Photothermal Detection of Circulating Tumor Cells Using Programmable Self-assembly of Enzyme-embedded DNA Nanoflowers
GAO Yu-fei,YAN Yong-cun,CAO Jing-yu,BI Sai
(1.Department of Hepatobiliary and Pancreatic Surgery,the Affiliated Hospital of Qingdao University,Qingdao 266003,China;2.College of Chemistry and Chemical Engineering,Qingdao University,Qingdao 266071,China)
Abstract:DNA nanotechnology has attracted a broad research interest in biosening field,in which various functional two-or three-dimensional DNA nanostructures have been constructed.Rolling circle amplification(RCA)as an isothermal amplification technique has provided a new way for the design and self-assembly of DNA nanomaterials.Herein,the horseradish peroxidase(HRP)-embedded DNA nanoflowers(HRP@DNFs)are one-pot synthesized by RCA,which are further applied to the fabrication of photothermal biosensor for highly sensitive and selective detection of circulating tumor cells(CTCs)in hepatocellular carcinomas(HCCs)based on dual-aptamer recognition.After encapsulation of HRP into DNFs,the catalytic activity of HRP is significantly enhanced due to the formation of highly ordered and hydrogen-bonded water environment.For HCCs detection,the aptamer TLS11a that can specifically recognize the HepG2 HCCs is designed in DNFs as the signal amplification labels,while the EpCAM aptamer-modified magnetic beads(EpCAM-MBs)are prepared as the carrieres to capture HepG2 cells.In the presence of HepG2,the sandwich structure of MBs/HepG2/HRP@DNFs is formed via specific dual-aptamer recognition.After magnetic separation and addition of 3,3',5,5'-tetramethylbenzidine(TMB)and H 2O2 into the system,the HRP@DNFs can catalyze the oxidation of TMB.Upon irradiated by 808 nm infrared laser,a strong photothermal conversion effect is produced by oxidized TMB,resulting in the photothermal detection of HepG2 cells with high sensitivity and specificity.Overall,the proposed strategy for one-pot self-assembly of enzyme-embedded DNA nanoflowers possesses a great potential in early diagnosis of diseases,especially cancers.
Key words:rolling circle amplification;DNA nanoflowers;photothermal biosensor;circulating tumor cells;aptamer
Nowadays,the liver cancer has become a major cause of cancer-related mortality worldwide.In particular,the hepatocellular carcinomas(HCCs)represent the most usual and fatal cases for liver cancers.Although the patients with early HCCs can be treated with resection or transplantation and the 5-year survival rate is more than 50%,the overall survival rate of liver cancer is only 20%because most patients are already in advanced stages when they are discovered[1].Hence,the rapid and accurate detection of HCCs is of great significance to promote the cure rate and long-term prognosis of liver cancers.Recently,it has been found that the circulating tumor cells(CTCs),the cancer cells that detach from tumor sites and invade into the peripheral blood during the tumor metastatic process,play an important role in the development and metastasis of tumors.Thus,the sensitive and selective analysis of CTCs is of great significance in prognosis prediction,patient stratification,and disease monitoring[2].So far,although a variety of methods have been developed of CTCs detection,there are still some challenges such as the rarity of CTCs,tumor heterogeneity,and so on.
Until now,DNA nanotechnology with the advantages of programmable sequence,easy modification,convenient synthesis,spatial addressability and good biocompatibility has been used to design and construct a variety of two-and three-dimensional DNA nanostructures,which is versatilely applied in biosensing,bioimaging and biomedicine.However,as the disadvantages of high cost,low yield,complicated design in sequence and inadequate control of self-assembly process often limit the further applications of DNA nanotechnology.In particular,the rolling circle amplification(RCA)has become a powerful tool in efficiently isothermal amplification,in which a circle DNA is used as the template to generate long single-stranded DNA or RNA with the assistance of DNA polymerase/dNTPs(or RNA polymerase/NTPs)[3-4].In recent years,RCA has been used to highly sensitive detection of a wide range of biomolecules,such as nucleic acids[5-6],multiple pathogens[7-8],proteins[9],and tumor cells[10].What′s more,through the performance of RCA,various DNA nanostructures have been constructed by liquid crystallization rather than Watson-Crick base-pairing,such as DNA nanotubes[11],DNA nanoclews[12],DNA nanoballs[13],and DNA nanoflowers(DNFs)[14-15].
