A Low-cost,Automated Nucleic Acid Extraction System Converted from the Open-Source Rep Rap 3D Printer
CHEN Yang-tian,WANG Zun-liang
(State Key Laboratory of Bioelectronics,School of Biological Science and Medical Engineering,Southeast University,Nanjing 210096,China)
Abstract:Automatic nucleic acid extraction is essential for nucleic acid detection such as DNA amplification and high-throughput sequencing.Currently,most commercial nucleic acid extractors are mainly based on magnetic separation technology,adopting a highly integrated structural design or a closed solution,which is too costly and not conducive to use in resource-limited areas.In addition,it is difficult to update the functionality of the instruments once the functional module is determined.To address these challenges,a desktop automated nucleic acid extraction system was built in this work,which was converted from a low-cost 3D printer based on the Self-Replicating Rapid Prototyping(RepRap)open-source project.With the RepRap open-source design,the module function of the system could be flexibly designed and programmable,thus allowing updates as needed,which significantly shortened the manufacturing and testing cycle of the system.In the developed system,the modular function of the heating,mechanical motion and multi-channel magnetic separation could be fully integrated with the open-source 3D printing hardware.an 8-channel magnetic separation based nucleic acid extraction module was designed,which could be assembled onto the original 3-axis motion platform by replacing the 3D printer extruder.The motion path planning required for automatic extraction was realized by G-code programming.The experimental protocols and control software were developed for the automated nucleic acid extraction in this work.The 3-axis motion platform and nucleic acid extraction module could be effectively controlled by the host computer and the module drive circuit,respectively.The heating module consisted of 4 custom-made aluminum heating bases and the heating plate on the 3D printer.The heating bases were adapted to a 96 deep-well plate,so that the deep-well plate is well attached to the heating block for high heating efficiency.By using theλDNA as the standard nucleic acid samples,the extraction performance of the system had been verified by evaluating extraction purity,and efficiency including consistency and stability.From the results,the system showed a better extraction purity and efficiency for the samples with high concentration than that with low concentration.Additionally,the 8-channel extraction experiment for the plasmid DNA from the cultured E.coli cells had been successfully carried out on the automated system in this study.This validation further demonstrated that the automated system can be utilized for extracting nucleic acids from real cell samples.Overall,the nucleic acid extraction system developed in this study is expected to provide a cost-effective means for rapid point-of-care molecular testing in resourcelimited environments outside the laboratory.
Key words:molecular diagnosis;magnetic separation;nucleic acid detection;open-source hardware
Molecular diagnosis is referred to as the detection of human genetic variants or the foreign,potentially pathogenic nucleic acids(NAs)in order to facilitate disease diagnosis,classification,prognosis,and assessment of treatment response[1-2].Continuous breakthroughs in sequencing and polymerase chain reaction(PCR)have greatly promoted molecular diagnostic applications into their golden era of rapid development[3-6].It is noteworthy that real-time PCR(qPCR),due to its high sensitivity and low cost,has become the golden standard for clinical pathogen detection.Particularly,as the current Coronavirus disease(COVID-19)pandemic has evolved,qPCR instruments serve as a most powerful tool for defense against COVID-19 in clinical and public health settings.
Effective extraction of NA from other components of the cell or sample background is critically important for molecular diagnosis methods including sequencing and qPCR[7].Solid-phase surface binding methods based on magnetic separation are now the most common method of NA extraction for the advantages of minimal use of hazardous chemicals,simplicity,and hence are widely use on automated platform.The paramagnetic beads coated with silica can bind to free NAs by hydrogen bonding under high ionic strength conditions,and release the NAs when the binding surfaces are hydrated[8-11].The paramagnetic beads can be moved through solutions by external magnetic force to achieve magnetic separation and purification of NAs.
