Morphological and phylogenetic analysis of a new Melosira species and revision of freshwater Melosira in China*
Lin YANG , Pan YU , Qingmin YOU , Guisheng LI , Quanxi WANG ,**
1 College of Environmental and Geographical Sciences, Shanghai Normal University, Shanghai 200235, China
2 College of Life Sciences, Shanghai Normal University, Shanghai 200235, China
3 Institute of Hydrobiology, Chinese Academy of Sciences, Wuhan 430000, China
Abstract Melosira is one of the most common diatom genera found in freshwater ecosystems. There are many freshwater species of Melosira, of which M. varians is the most common. In our investigation of periphytic diatoms in the Jinsha River, China, a new species, M. capsularum sp. nov, was characterized in combined morphological and molecular approaches. M. varians was also analyzed using molecular data.The new species is similar to M. varians, M. moniliformis, M. nummuloides, and M. lineata in morphology.The cells are capsular and join to f ilaments by mucilage pads that are secreted on the valve face and united into pairs by their cingula. The valve face is domed and covered with small granules, and the valve mantle edge has a milled appearance. The two strains of M. capsularum are in a single clade obviously away from other Melosira species, as determined in phylogenetic analysis based on nuclear small subunit (SSU) rDNA sequences and the chloroplast-encoded rbc L gene. Thus, the results of morphological comparisons and phylogenetic analysis based on molecular data provide strong evidence that M. capsularum is a new species,thereby increasing the total number of recognized freshwater diatom species in China. In addition, we have systematically reclassif ied the freshwater Melosira that have been recorded in China.
Keyword: Melosira; morphology; molecular phylogeny; new species; revision
1 INTRODUCTION
During the identif ication of periphytic diatoms in the Jinsha River, China, we discovered a new species,Melosiracapsularumsp. nov. When identifying this new species, we were confused in taxonomy concerning freshwaterMelosirain China, and thus it seemed necessary to clarify the taxonomic status of the genus.Melosirawas established by Agardh(1824), withM.nummuloidesC. Agardh as the type species.M.nummuloideswas described as having subspherical cells forming moniliform f ilamentous diatom communities. Since then, species that possess these characteristics have been placed in the genusMelosira. The genus comprised more than 800 species and varieties, including both freshwater and marine species, and other varieties (Guiry and Guiry, 2020).In China, a total of 48 taxa of freshwaterMelosirahave been reported (Qi, 1995).
In the 1970s, it was proposed by some studies(Florin, 1970; Ross and Sims, 1973; Gasse, 1975;Crawford, 1975a, 1979) thatM.granulataand species conforming to its characteristics should be separated fromMelosiraas a separate genus, as they were not in accordance with the original description; the characteristics of the type species specif ied that the mantle should be highly ornamented and that the cells should often form colonies. Moreover, depending on the species, cells may be joined by linking spines. On which genera the separated species should be classif ied into, there is disparity among researchers.The genusAulacoseirawas established by Thwaites(1848), withA.crenulata(Ehrenberg) Thwaites as the type species. The genus was characterized as having cylindrical cells with siliceous cell walls and cells connected by small spines forming straight, curved,or even coiled f ilaments. However, since its establishment in 1848,Aulacoseirahas largely been ignored by researchers and has long been in a state of disuse. It was not until 1979 that Simonsen resurrected the genus name and associated it with 59 common species, includingM.distans(Ehrenberg) Kützing,M.granulataEhrenberg, andM.italica(Ehrenberg)Kützing (Simonsen, 1979). Subsequently, some researchers successively revised species withinMelosirathat conformed to the characteristics ofAulacoseira(Hartley et al., 1986; Haworth, 1990;Krammer, 1991; Trifonova and Genkal, 2001; Houk,2010; Ognjanova-Rumenova and Crawford, 2012).Thus, most species that formerly belonged to the genusMelosirahave been reassigned toAulacoseira.Additionally,Melosiraincludes an extensive range of taxa, some of them have been reclassif ied into other genera, includingParalia,Orthoseira,Stephanodiscus,andEllerbeckia(Guiry and Guiry, 2020).
At present, the freshwaterMelosirareported in China have not been systematically reclassif ied, and the taxonomic status of some species remains unclear.Therefore, while describing a new species ofMelosirafrom the Jinsha River, we attempted to clarify the taxonomic position of freshwaterMelosiragenus of China.
2 MATERIAL AND METHOD
2.1 Sample collection, isolation, and cultures
The Jinsha River is the upper part of the Changjiang(Yangtze) River. Jinsha River runs through Sichuan and Yunnan provinces, with total length of 2 326 km and big drop of ~3 280 m, water area of 473 000 km2,and annual average f low of 4 750 m3/s (Gao et al.,2019). The Jinsha River accounts for ~26% of the Changjiang River basin area (Gao et al., 2019).