In this study,the horseradish peroxidase-embedded DNA nanoflowers(HRP@DNFs)are synthesized by RCA,which is further used to fabricate photothermal biosensor for the detection of CTCs(Fig.1).For the preparation of HRP@DNFs,the complementary sequence of TLS11a is designed in the padlock probe of RCA.The circular template is formed by connecting the padlock probes with primers in the presence of T4 DNA ligase.During the RCA reaction,a large number of DNA blocks and inorganic magnesium pyrophosphate(Mg2PPi)crystals are generated.With the rapid increase of local concentration of DNA products and Mg2PPi,the high-density and graded DNA-Mg2PPi complex crystals will precipitate and grow continuously through the crystallization of isotropic liquid,eventually forming the porous DNFs to provide adequate sites for loading.When HRP is introduced in the system,the multifunctional HRP@DNFs containing repeated TLS11a sequences and a large number of HRP molecules are obtained during RCA,which can specifically recognize the liver CTCs HepG2.Meanwhile,the EpCAM aptamer-modified magnetic beads(EpCAM-MBs)are fabricated via amide reaction to capture the HepG2 cells.Thus,upon the introduction of HepG2,the sandwich structure of MBs/HepG2/HRP@DNFs is formed via specific dual-aptamer recognition.It has been found that the catalytic activity of HRP can be improved after encapsultating in DNFs,which can efficiently catalyze the oxidation of TMB in the presence of H2O2.Since the oxidized TMB has a strong photothermal conversion effect,the temperature of the system is increased after irradiated by 808 nm infrared laser,leading to the photothermal detection of HepG2 with high sensitivity and specificity.Compared with the other sensors,the outstanding advantages of photothermal sensor are convenient and easy popularization.In this work,a common thermometer,as one of the simplest and the most widely used household tools,can convert the photothermal signal to temperature changes sensitively,which make it possible to realize extensive application in early and accurate diagnosis of cancers.
1 Experimental section
1.1 Chemicals and materials
The oligonucleotides were synthesized and HPLC-purified by Sangon Biotechnology Co.,Ltd.(Shanghai,China),and the sequences are listed in Table 1.The T4 DNA ligase,exonuclease I(Exo I),2-(N-morpholino)ethanesulfonic acid(MES),phi29 DNA polymerase,PicoGreen dsDNA assay kitwas purchasedfrom Yeasen Biotech Co.,Ltd.(Shanghai,China).TMB,and 4S Red Plus solution were purchased from BBI Life Sciences Co.,Ltd.(Shanghai,China).The Bradford protein assay kit and deoxynucleotides(dNTPs)were purchased from Sangon Biotechnology Co.,Ltd.(Shanghai,China).The HRP,H2O2,and HCl were provided by Sinopharm Chemical Reagent Co.,Ltd.(Shanghai,China).The 1-ethyl-(3-3'-dimethylaminopropyl)carbodiimide(EDC)and N-hydroxysuccinimide(NHS)were ordered from Aladdin Reagent Co.,Ltd.(Shanghai,China).The MBs(1~2μm,1%W/V)were obtained from BaseLine ChromTech Research Centre(Tianjin,China).

Table 1 Sequences of oligonucleotides used in this work*
1.2 Preparation of circular template
A 20μL of primer(100μmol/L)was hybridized with 10μL of 5′-phosphorylated padlock probe(100μmol/L)in 1×T4 ligase reaction buffer(pH 7.8),and the solution was heated at 95℃for 5 min and slowly cooled to 25℃followed by stabilized for 3 h.Then,10μL of T4 DNA ligase(5 U/μL)was added and the mixture was incubated at 16oC overnight.Finally,the solution was transferred to 1×Exo I buffer,and 5μL of Exo I(20 U/μL)was added and incubated at 37℃for 3 h to hydrolyze the uncircularized padlock probes and primers,resulting in the production of circular template for RCA.