Nowadays,many automated NA extraction systems based on magnetic separation are commercially available,most of which are mainly restricted to centralized research and clinical laboratories[12].Due to their complex structure and high price,these existing commercial systems cannot be provided for molecular diagnostic applications in resource-limited settings or poor areas.In recent years,point-of-care testing(POCT)has become a trend towards the decentralized diagnosis and analysis,and has gained more and more attention due to its progress in reducing both the size and complexity of the system[13-14].A simple,affordable and miniaturized automatic extraction system is still a key challenge for promoting POCT molecular diagnosis applications.
The Self-Replicating Rapid Prototyping(RepRap)project aims to create self-replicating machines that can be used for rapid prototyping and manufacturing[15].One of the core advantages of RepRap is the ability to fully control the system and make changes and enhancements to the system.Herein,based on the magnetic separation method,we built a desktop automated NA extraction system,which was efficiently converted from the open-source RepRap 3D printer.Owing to the open source design,the manufacturing and testing cycle of the system can be significantly shorted,thus allowing for the system to be developed at a lower cost and faster speed than most commercial system.Moreover,the RepRap system for automated NA extraction can be conducted with simple settings or alter virtually any function of the machine.This flexibility is important especially for POCT application since the function of the system cannot be limited by specified settings from manufacturers,and can easily keep up with demand and upgrade,repair,or self-replicate.
To convert the RepRap 3D printer to our desired extraction system,we designed an 8-channel nucleic acid extraction module,and integrated it onto the original 3D printing platform by replacing the extruder of 3D printer.The motion path planning required for automatic extraction is realized by G-code programming.In our work,theλDNA were used as the standard nucleic acid samples for the verification of the performance of the NA extraction system.The experimental results demonstrated the our designed low-cost open-source system can provide a powerful tool for automatic extraction of NA samples.
1 Method and materials
1.1 Module design of the nucleic acid extraction system
As shown in Fig.1,our developed NA extraction system consists of the NA extraction module,and the 3-axis motion platform.The NA extraction can be further divided into the magnetic separation and heating module.
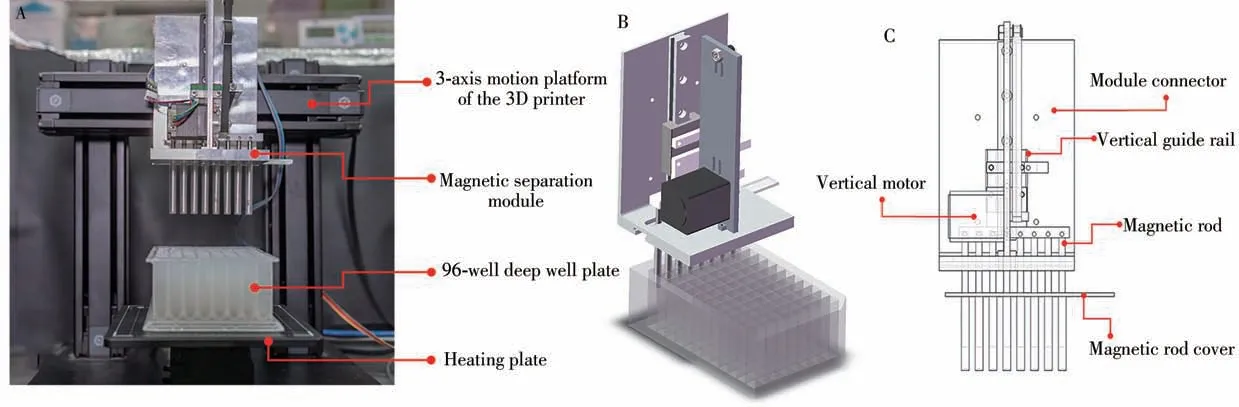
Fig.1 The automated NA extraction system based on the RepRap open-source 3D printer(A);Structure design(B);The key parts of the magnetic separation module(C)
The magnetic separation module is composed of 8 permanent magnet magnets,Computer Numerical Control(CNC)machined accessories,stepper motors and belts,which can be used for extracting 8 samples at the same time.The 3-axis motion platform is directly derived from the dobot3D printer(Shenzhen Yuejiang Technology Co.,Ltd.),which integrated NA extraction module seamless based on the RepRap open source project[15].The movement of the three-axis platform,up and down movement of magnetic separation module,and heating control can be driven and controlled by reprogramming the G-code for the 3D printer.
In this study,the structure of the magnetic separation module needs to be adapted to a 96-well deep-well plate.The size of the deep-well plate is 8×12,so that the NA extraction module can perform 8-channels samples purification in parallel.At the same time,a single sample processing requires a total of 6 wells at a time,so a deep-well plate can process up to 16 samples and make full use of each well in the deep-well plate.The nucleic acid extraction module needs to perform 3 functions:①the magnetic rod movement in the vertical direction;②the entire module movement in the vertical direction;③liquid heating.So,the module can be further divided into the magnetic separation module and the heating modules.As shown in Fig.2,the entire NA extraction module consists of a vertical motor,a vertical guide rail,a set of 8-channel magnetic rods and their cover.Among them,the 8-channel magnetic rod is connected to the vertical guide rail through the connecting piece,and the vertical motor drives the magnetic rods to move vertically along the guide rail through the rubber crawler belt.When the magnetic rod moves down to the magnetic rod and the magnetic rod cover are attached,the magnetic beads are adsorbed on the surface of the magnetic rod cover.When the magnetic rod is moved up to the magnetic rod and the magnetic rod cover are separated,the magnetic beads are dispersed into the liquid in the deep-well plate.
The heating module is composed of four custom-made aluminum heating bases and the heating plates of the 3D printer,which is illustrated in Fig.2A.The heating base can transfer the heat of the heating plate to the deep hole plate.As shown in Fig.2B,the heating base can be adapted to a 2.2 mL 96 deep-well plate,so that the deep-well plate is attached to the heating block to ensure the stability and heating efficiency of the deep-well plate during the movement of the platform.