Periphytic samples were collected on May 2, 2020,from the Jinsha River (28°19′11″N, 103°55′2″E) from the surfaces ofCladophorasp. Samples were analyzed for total phosphorus (TP) using the ammonium molybdate spectrophotometric method (GB 11893-1989) and for total nitrogen (TN) using alkaline potassium persulfate digestion and the UV spectrophotometric method (HJ636-2012). Water temperature, conductivity, salinity, and pH were measured on-site with an YSIPro Plus multiparameter meter (YSI, Yellow Springs, OH, USA). Single diatom cells derived from clone cultures of subsamples were isolated using a Pasteur pipette and the capillary method under a Nikon Ts2 inverted microscope(Nikon, Tokyo, Japan). Cells were isolated and cultured in 24-well cell plates and each well contained 2-mL CSI medium. Nonaxenic unialgal cultures were maintained in CSI medium at 24 °C in a growth chamber under a 12-h:12-h light/dark photoperiod. A list of all of the strains examined in this study with their GenBank accession numbers, geographic locations of the sample areas, and ecological parameters are presented in Table 1.
2.2 Light microscopy (LM) analysis
Field samples were treated with concentrated nitric acid using a microwave accelerated reaction system(MARS) (CEM Corporation, Charlotte, NC, USA)and a preprogrammed digestion scheme (temperature,180 °C; ramp, 15 min; hold, 15 min) (Luo et al., 2018).Cultured samples were treated with an alcohol gradient(30% → 50% → 70% → 80% → 100%). To remove the acid from the oxidized cultures, the samples were washed six times with distilled water, and the cleaned diatoms were then mounted in Naphrax®to obtain permanent slides for the LM analysis.
2.3 Scanning electron microscopy (SEM) analysis
Cleaned specimens were air-dried on glass cover slips and attached to copper stubs, coated with ~15-nm gold-palladium using a sputter coater (HITACHI E-1045), and examined using a Hitachi SU 8010 SEM(2 kV) at Shanghai Normal University, Shanghai,China. Both the cleaned material and slides were stored at the Laboratory of Algae and Environment of Shanghai Normal University. Diatom images were compiled with Photoshop CS4 (Adobe Photoshop CS4 Extended), and the terminology and identif ications were based on previous publications.
2.4 Extraction of DNA and amplif ication
The total DNA of monoclonal culture strains was extracted using InstaGeneTM Matrix following the manufacturer’s protocol. Fragments of SSU rDNA(~1 750 nt) andrbcL (~1 470 nt) were amplif ied by PCR using primers 66F and 1255R for therbcL fragments and 11F and 1174R for the SSU rDNA fragments (Alverson et al., 2007).
Amplif ications of the SSU rDNA fragments and partialrbcL gene fragments were carried out using premade ScreenMix (Evrogen, Moscow, Russia) for the PCR assays. The amplif ication conditions for the SSU rDNA fragments were as follows: initialdenaturation of 3 min 30 s at 94 °C, denaturation for 35 cycles at 94 °C for 50 s, annealing at 58 °C for 50 s, extension at 72 °C for 60 s, and a f inal extension at 72 °C for 10 min. The amplif ication conditions for the partialrbcL fragments were as follows: initial denaturation of 3 min 30 s at 94 °C, denaturation for 35 cycles at 94 °C for 50 s, annealing at 53 °C for 50 s, extension at 72 °C for 60 s, and a f inal extension at 72 °C for 10 min. Each PCR mixture (50 μL)contained 17-μL ddH2O, 25-μL 2×EasyTaq PCR SuperMix (TransGen Biotech, Beijing, China), 2 μL of each primer (10 mmol/L) (BGI, Shanghai, China),and 4-μL DNA template.