1.3 Synthesis of HRP@DNFs
Firstly,2μL of circular template(1.8μmol/L),5μL of Phi29 DNA polymerase(10 U/μL),10μL of dNTPs(10 mmol/L)and 10μL of HRP(5 mg/mL)were added to 1×DNA polymerase buffer(pH 7.9).The mixture was incubated at 30℃for 30 h,and the obtained HRP@DNFs were washed with ultra-pure water for three times via centrifugation(at 8 000 r/min for 10 min each time).The resulting HRP@DNFs were stored at 4℃before use.
1.4 Non-denaturing polyacrylamide gel electrophoresis(PAGE)
A 2.7 mL of 30%acrylamide/bis-acrylamide gel solution,6.2 mL of ultrapure water,1 mL 50×TAE buffer,90μL of 10%ammonium persulfate,and 10μL of N,N,N′,N′-tetramethylethylenediamine were added into theglue tank in sequence and mixed well.Then,the gel was polymerized for~30 min at room temperature and soaked in 1×TAE/Mg2+buffer(pH 8.0).Subsequently,12μL of each sample was mixed with 2μL of 10×loading buffer and added into the gel.The electrolysis was proceeded at room temperature with 170 V for 5 min,followed by 110 V for 40 min.After taking the gel,it was placed in 4S Red Plus and stained for 30 min,followed by photographed with gel imaging system(Tanon 2 500R).
1.5 Quantification of DNA and HRP in HRP@DNFs
To obtain a standard DNA curve,theλ-DNA was dissolved in TE buffer(pH 8.0)at different concentrations(0,10,20,40,60,100,150,and 200μg/mL).A 5μL ofλ-DNA at different concentrations and 195μL of Picogreen reagent were mixed in a 96-well plate for 2 min in darkness.Then,the plate was placed into a microplate detector(BioTek)to measure the fluorescence intensity(λex=485 nm,λem=530 nm).Next,the DNA concentration in HRP@DNFs was measured according to the standard curve.To obtain the protein standard curve,HRP with different concentrations(0,10,20,40,60,100,150,and 200μg/mL)was prepared in PBS buffer(pH 7.4).Then,5μL of HRP with different concentrations was added to the well plate and mixed with 195μL of Coomassie Brilliant Blue(CBB)solution.The mixture was incubated at room temperature for 5 min,and the absorbance at 595 nm was determined using a microplate reader.Finally,the amount of HRP in HRP@DNFs was determined according to the standard curve.
1.6 Determination of the catalytic activity of HRP@DNFs
The catalytic activity of HRP encapsulated in DNFs was detected through the oxidation rate of TMB by H2O2.Firstly,83μL of MES buffer(pH 5.5)was added into each well of 96-well plate.Then,2μL of HRP@DNFs and 10μL of TMB at different concentrations(0,25,50,100,200,300,400,500μmol/L)were added into the solution.Finally,5μL of H2O2(5 mmol/L)was introduced to each well,and the absorbance of each well at 652 nm was measured every 30 s with a microplate reader for 20 min.The obtained absorbance value was used to fitted the Michaelis-Menten equationv0=Vmax[S]/(Km+[S])to obtain the Michaelis constant(Km)and the maximum reaction rate(Vmax)of HRP@DNFs.The catalytic activity of free HRP and its mixture with DNFs were also determined in the same way.
1.7 Preparation of Ep CAM aptamer-MBs
Firstly,60μL of carboxyl-modified MBs were washed three times with 200μL of MES buffer(0.1 mol/L,pH 5.5)and re-dispersed in 150μL of MES buffer.To activate the carboxyl groups on MBs,500μL of EDC(0.1 mol/L)and NHS(0.01 mol/L)were added and reacted at 37℃for 30 min.Then,the activated MBs were washed twice with PBS buffer and re-dispersed in 150μL of PBS buffer.Subsequently,300μL of aminomodified EpCAM aptamer(1μmol/L)was mixed with MBs and reacted at 37℃for 12 h.Finally,the asprepared EpCAM-MBs were washed with PBS(pH 7.4)for three times and then dispersed in 150μL of PBS for later use.