Fig.2 Heating module
1.2 Automated NA extraction protocols
In this work,the magnetic separation method was employed for isolation and purification of NAs,which can be achieved by use of the magnetic rod to transfer the magnetic beads.In each experiment,a 96-well deep well plate is used to hold reagents and samples,and a total of 6 wells are required for NA extraction of a single sample.Solid phase extraction is often described in four basic steps:lysis,binding,washing,and eluting.As clearly illustrated in Fig.3,the experimental protocols designed for automated NA extraction are described as follows:

Fig.3 The experimental protocol for the automated NA extraction
(1)Before extraction,the required reagents need to be added into all wells from Well 1 to Well 6,where Well 1 is added to lysis solution,sample solution,binding solution and proteinase K for cell lysis to release nucleic acids,Well 2 and Well 3 are added to washing solution I and II respectively for nucleic acid purification,absolute ethanol and magnetic beads are added to Well 4 for storage of magnetic beads,Well 5 is prepared for subsequent drying of the magnetic beads binding with nucleic acids,and Well 6 is added with eluent solution,which is used to elute nucleic acid from the magnetic beads.
(2)In the lysis step,move the magnetic rod and its cover down to Well 1,and move the magnetic rod up and down for a few minutes to fully mix the lysis reagent with the sample,thus leading to release of the NAs from the sample.
(3)The magnetic rod and its cover are moved to the bottom of Well 4,and stand for 10 seconds to adsorb the magnetic beads.
(4)Move the magnetic rod with magnetic beads into Well 1.After the magnetic rod is separated from the cover for a certain distance,move the rod cover up and down for a few minutes to make the NAs binding with magnetic beads.
(5)In the washing step,the magnetic rod is moved down to adsorb the magnetic beads,then the magnetic rod and its cover absorbed with the magnetic beads are transferred to the bottom of Well 2,and the magnetic rod is moved up to release the magnetic beads from the surface of the magnetic rod cover.Move the rod cover up and down repeatedly for a few minutes,so that the magnetic beads are fully washed from the magnetic rod cover in Well 2.
(6)Repeat step 5 and wash the magnetic beads for a few minutes.
(7)In the air-drying step,move the magnetic rod and the magnetic rod cover with the magnetic beads into Well 5,and heat it at 56℃for a few minutes to keep the surface of the magnetic beads dry.
(8)In the elution step,move the magnetic rod and the magnetic sleeve to Well 6.After moving the magnetic rod up a certain distance,move the magnetic sleeve up and down for a few minutes,so that the absorbed nucleic acids can be eluted from the magnetic beads in the eluent solution.
(9)Move the magnetic rod down to Well 6 to adsorb the magnetic beads for 10 seconds,then transfer the beads to Well 4,thus the purified nucleic acids are obtained in Well 6.
1.3 System driver control
The 3-axis movement platform and nucleic acid extraction module was controlled by the host computer and the module drive circuit,respectively.The basic steps can be described as follows:
(1)The host configures the control parameters for the platform,and generates G-code control scripts according to the parameters.
(2)The host sends the generated G-code control script to the drive motherboard in real time via USB communication.
(3)The drive mainboard can control the NA extraction module and the 3-axis linear motion platform according to the G-code script,respectively.
(4)According to the G-code script,complete the heating and cooling of the heating plate and the temperature collection,and send it back to the host.
1.4 Software control flow
According to the above extraction protocols and drive control method described above,the software control flowchart of the system is illustrated in Fig.4.