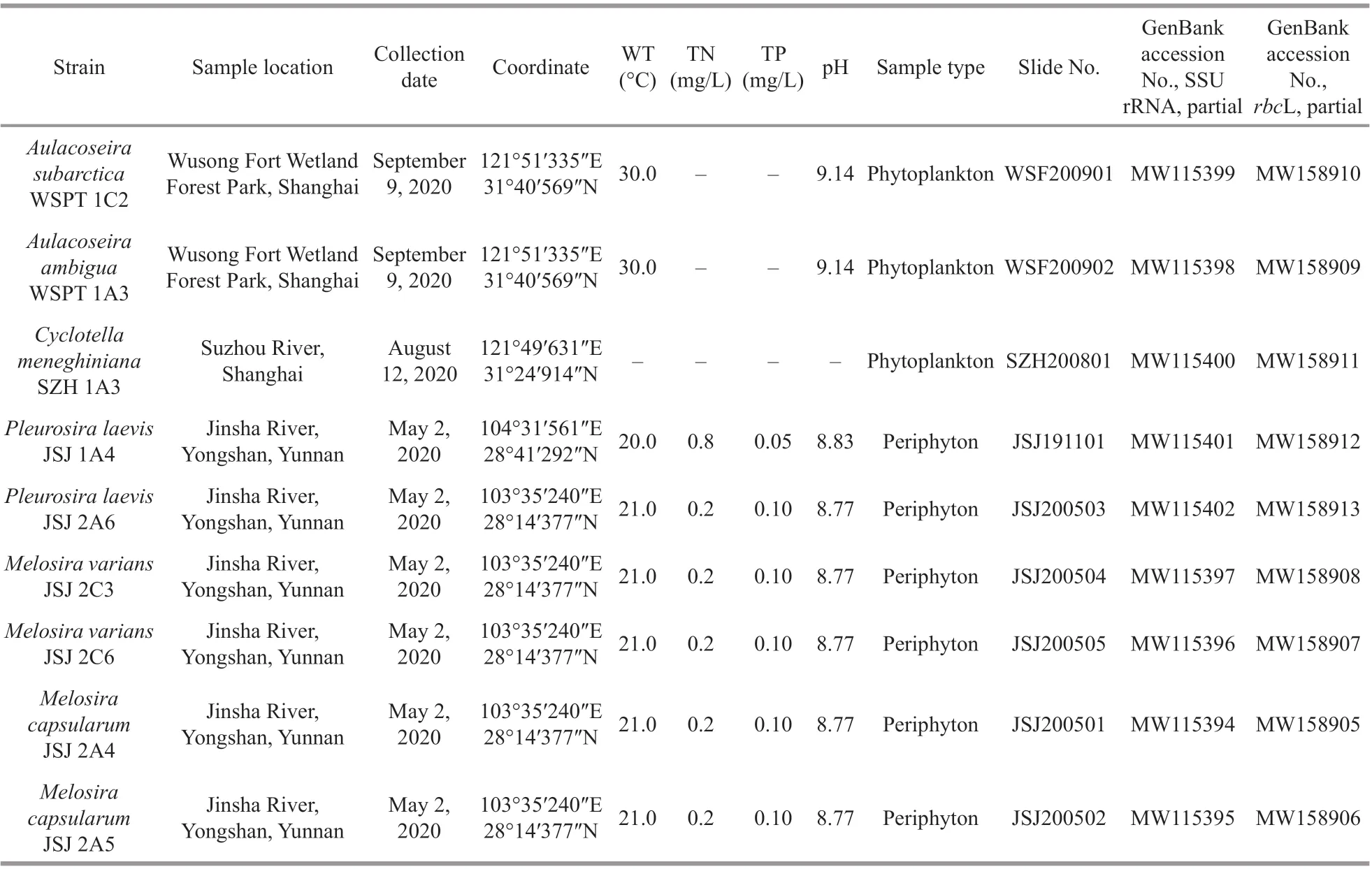
Table 1 List of strains examined in this study and their GenBank accession numbers
PCR products were purif ied using a SanPrep column DNA gel purif ication kit (Sangon, China).Then, PCR products, SSU rDNA fragments, and partialrbcL genes (decoded from two sides using forward and reverse PCR primers) were sent to the BGI Tech Corporation (Beijing, China) for sequencing on an ABI 3730XL sequencer. Sequences were submitted for a BLAST search of the National Center for Biotechnology Information (NCBI) database to f ind closely related sequences.
2.5 Tree construction
Newly determined sequences and GenBank sequences of 59 separate centric diatoms from diff erent morphological groups were included in the alignments. One araphid diatom,Fragilariacrotonensis, was selected as outgroup taxon. The obtained sequences and their data were downloaded from GenBank (Supplementary Table S1).
Sequences were aligned using the Clustal W option in the BioEdit sequence analysis software (Thompson et al., 1997; Hall, 1999). Untrimmed bases from both ends were deleted to produce identical length alignments. Using the ModelTest v3.7 Software, the GTR model of nucleotide substitutions with gamma(G) distribution rates and equal proportions across invariable sites (I) was the most appropriate evolutionary model for the SSU rDNA andrbcL alignments individually, and for therbcL-SSU rDNA alignments in general (Posada, 2006) (Table 2).Finally, concatenated SSU rDNA +rbcL alignments of 59 taxa were constructed for which both SSU rDNA andrbcL sequences were available. Phylogenies were constructed based on this model using Bayesian inference (BI) and maximum likelihood (ML)analyses. PHYML software was used to generate ML trees and the bootstrap analysis was conducted using 1 000 replicates (Felsenstein, 1981; Guindon and Gascuel, 2003). Bayesian analyses were conductedusing MrBayes v3.1.2 (Ronquist and Huelesenbeck,2003). The Markov chain Monte Carlo (MCMC)algorithm running three hot Markov chains simultaneously and one cold Markov chain was used to estimate the posterior probabilities of phylogenetic trees. The Markov chains started from a random tree and ran for 2 000 000 generations with sampling every 1 000 generations for a total of 2 000 samples for each run. FigTree v1.4.2 and Adobe Illustrator CS6 were used to edit all resulting phylogenetic trees.
