration of PDGF-BB was between 1.0×10-7mol/L and 1.0×10-18mol/L,the fluorescence signal intensity decreased with the decrease of PDGF-BB concentration.The fluorescence signal at 518 nm and the PDGF-BB concentration showed a good linear relationship between 1.0×10-17mol/L and 1.0×10-10mol/L.The linear regression equation of the standard curve wasY=306.897 8X+6 849.153 4,r2=0.999 4.The detection limit of PDGF-BB in the enzyme circulation system was 1.0 amol/L.Compared with the enzyme-free system,the linear correlation was better and the range of linear correlation was wider.
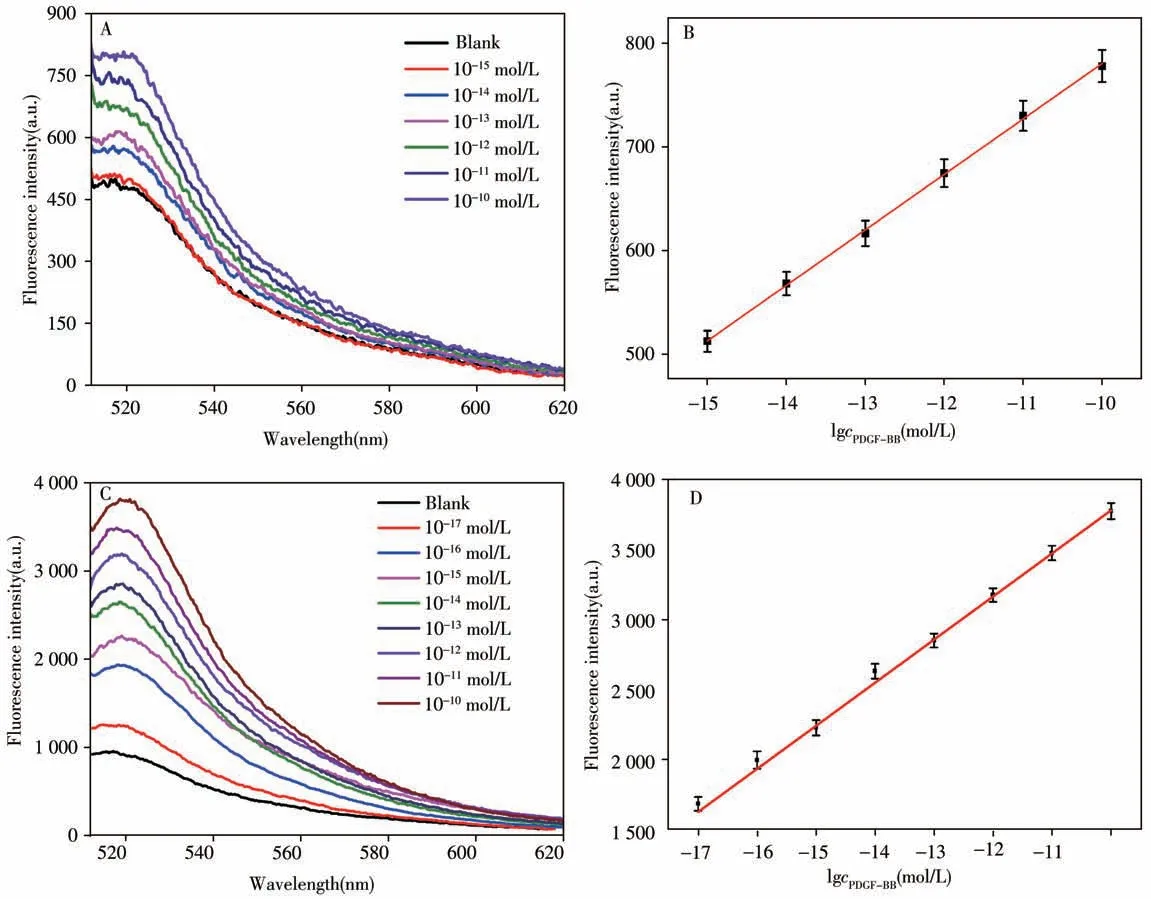
Fig.6 Fluorescence signal spectra of different concentrations of PDGF-BB(A,C)and the standard curves of fluorescence intensity at 518 nm and PDGF-BB concentration(B,D)enzyme-free systems(A,B)and enzyme cycle system(C,D)
Compared with other reported detection methods,the detection limit of the established method was reduced from nmol/L or pmol/L level to amol/L level(Table 2).In addition,the method has a good linear correlation in the range of 1.0×10-17mol/L to 1.0×10-10mol/L.In other words,this method has higher sensitivity and lower detection limits.The traditional method has the disadvantages of being time-consuming,complex,and having low sensitivity.Therefore,this method can be used as a useful substitute or supplement to other assays.