Fig.4 The software control flowchart of the extraction system
2 Results and discussion
The purification performance of the nucleic acid extraction system can be verified by the purity of the sample extraction,and the sample extraction efficiency can be verified by the nucleic acid concentration changes before and after the nucleic acid extraction,including the consistency and stability verification of the nucleic acid extraction module.Furthermore,the 8-channel extraction of the plasmid DNA from the cultured E.coli cells had been performed on the system in this study.
2.1 Purification quality
For the automated NA extraction,it is crucial to extract high-purity nucleic acids from samples stably and reliably.The robustness and reliability of the NA extraction system largely depends on whether the system can stably extract high purified NAs and the purity of the extraction is consistent in the same batch and between batches experiments.
The absorbance ratio OD260/OD280and OD260/OD230were used to evaluate the purity of the NA extraction.The stability and consistency of the NA extraction can be assessed by calculating the average value((OD260/OD280)within,(OD260/OD230)within)and the standard deviation(S260/280,within,S260/230,within))of the purified samples in each channel from the same batch of experiments using Equation(1)-(2),respectively.A total of 10 batches of experiments had been performed for the consistency verification of the purity of the NA extraction.In each batch of experiments,the samples from the 8 channels were extracted and purified,simultaneously.

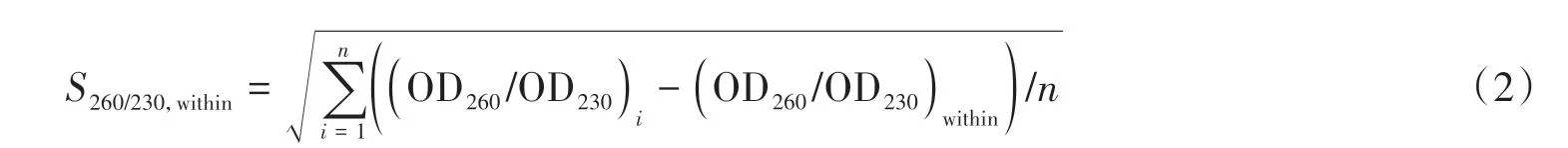
where,n=8,denoting number of channels in a single batch,andirepresentsi-th channel.We also counted the average of all batches((OD260/OD280)between,(OD260/OD230)between))using Equation(3),and the standard deviation((S260/280,between,S260/230,between)using Equation(4).

wherem=10,denoting the total batches,andjrepresents thej-th batch.

UsingλDNA as the samples for purity verification,the initial concentrations of the samples were measured by UV spectrophotometry,and the sample is divided into two ranges of concentrations:95-110 ng/μL and 45-55 ng/μL.The experimental reagents for the verification experiments are as follows:λDNA(Sangon Biotech(Shanghai)Co.,Ltd.),PCR product purification kit(Nanjing Nanoeast Biotech Co.,Ltd.),ethanol(Nanjing Chemical Reagent Co.,Ltd.),deionized water(Laboratory preparation).
In the specific experimental steps,the reagents listed in Table 1 was firstly added into their corresponding deep wells,then the NA extraction was performed,and finally the purified NAs from the eluted solution were measured by using UV spectrophotometry.

Table 1 The reagent in the deep well plate before the automated extraction
As shown in Fig.5A,the average ratios(OD260/OD280)within each batch of experiment are between 1.8 and 2.0 with the minimum value of 1.808±0.0373,indicating that RNA or protein molecules are not in the purified sample.Meanwhile,it can be seen from Fig.5B that only one(D260/OD230)within<2.0,for the 95-110 ng/μL samples,the minimum value is 1.905±0.0517,and the maximum value in all batches is 3.103±0.1982.
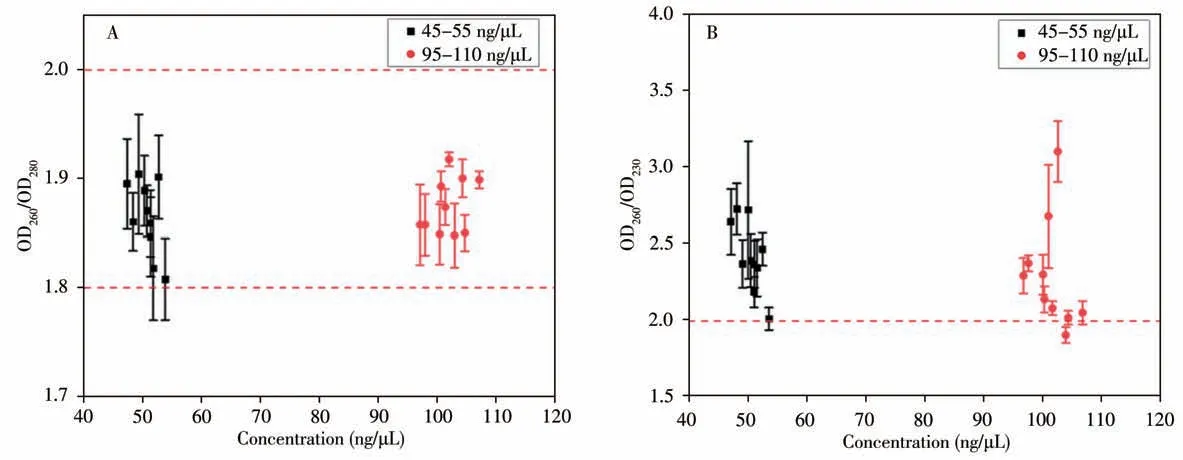
Fig.5 Distribution for the absorbance ratio of OD260/OD280(A)and OD260/OD230(B)for different concentrations of the purified NAs
TheS260/280,withinof all samples in the two concentration ranges are less than 0.1,which maximum value is 0.047 6,indicating that the purity of NA extracted from all batches of experiments shows good consistency.In a total of 20 batches of purification experiments with two concentrations of samples,there are 12 batches withS260/230,within>0.1,which the maximum value is 0.45,and the maximum of(OD260/OD230)withinis 3.67 and its minimum value is 2.33,indicating that the purified NAs can meet downstream detection requirements.
From Table 2,it can be seen that the absorbance ratios(OD260/OD280)betweenare between 1.8 and 2.0.This result indicates that the purity of the nucleic acid obtained in different batches of experiments is sufficiently high.The value(OD260/OD230)betweenshows a larger standard deviation,indicating there are certain impurity differences in the NAs purified under different batches,but it is still greater than 2,indicating that the purity of the NAs meets the purification requirements and will not affect downstream detection.