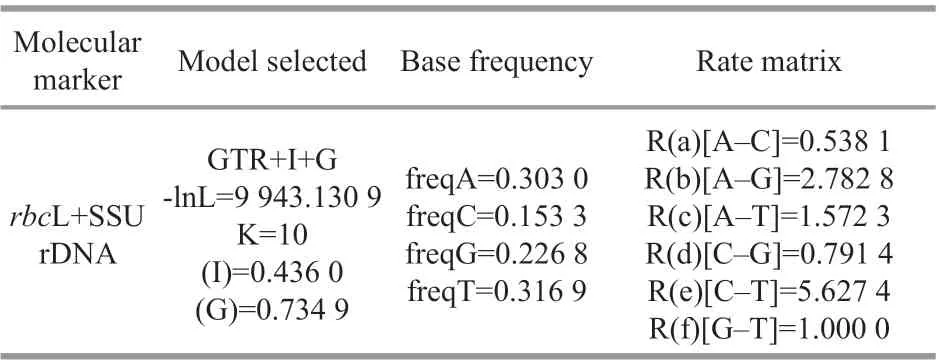
Table 2 Parameters of the nucleotide substitution model estimates using Modeltest v3.7
3 RESULT
3.1 Morphological investigations
MelosiracapsularumL. Yang & Q. X. Wang sp.nov. (Figs.1-6)
Description: Cells are capsular and joined by mucilage pads secreted on the valve face consisting of paired groups united by the cingula. Diameter: 13.0-16.0 μm; mantle height: 10.0-14.5 μm. Based on the SEM observation, the valve face is convex hemispherical with small granules. Areolae are arranged randomly or in rows radiating from the center of the valves. The valve mantle is deep; the edge of the mantle in both valves has the same milled edge appearance (arrow in Fig.4d). In addition,numerous rimoportulae are scattered over the valve face and mantle and occur in a ring near the mantle edge (arrow in Fig.4e). From the internal valve view,rimoportulae are with openings rounded or elongated pores, 0.2 μm in diameter, and there is a narrow groove around the rimoportulae (arrow in Fig.4b &4f). There are two to eight open cingular bands, and the pores in the bands are in distinct rows nearly parallel to the pervalar axis or slightly curved. All cingular bands but the valvocopula (black arrow in Fig.5) are ligulate; the ligulae are triangular (white arrow in Fig.5). Cingular bands are covered on the valve mantle. Each band is composed of one row of elongated pores that is close to the valvocopula, an unornamented area, and transverse 1-6 rows of circular or elongated pores. It is precisely due to the existence of the cingula that cells are distinctly united into pairs or triplets.
Holotype: Slide No. JSJ200501-1, Lab of Algae and Environment, College of Life Sciences, Shanghai Normal University, Shanghai (SHNU), China.
Etymology: The specif ic epithet refers to the shape of the frustule.
3.2 Molecular analysis
Pairwise comparisons of putatively related taxa showed thatM.capsularumstrains exhibited 100%similarity. However, they had an evolutionary distance of 0.06-0.08 with otherMelosiradiatoms and >0.09p-distances withAulocoseirabased on the partial ribulose-1,5-bisphosphate carboxylase large subunit.Thep-distances betweenAulocoseiraspecies ranged from 0.00-0.06 (Table 3). The pairwise uncorrectedp-distances based on the partial SSU rDNA gene showed that twoM.capsularumstrains had ap-distance of 0.00, indicating that they exhibited 100% similarity. However, these strains hadp-distances that ranged from 0.09-0.16 with otherMelosiraspecies and from 0.14-0.17 withAulocoseiraspecies. Thep-distances betweenAulocoseiraspecies ranged from 0.00-0.05 (Table 4).