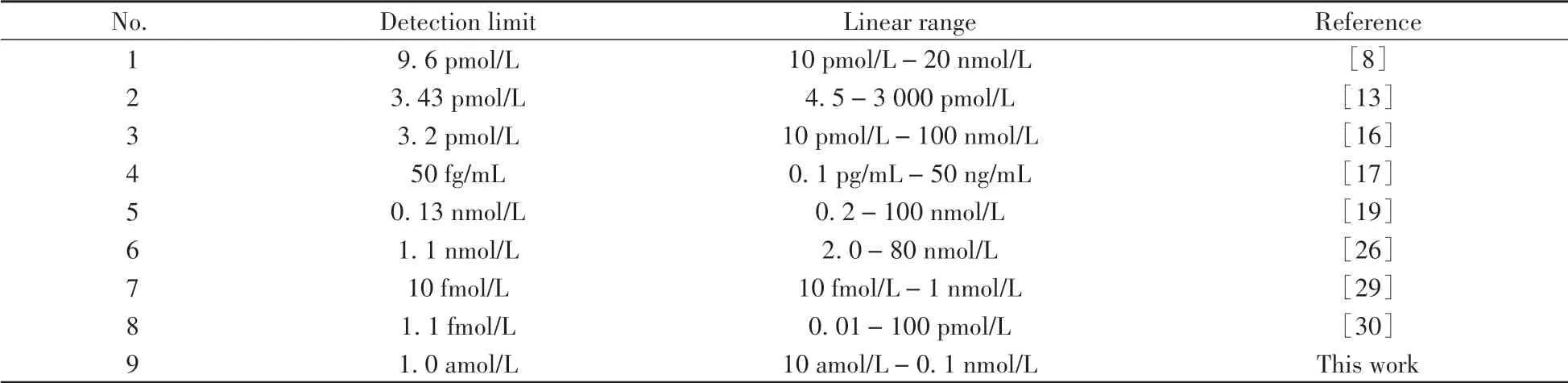
Table 2 Comparison of this work with the other studies for determination of PDGF-BB
2.5 Detection of PDGF-BB in blood samples
Finally,the DNA biosensor based on enzyme cycle amplification was used to detect the blood samples of some patients with ESCC.The average value of detected fluorescence was substituted into the working curveY=306.897 8X+6 849.153 6,and the content of PDGF-BB in the blood samples was calculated.The lowest level of PDGF-BB in blood samples from these patients was 3.89×10-16mol/L and the highest level was 3.16×10-13mol/L.In the enzyme-free system,PDGF-BB in blood of the lowest patient could not be detected,resulting in the patient missing the optimal treatment.The DNA biosensor we designed by amplifying the signal,improves the detection limit by three orders of magnitude on the original basis,and has high sensitivity,which is expected to further test its performance and achieve biological applications.
3 Conclusions
In this study,a new method was developed for the detection of PDGF-BB based on enzyme cycle amplification.The method made full use of the characteristics of PDGF-BB and used a shear enzyme to amplify the fluorescence signal,which greatly improved the detection sensitivity and reduced the detection limit.According to the above experimental results,the detection limit of PDGF-BB in the detection system without enzyme cycle amplification was 1.0 fmol/L.The detection limit of PDGF-BB was reduced to 1.0 amol/L in the enzyme cycle amplification system with the shearing enzyme.It could be shown that the detection limit of PDGF-BB was reduced 1 000 times by cyclic amplification of fluorescence signal using Nb.BbvCI shearing enzyme.This method is expected to simplify the diagnosis and detection of esophageal squamous cell carcinoma and is of great significance in biochemistry and medical clinic.
- 分析测试学报的其它文章
- Research Progress of Hemicyanine Dye for Molecular Imaging
- 碱性磷酸酶的体外检测和体内成像研究进展
- Recent Progress in Nanoscale MOFs for Biological Imaging of Tumors and Tumor Markers
- I-Motif-based Nanosystems for Biomedical Applications:p H Imaging,Drugs Controlled Release and Tumor Theranostics
- A Low-cost,Automated Nucleic Acid Extraction System Converted from the Open-Source Rep Rap 3D Printer
- Research Progress on Analytical Methods for Deciphering Adenosine-to-inosine RNA Editing