Table 2 The mean and standard deviations of the absorbance ratio between batches
Overall,the experimental results show that the automated NA extraction system can provide high-quality purified nucleic acids for downstream molecular detection.
2.2 NA extraction efficiency
The extraction efficiency in this work refers to the ability to extract nucleic acids from samples at a certain concentration and the integrity of nucleic acid fragments.If the concentration of nucleic acid is too low or fragments of nucleic acid are broken after extraction,it will directly affect downstream detection.Moreover,for the automated extraction system,it is important to ensure stable extraction in different channels in the same batch and between different batches.Therefore,the efficiency of nucleic acid extraction can be verified by counting the nucleic acid extraction concentration within and between batches.
The NA extraction efficiency for each sample channel within the same batch is proposed(E,Equation(5)),and the average value of the extraction efficiency within the same batch and between different batches(Ewithin,Ebetween,Equation(6),(9)),standard deviation(Se,withinEquation(7),Se,betweenEquation(10)),dispersion degree(C Vwithin,Equation(8),CVbetween,Equation(11))were statistically analyzed,where the analysis of dispersion degree can reflect the dispersion of the extraction efficiency relative to the average value within and between batches.

whereCeis concentration of the purified NAs,Csis the original concentration of the NA sample.

whereEirepresents the extraction efficiency in the same batch.
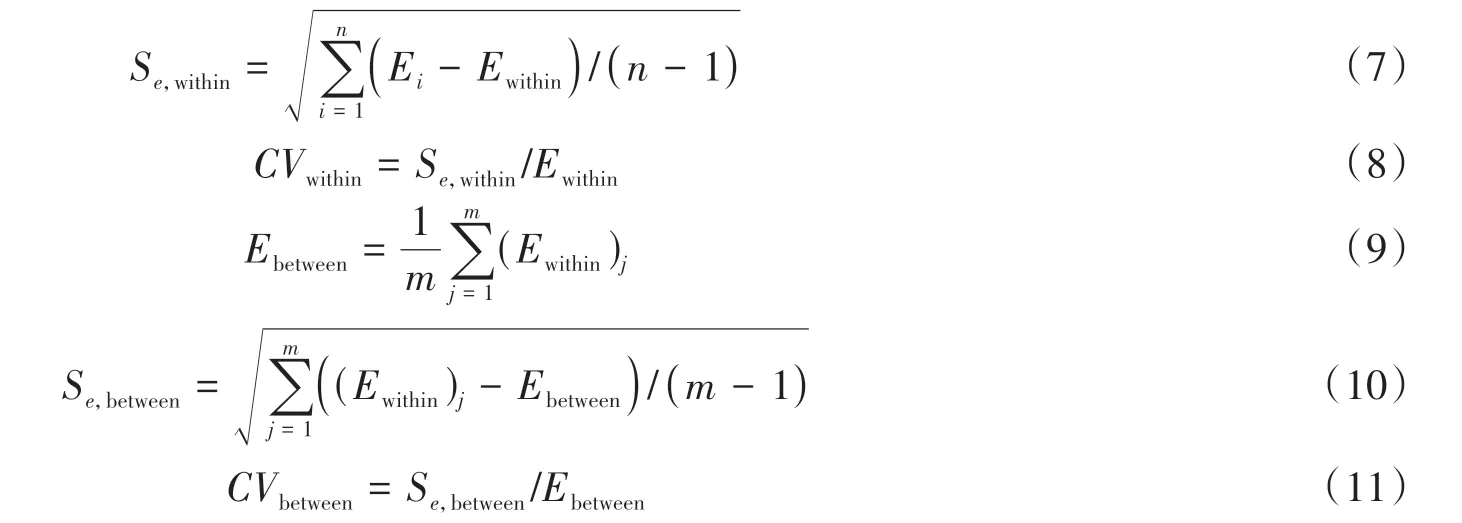
In order to verify the efficiency of nucleic acid extraction,the initial samples were divided into 4 concentration ranges:95-110 ng/μL,45-55 ng/μL,1-10 ng/μL and 0.1-1 ng/μL.Here,two methods with different detection limits were conducted to determine concentration of the 4 groups of samples.The concentrations of the 95-110 ng/μL and 45-55 ng/μL samples were measured by UV spectrophotometry.While,the fluorescence method was used to measure the samples with the concentration of 1-10 ng/μL and 0.1-1 ng/μL.
In this work,SYBR Green dye was used for fluorescence measurement.The maximum excitation wavelength of SYBR Green dye is 497 nm and its maximum emission wavelength is 520 nm.In the verification process,the standard curve of concentrationvs.fluorescence intensity is firstly calibrated by a group of gradient concentration experiments.The minimum calibrated concentration of the curve is 0.002 ng/μL and the maximum concentration is 20 ng/μL.Upon the concentration is higher than 0.02 ng/μL,the emitted fluorescence value is significantly different with that of negative controls.Thus,the linear fitting formula of nucleic acid concentration and fluorescence value from 0.02 ng/μL to 20 ng/μL can be defines as:

whereIdenotes the fluorescence intensity,Cdenotes the sample concentration(ng/μL).
2.2.1 Consistency of NA extractionThe consistency of nucleic acid extraction can be reflected mainly by the uniformity of the extraction efficiency of the sample from each channel within the same batch of experiments.The nucleic acid extraction efficiency is shown in Fig.6.