The concatenated alignments of therbcL and SSU rDNA matrix included 59 centric diatom strains and one outgroup taxon, and consisted of 1 307 characters,of which 487 (37.4%) were variable and 403 (30.9%)were parsimony-informative. There were 815 (62.6%)conserved sites. The average percentages of T, C, A,and G were 32.0%, 15.4%, 30.0%, and 22.6%,respectively. Two molecular markers, SSU rDNA andrbcL, were included in the ML and BI analyses for phylogenetic inference (Fig.7). Both analyses produced similar topologies. All of the centric species were subdivided into three main lineages with diff erent levels of statistical support, namely,Melosirales, Biddulphiales, and Thalassiosirales.Melosiraformed a lineage next toAulocoseira(1.00/98.5). Another lineage consisted ofPleurosira,Terpsinoe,Hydrosera,Urosolenia, andOrthoseira(0.51/-). Within the remaining centric diatom group,there were two main clades: one included the generaCyclotellaandThalassiosira, and the other includedSkeletonema, which is a sister genus toStephanodiscus,Cyclostephanos,Lindavia, andDiscostella.
Within theMelosiraspecies,M.nummuloidesformed a single branch in the clade with the otherMelosiraspecies included in the analysis (1.00/100.0).The otherMelosiraspecies subdivided into three lineages. The f irst lineage consisted ofM.dubiaandM.moniliformis(1.00/98.9). Two strains ofM.capsularumformed the second linage next toM.varianswith low statistical support (0.82/59.6),but this single branch had high statistical support(1.00/100.0). The third lineage was comprised of three strains ofM.varians(1.00/100.0).
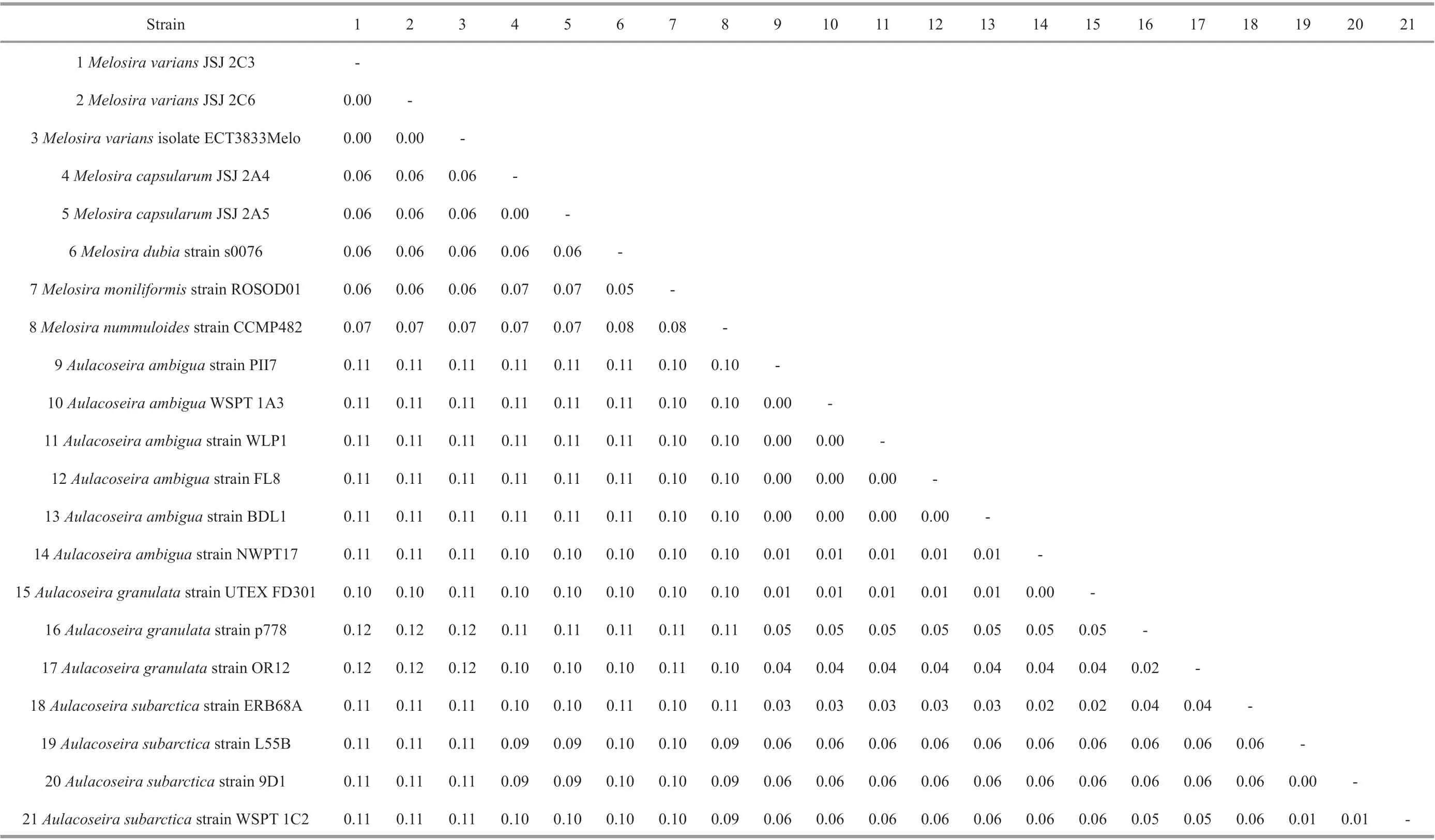
Table 3 Pairwise uncorrected p-distances of 21 strains based on the partial ribulose-1,5-bisphosphate carboxylase large subunit
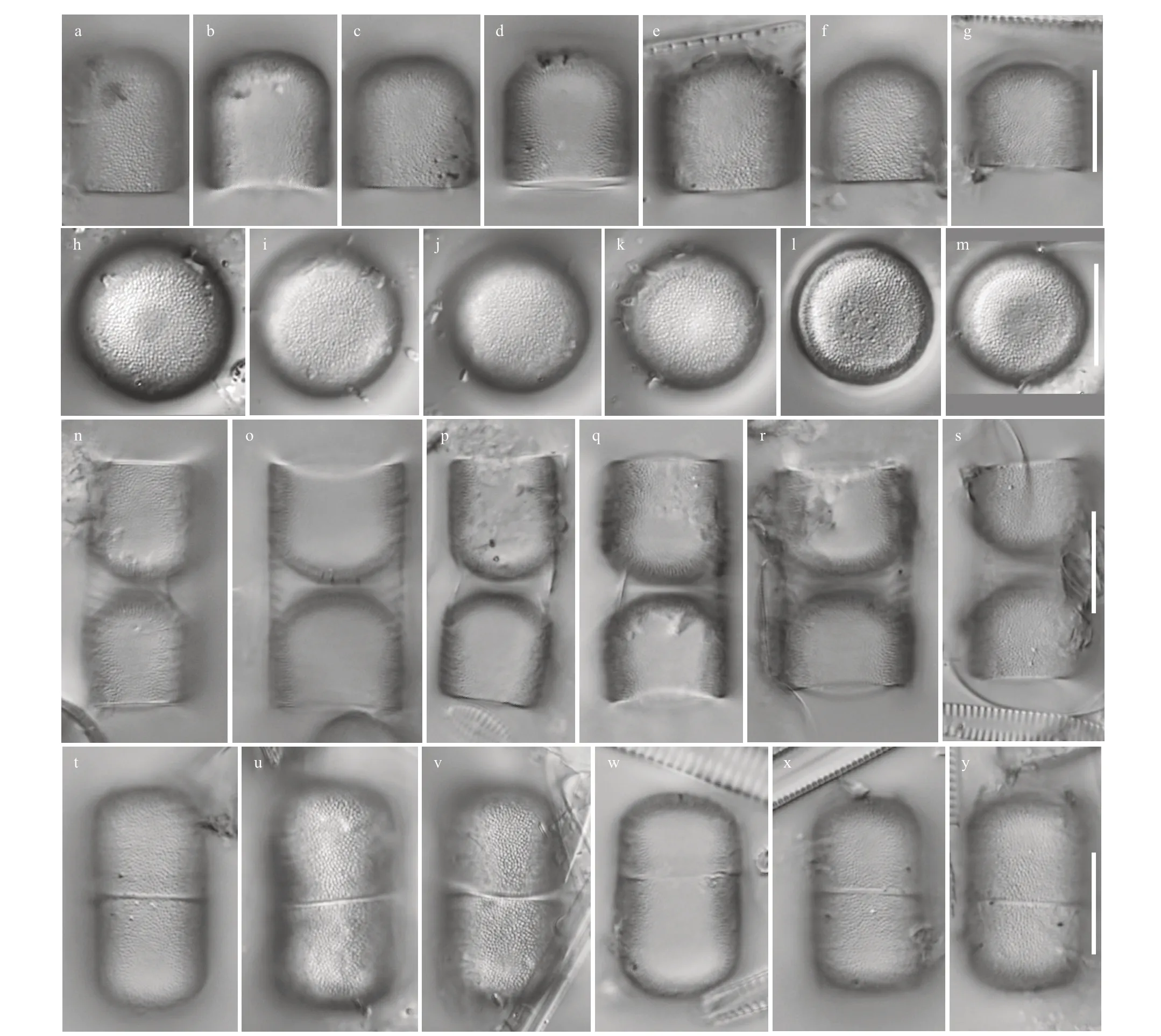
Fig.3 LM micrographs of Melosira capsularum sp. nov.