1.8 Cell culture
The involved cell lines were cultured in a DMEM flask containing 10%(by volume)fetal bovine serum and penicillin-streptomycin(100 U/mL)in a humidified atmosphere of 5%CO2at 37℃.The culture medium was changed daily.When the cell density reached to~106cells m/L,the cells were isolated with 0.25%trypsin-EDTA solution trypsin,followed by centrifuged at 1 500 r/min for 5 min and then dispersed in PBS.
1.9 Confocal fluorescence imaging
The HepG2 and L-02 cells were seeded into a confocal well at a density of 3×105cells/well and incubated overnight in DMEM at 37℃under 5%CO2atmosphere.After washing,the cells were incubated with 10μL of Cy3-labeled EpCAM-MBs for 3 h,and the nuclei were stained with H33342.Finally,a confocal laser scanning microscope(CLSM,Nikon Confocal Microscope A1)was used to monitor the fluorescence imaging of HepG2 and L-02 cells.
1.10 Photothermal detection of Hep G2
A 10μL mixture of EpCAM-MBs and HRP@DNFs was added to 96-well plate containing HepG2 cells with different amounts(0,1×102,2×102,4×102,6×102,8×102,and 1×103cells)and incubated at 37℃for 1 h.After magnetic separation,the resulting sandwich structures of MBs/HepG2/HRP@DNFs were washed twice with PBS(pH 7.4).Then,5μL of H2O2(5 mmol/L)and 10μL of TMB(5 mmol/L)were added into the solution.The TMB oxidation reaction was performed at 37℃for 5 min and then irradiated vertically with an 808 nm laser(MDL-Ⅲ-808)for 5 min at a power density of 2 W·cm-2.The temperature change after the irradiation was immediately recorded using an infrared thermal imager(Fotric 225 s).
2 Results and discussion
2.1 Characterization of HRP@DNFs
To prove the formation of circular template and the process of RCA reaction,non-denatured PAGE was carried out(Fig.2).Compared to the primer(Lane 1)and linear padlock probe(Lane 2),the annealed hybridization product of primer/padlock probe shows a band with a significantly reduced migration rate(Lane 3).No significant change is observed after addition of T4 DNA ligase(Lane 4).Due to the Exo I can only hydrolyze ssDNA in the 3'to 5'direction,the excessive padlock probe and primers can be degraded upon addition of ExoⅠ,resulting in the only one band corresponding to the circular template(Lane 5).Next,the DNFs(Lane 6)and HRP@DNFs(Lane 7)with large molecular weight are synthesized by RCA reaction.Therefore,the electrophoresis results confirm the successful circularization of padlock probe into a circular template,and the feasibility of one-pot preparation of DNFs and HRP@DNFs via RCA reaction.
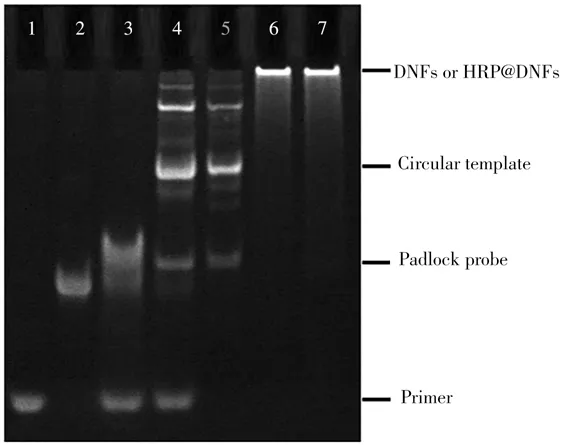
Fig.2 Non-denaturing PAGE image
The geometric morphology of DNFs and HRP@DNFs are characterized by scanning electron microscopy(SEM,JEOL JSM-6390LV)and transmission electron microscopy(TEM,Hitachi HT-7700),respectively.From the SEM images(Fig.3A),the DNFs and HRP@DNFs are successfully prepared with a good dispersibility with the average sizes of~900 nm and 910 nm,respectively.The heterogeneous flowerlike surfaces of DNFs provide a larger specific surface area and abundant binding sites for efficient loading biomolecules.The TEM study is preformed to further verify the scale and structure of DNFs and HRP@DNFs.From Fig.3B,the densities of DNA and Mg2PPi in the core area of DNFs and HRP@DNFs are higher.As the crystals are further assembled,the outer layers of DNFs and HRP@DNFs become more porous and have fewer layers in the enlarged TEM images(Fig.3B),proving the hierarchical internal structure of DNFs and HRP@DNFs.