From Fig.6A,it can be seen that the maximum extraction efficiency within the same batch with concentration of 95-110 ng/μL is 102.5%±3.40%,and the minimum is 76.50%±6.45%,The extraction efficiency of 70%of the batches is greater than 85%;among the batches with a sample concentration of 45-55 ng/μL,the maximum intra-batch extraction efficiency is 86.00%±6.62%,the minimum is 64.90%±14.92%,and the extraction efficiency of 60%of the batches is greater than 75%.The extraction efficiency of each batch of samples in both concentration ranges is higher than 60%,showing most of the nucleic acid samples can be efficiently extracted from the samples.The maximum extraction efficiency in the batch with the concentration of 1-10 ng/μL is 44.89%±5.76%,and the minimum is 19.12%±2.11%,where 50%of the batches exhibited an extraction efficiency greater than 25%.In the batches with a sample concentration of 0.1-1 ng/μL,the maximum extraction efficiency is 19.53%±1.97%,and the minimum is 9.19%±2.86%.Compared with the samples with 95-110 ng/μL and 45-55 ng/μL concentrations,the extraction efficiencies of samples with concentrations of 0.1-1 ng/μL and 1-10 ng/μL are significantly reduced.
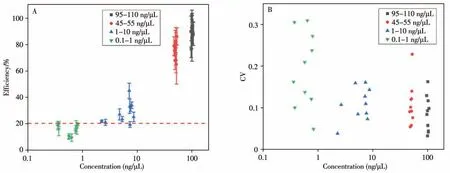
Fig.6 Distribution of extraction efficienies(A)and dispersion degrees(B)of different concentrations of the purified NAs
It can be seen from Fig.6B that 70%of the 95-110 ng/μL samples showed a CV<0.1,which maximum is 0.164;50%of the 45-55 ng/μL samples showed the CV<0.1,which maximum value is 0.230;40%of the 1-10 ng/μL CV samples is less than 0.1,and the maximum value is 0.162.For the samples with three different concentration ranges,only one batch of experiments showed a CV>0.2.The results of the above multiple extraction experiments show that the efficiency of automated nucleic acid extraction is highly consistent.Differently,when the sample concentration was reduced to the range of 0.1-1 ng/μL,only 10%of the samples showed a CV<0.1,and the maximum value reached to 0.312.The extraction efficiency of NAs between the channels shows more significant differences.
The result shows that the lower sample concentration can affect the NA extraction efficiency,which will further lower the reliability and accuracy of downstream molecular detection.
2.2.2 Stability of NA extractionThe extraction stability is mainly determined by the integrity of extracted nucleic acid fragments and the uniformity of extraction efficiency between batches at different concentrations.It can be seen from Table 3 that the extraction efficiency between the different batches of the samples with a concentration in the range of 95-110 ng/μL and 45-55 ng/μL is larger than 75%,and the CV value between different batches is less than 0.1,indicating that the extraction efficiency is higher and the difference between batches is small.While,the extraction efficiency of the sample with the concentration of 1-10 ng/μL and 0.1-1 ng/μL is smaller than 30%,and the inter-batch CV value is greater than 0.25,showing much lowered extraction efficiency and large differences between different batches.Therefore,it can be found that as the concentration of sample nucleic acid decreases,the uniformity of extraction efficiency between batches decreases,and the dispersion degree of the extraction efficiency between batches increases.