3.3 The revision of the freshwater Melosira in China
At present, of the 48 taxa of the genusMelosirathat have been reported in China, 17 remain within the genusMelosira, 18 taxa have been transferred intoAulacoseira, and 5 taxa have been transferred into other genera (Ellerbeckia,Stephanodiscus,Orthoseira,andParalia). In addition, there are seven varieties and forms for which their classif ication status needs to be reassessed. Therefore, according to the detailed morphological description and hand drawings, we have proposed seven new combinations and have rearranged the freshwaterMelosirain China. The details required for seven new combinations are therefore given below.The arrangement of 48 freshwaterMelosirataxa reported in China is shown in Table 5.
1Aulacoseiraliratavar.seriata(Müller) L. Yang &Q. X. Wang, comb. nov.
Basionym:Melosiraliratavar.seriataMüller 1898. Bacillariales aus den Hochseen des Riesengebirges. Forschungsberichte aus der Biologischen Station zu Plön 6: 8, pl. 3: Fig.34.
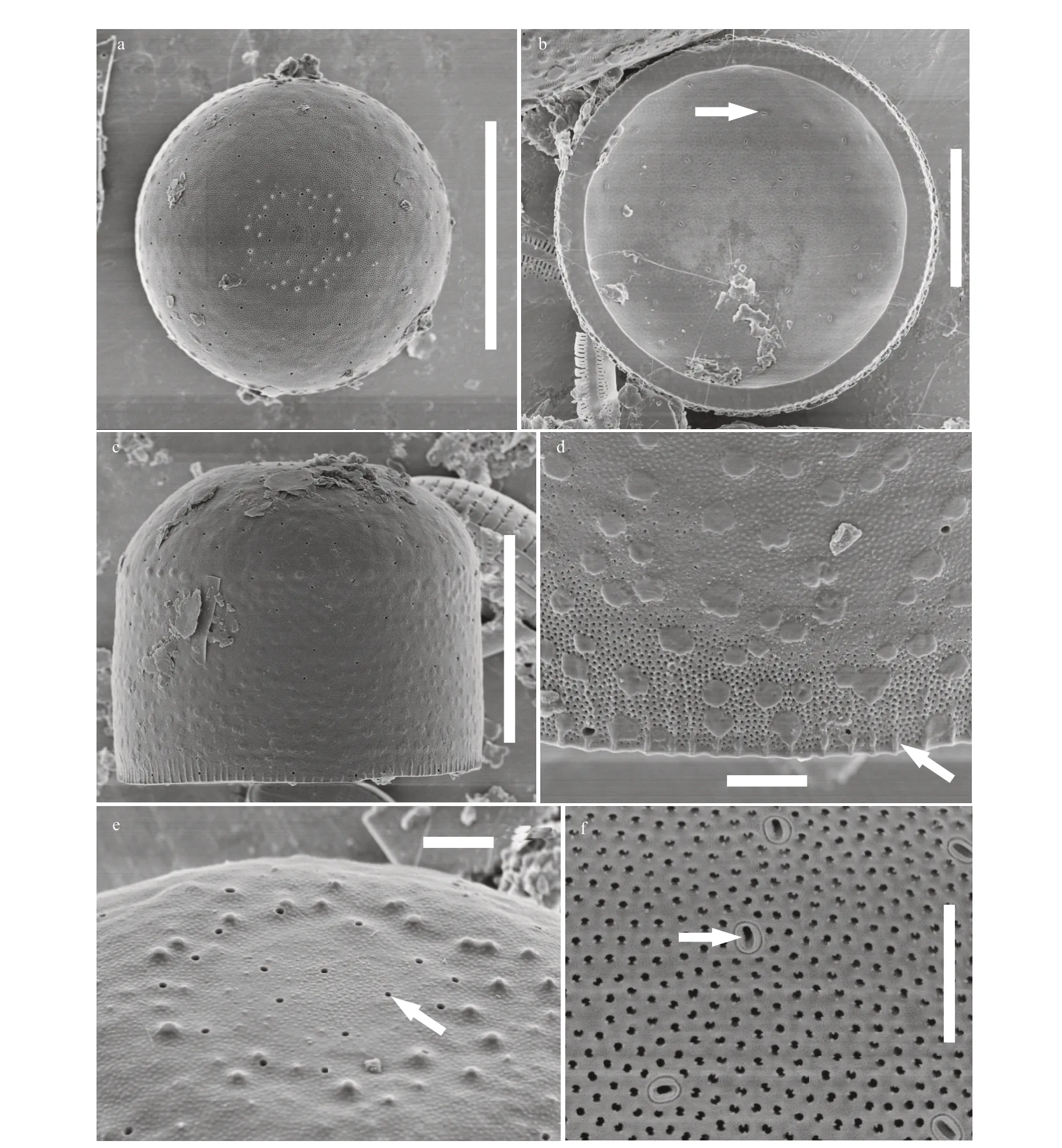
Fig.4 SEM micrographs of Melosira capsularum sp. nov.