The composition of HRP@DNFs is further confirmed by scanning transmission electron microscopy-Xray energy spectrum(STEM-EDS,ZEISS Sigma 500)(Fig.3C).The high-angle annular dark-field STEM(HAADF-STEM)images show that both DNFs and HRP@DNFs have porous and layered structures,which are consistent with the results of SEM and TEM images.The element mapping and spectrogram show that the C,N,O,Mg and P elements are uniformly exist in HRP@DNFs,which are the main elements of DNA and Mg2PPi(Fig.3C,3D).In addition,S element in HRP@DNFs is increased compared with that in DNFs,which further verify the successful encapsulation of HRP in DNFs.
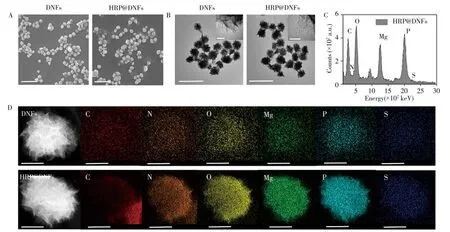
Fig.3 SEM(A)and TEM(B)images of DNFs and HRP@DNFs(scale bar:2μm),EDC spectrum of HRP-DNFs(C),HAADF-STEM images of DNFs and HRP@DNFs(D)(scale bar:500 nm)
To determine the loading capacity of HRP@DNFs,the amounts of DNA and HRP in DNFs are quantitatively determined by the stand curve method(Fig.4A,4B).The curves are obtained from three independent measurements.As a result,the amount of DNA in HRP@DNFs(61.28±2.51μg/mL)is less than that in DNFs(65.71±1.82μg/mL),which can be attributed to the inhibition of RCA by HRP molecules(Fig.4C)[16].Through using cell counter to obtain the particle concentration of HRP@DNFs(3.71×108±4.32×106particles m/L),the amount of HRP in each HRP@DNF particle is calculated to be 4.56×106±1.36×105molecules,which demonstrate the high load capability of HRP molecules in DNFs.
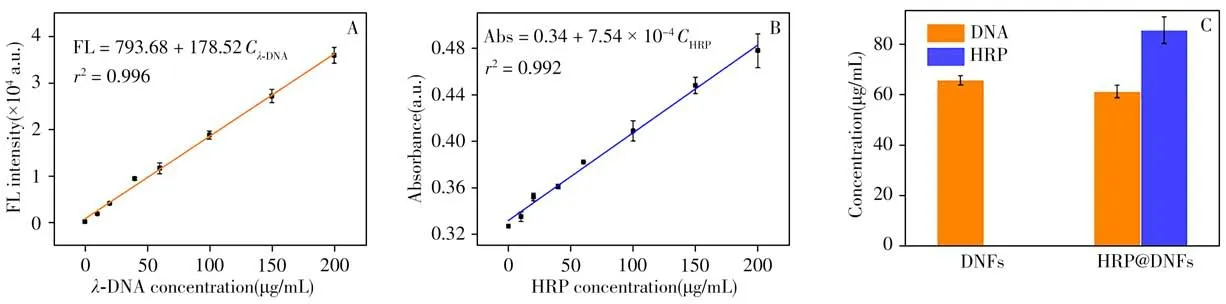
Fig.4 Loading capacity of HRP@DNFs
2.2 Catalytic activity of HRP@DNFs
The catalytic activity of HRP encapsulated in DNFs is measured by the reaction rate of TMB-H2O2catalyzed by HRP@DNFs.When free HRP is at the same concentration,the slope of HRP@DNFs catalyzed TMB oxidation is higher than that of free HRP(Fig.5A).For the HRP@DNFs catalyzed TMB-H2O2reaction,although two obvious UV-Vis absorption peaks at 370 nm and 652 nm are detected(Fig.5B),the following photothermal conversion effect is generated by the near-infrared absorption peak at 652 nm[17].Therefore,the peak at 652 nm is chosen to represent the reaction.Then,we fit the Michaelis-Menten equation(Fig.5C)and the kinetic parameters are shown in Table 2.The maximum velocity(Vmax)of HRP@DNFs shows a 1.3-fold increase compared to that of free HRP.However,there is little difference between theKmof the above two sample,which indicate that the affinity of HRP with the substrates is not impeded by the porous structures.The enhanced catalytic