Table 3 Extraction efficiencies between batches in different concentration ranges
To identify the integrity of the nucleic acid fragments after purification,the DNA gel electrophoresis had been used to analyze the DNA fragments from a batch of experiment in each concentration range used in this study.The gel electrophoresis images were shown in Fig.7,where the electrophoresis patterns of the purified NAs from the 8 channels were represented by the band 2-9,respectively,and the band 10 represented result of the original sample.

Fig.7 Gel electrophoresis images of the purified NAs with concentration of 95-110 ng/μL(A),45-55 ng/μL(B),1-10 ng/μL(C),and 0.1-1 ng/μL(D)
It can be seen from Fig.7A-7D that as the sample concentration decreases,the brightness of the band becomes darker,while the band 2-9 showed a similar band pattern with the band 10,and no other obvious band.This shows that the integrity of the nucleic acid fragments was well maintained during the entire extraction process.
From the above experimental results,it can be noted that our extraction system demonstrated much higher extraction efficiency and better extraction consistency for high-concentration sample.For low-concentration sample,however,the NA extraction efficiency was lowered.
2.3 Experimental validation of nucleic acid extraction for E.colicells
To validate the usability of the system for NA extraction from real cell samples,the 8-channel extraction experiments for the plasmid DNA from the cultured E.coli cells had been performed on the automated system in this study.The preparation steps of the cell samples used for NA extraction are as follows:
(1)Add the corresponding reagents shown in Table 4 into the 96-deep well plate;
(2)The 18 mL of E.coli samples cultured for 24 h need to be equally divided into 8 centrifuge tubes and centrifuged at 1 200 r/min for 2 min.Then remove the supernatant from the centrifuged samples,leaving only the cells in the centrifuge tube;
(3)Add 250μL of lysate R1 in the kit to the above 8 centrifuge tubes respectively to suspend the E.coli cells after centrifugation.At this time,add 250μL of lysate R2 and 250μL of lysate R3;
(4)Add 350μL of the sample solution in each centrifuge tube to the corresponding 8 deep wells in the first column of the 96-deep well plate to complete the sample loading.
The purity and concentration of the extracted NAs were evaluated by measuring the OD260/OD280and OD260/OD230ratios by a UV spectrometry(OneDrop ultra-micro spectrophotometer,Nanjing Wuyi Technology Co.,Ltd.).The quality and integrity of the extracted DNA fragments from each channel were further analyzed by using the agarose gel electrophoresis.The reagents for the automatic NA extraction are illustrated as follows:plasmid extraction kit(Nanjing Dongna Biotech Co.,Ltd.),ethanol(Nanjing Chemical Reagent Co.,Ltd.),deionized water(Laboratory preparation).After loading the sample,the system was started up to execute the automatic nucleic acid extraction according to the setup parameters shown in Table 5.
The purified nucleic acids from each channel had been measured by using the measured using UV spectrometry.As shown in Table 6,the extracted NAs in each channel have an OD260/OD280ratio of>1.8,and an OD260/OD230ratio of>3.0,indicating a high purity of NAs without contamination with protein.Moreover,three bright bands clearly occurred in the agarose gel electrophoresis image(Fig.8),which reflected 3 conformations of plasmid DNA:opencircular(oc),linear(l)and covalently closed circular(ccc).This indicates that larger DNA fragments(>2 500 bp)maintained good integrity. While,the shorter DNA segments(<250 bp)exhibited smeared bands in the gel electrophoresis image,indicating a certain degree of fragmentation.Therefore,in the future work,it is necessary to further optimize the processing time and intensity of the key operations of the system in combination with purification reagents.Nevertheless,the verification results demonstrated that our system can be utilized for extracting NAs from real cell samples.

Table 4 The reagents in each deep well for the automatic NA extraction

Table 5 System parameter setup
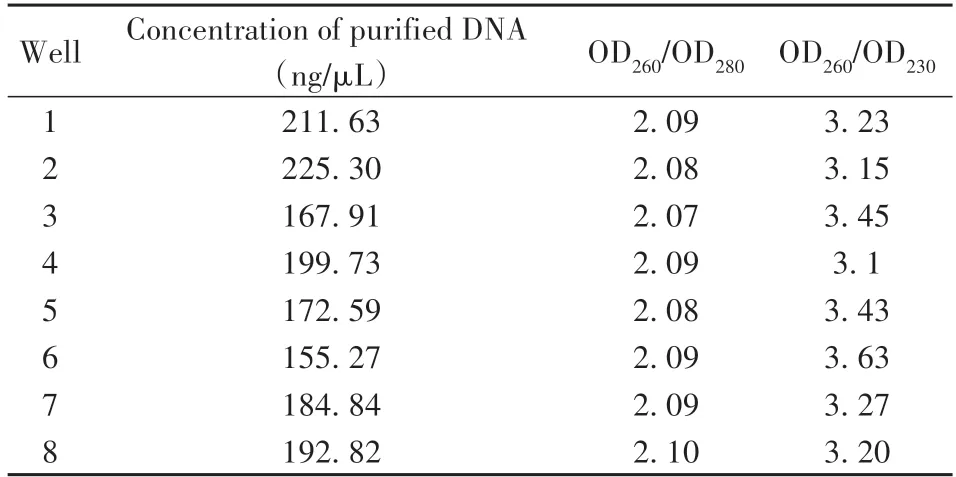
Table 6 Concentration and purity of extracted plasmid DNA
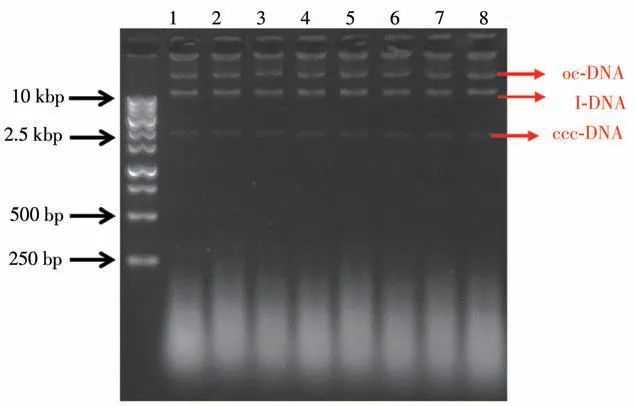
Fig.8 Agarose gel electrophoresis image of the plasmid DNA fragments extracted from the 8 channels of the E.coli cells
3 Conclusions
In the present study,we have developed an automated NA extraction system that seamlessly integrated the designed NA extraction module on the RepRap open source 3D printing platform.The NA extraction module consists of the magnetic separation module and heating module,both of which can be driven and controlled by reprogramming the G-code script of the open source 3D printer.Our study shows that this RepRap open source hardware design can greatly shorten the development cycle and test time of the system,thereby significantly reducing the development cost of the entire system.
The purification quality and extraction efficiency of our system had been well verified through 10 batches of systematic extraction experiments.The system had also been utilized for extraction of plasmid DNA from E.coli cells,which further demonstrated the effectiveness of the system for the purification of real cell samples.In future work,further optimization of the extraction protocol of the system is needed for different sample species and extraction reagents.In conclusion,the extraction system developed in this work is expected to offer an affordable tool for point-of-care molecular testing in resource-limited settings.
- 分析测试学报的其它文章
- Research Progress of Hemicyanine Dye for Molecular Imaging
- 碱性磷酸酶的体外检测和体内成像研究进展
- Recent Progress in Nanoscale MOFs for Biological Imaging of Tumors and Tumor Markers
- I-Motif-based Nanosystems for Biomedical Applications:p H Imaging,Drugs Controlled Release and Tumor Theranostics
- Research Progress on Analytical Methods for Deciphering Adenosine-to-inosine RNA Editing
- CRISPR/Cas Based Biosensing Platforms for Molecular Diagnosis