activity of HRP@DNFs can be attributed to the DNFs with high density of negatively charged phosphate groups,which could improve the catalytic activity of HRP[18]In addition,the negatively charged phosphate skeleton of DNA scaffolds increases the degree of hydrogen-bonded water structure,and a highly ordered hydrogen-bonded water environment is formed in the lacuna of DNA structure[18-19].Moreover,the catalytic time on the reaction system is optimized as 5 min.
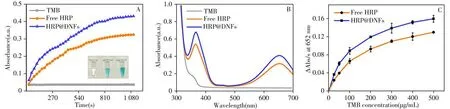
Fig.5 Dynamic UV-Vis absorbance of TMB at 652 nm catalyzed by free HRP and HRP@DNFs,respectively(A).UV-Vis spectra of TMB-H2O2 mediated by different substance(B).Michaelis-Menten curves of free HRP and HRP@DNFs,the curves are obtained from three independent experiments(C)

Table 2 Kinetic data of free HRP and HRP-DFs*
2.3 Photothermal detection of Hep G2 cells
To quantitatively detect HepG2 cells,we construct a photothermal biosensor based on the HRP@DNFscatalyzed TMB-H2O2reaction.It has been found that the oxidized TMB has photothermal property,which can be attributed to its redshift of absorption peak to near-infrared(NIR)region(652 nm)after oxidation[20-21].As shown in Fig.6A,the temperature of control group without HepG2 cells keeps 25.0℃no matter how long the infrared laser irradiation is,which indicates that the TMB molecules do not have the ability to convert infrared light energy into heat energy.Then,HepG2 cells with different amounts are introduced and incubated with HRP@DNFs and EpCAM-MBs.The sandwich structure of MBs/HepG2/HRP@DNFs are magnetically separated,followed by addition of TMB and H2O2.After irradiated by 808 nm laser(2.0 W·cm-2)for 2 min,the maximum temperature shown by an infrared thermograph is recorded.As expected,the maximum temperature increases with the increase of HepG2 cells.As shown in Fig.6B,the photothermal reactions in these groups can be finished in 2 min.In the end,the temperature change of each group is calculated.The temperature increasement corresponding to each group is demonstrated in Fig.6B,in which 1×103cells group generates an significant increasement as much as 17.7℃.The temperature increase is well linear correlated with the amount of HepG2 cells(Fig.6C).The limit of detection(LOD)of this photothermal biosensor is calculated as 59 cells.Therefore,a sensitive photothermal sensing method is successfully constructed based on the efficient catalysis of HRP@DNFs,which provide a new insight into clinical analysis of CTCs and early diagnosis of cancers.
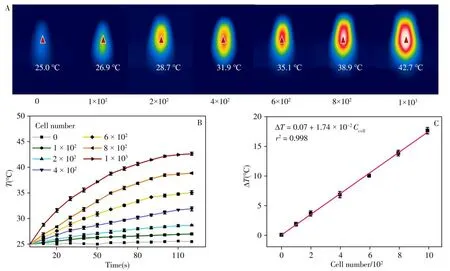
Fig.6 Photothermal effect of HRP@DNFs-based biosensor for HepG2 detection
To study the specificity of this photothermal biosensor,different cells with same amounts as HepG2 cell(1×103cells),including white blood cell(WBC),MCF-7,4T1,Hela,SW480 and LNCaP are detected in the same way.As shown in Fig.7,only HepG2 group can produce a dramatic temperature increasement of 17.7℃,while almost no temperature increase is recorded for other cells,demonstrating the high specificity of the photothermal biosensor towards HepG2 cells.The good selectivity could be attributed to the high expression level of EpCAM on the membrane of HepG2 cells.
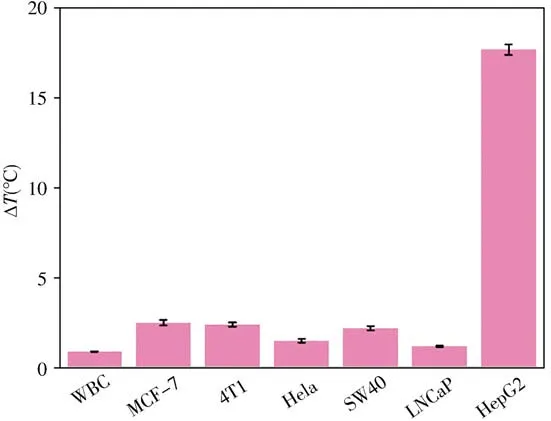
Fig.7 Temperature increasement corresponding to different cells with same amounts(1×103 cells)after laser irradiation
2.4 Real sample assays
To evaluate the availability and practicability of the photothermal biosensor,the recovery experiments are carried out by spiking a series of HepG2 cells with different amounts into human serum to simulate the complicated biological samples.The acceptable recoveries shown in Table 3 ranged from 92.7%to 103%,and the relative standard deviations(RSDs)are obtained from 5.3%to 8.9%,demonstrating the significant potential of this biosensing platform in real sample analysis and clinical research.

Table 3 Recoveries of HepG2 cells detected in human serum samples by the proposed photothermal biosensor
3 Conclusions
In summary,we have developed a one-pot method for self-assembly of enzyme-embedded DNA nanoflower via RCA,achieving the enhanced catalytic activity and the fabrication of photothermal biosensor for CTCs detection based on dual-aptamer recognition with high sensitivity and specificity. The present HRP@DNFs-based photothermal biosensor has displayed many advantages:(1)The RCAbased DNFs with porous and hierarchical structure,good programmability and high loading efficiency can be easily prepared using only two ssDNA,which facilitate the high encapsulation efficiency of HRP molecules.(2)The catalytic activity of HRP is dramatically enhanced by 1.3-fold after embedding into DNFs because of the formation of highly ordered,hydrogen-bonded water environment in the cavity of DNA structure.(3)The sandwich structure of MBs/HepG2/HRP@DNFs are formed through dual-aptamer recognition,which endow the biosensing strategy with excellent specificity.(4)The oxidization of TMB catalyzed by HRP@DNFs has demonstrated high photothermal efficiency after irradiated by 808 nm infrared laser.Combined with magnetic separation to markedly reduce the background signal,the constructed photothermal biosensor is versatilely applied to the ultrasensitive detection HepG2 cells with a LOD as low as 59 cells.Overall,our proposed enzyme-embedded DNA nanoflower strategy offers a powerful tool for the development of biosensing platforms,which is of great significance to early and accurate diagnosis of cancers.
- 分析测试学报的其它文章
- Research Progress of Hemicyanine Dye for Molecular Imaging
- 碱性磷酸酶的体外检测和体内成像研究进展
- Recent Progress in Nanoscale MOFs for Biological Imaging of Tumors and Tumor Markers
- I-Motif-based Nanosystems for Biomedical Applications:p H Imaging,Drugs Controlled Release and Tumor Theranostics
- A Low-cost,Automated Nucleic Acid Extraction System Converted from the Open-Source Rep Rap 3D Printer
- Research Progress on Analytical Methods for Deciphering Adenosine-to-inosine RNA Editing