Synonym:Melosiradistansvar.lirataf.seriata(Müller) Hustedt 1927a. Die Kieselalgen Deutschlands, Österreichs und der Schweiz unter Berücksichtigung der übrigen Länder Europas sowie der angrenzenden Meeresgebiete. Bd. VII: Teil 1: 264.
Aulacoseiradistansf.seriata(Müller) Davydova in Glezer et al. 1992. The diatoms of the USSR fossil and recent. Vol. II. fasc. 2: Stephanodiscaceae,Ectodictyonaceae, Paraliaceae, Radialiplicataceae,Pseudopodosiraceae, Trochosiraceae, Melosiraceae,Aulacosiraceae. pl. 80.
2Aulacoseiragranulatavar.curvata(Grunow)L. Yang & Q. X. Wang, comb. nov.
Basionym:Melosiragranulatavar.curvataGrunow in Van Heurck 1882. Synopsis des Diatomees de Belgique: 87: Fig.18.
Synonym:Melosiragranulataf.curvata(Grunow)Hustedt 1927b. Fossile Bacillariaceen aus dem Loa-Becken in der Atacama-Wüste, Chile.ArchivfürHydrobiologie18(2): 250.
Aulacoseiraambiguavar.curvata(Grunow)Simonsen 1979. The diatom system: ideas on phylogeny.Bacillaria2: 56.
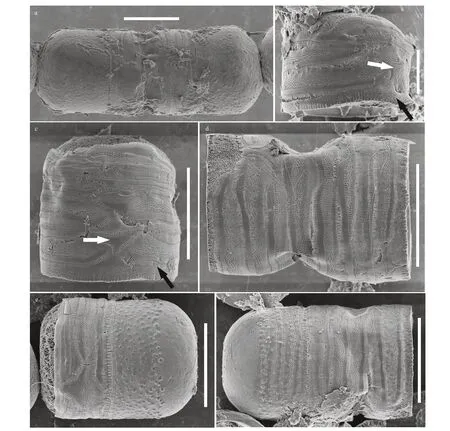
Fig.5 SEM micrographs of Melosira capsularum sp. nov.
3Aulacoseiraitalicaf.curvata(Pantocsek) L. Yang& Q. X. Wang, comb. nov.
Basionym:Melosiracrenulataf.curvataPantocsek 1902. Kieselalgen oder Bacillarien des Balaton.Resultate der Wissenschaftlichen Erforschung des Balatonsees, herausgegeben von der Balatonsee-Commission der Ung. Geographischen Gesellschaft.Bd 2(2): 103; pl. 15, Fig.327.
Synonym:Melosiraitalicaf.curvata(Pantocsek)Hustedt 1927a. Die Kieselalgen Deutschlands,Österreichs und der Schweiz unter Berücksichtigung der übrigen Länder Europas sowie der angrenzenden Meeresgebiete. Bd. VII: Teil 1: Liefrung 1. In:Rabenhorst's Kryptogamen Flora von Deutschland,Österreich und der Schweiz: 260.
4Aulacoseiraitalicavar.hankensis(Skvortzov)L. Yang & Q. X. Wang, comb. nov.
Basionym:Melosiraitalicavar.hankensisSkvortzov 1929. Freshwater diatoms from Amoy,South China.TheChinaJournal11(1): 42, pl. 1: Fig.8.
5Orthoseiraroeseanaf.spinosa(Skvortzov)L. Yang & Q. X. Wang, comb. nov.
Basionym:Melosiraroeseanavar.epidendronf.spinosaSkvortzov 1938. Subaërial diatoms from Pin-Chiang-Sheng Province.PhilippineJournalofScience. Section C 65(3): 265, pl. 3: Fig.2.
6Orthoseiraroeseanavar.xizangensis(Chen)L. Yang & Q. X. Wang, comb. nov.
Basionym:Melosiraroeseanavar.xizangensisChen in Chen & Zhu 1985. Studies on the freshwater Centricae of China.ActaHydrobiologicaSinica9(1):81, Figs.1, 2.
7Aulacoseirayoungiivar.tenuissima(Skvortzov)L. Yang & Q. X. Wang, comb. nov.
Basionym:Melosirayoungiivar.tenuissimaSkvortzov 1937. Neogene diatoms from Eastern Shantung.BulletinoftheGeologicalSocietyofChina17(2): 195, pl. 1: Figs.1-3.
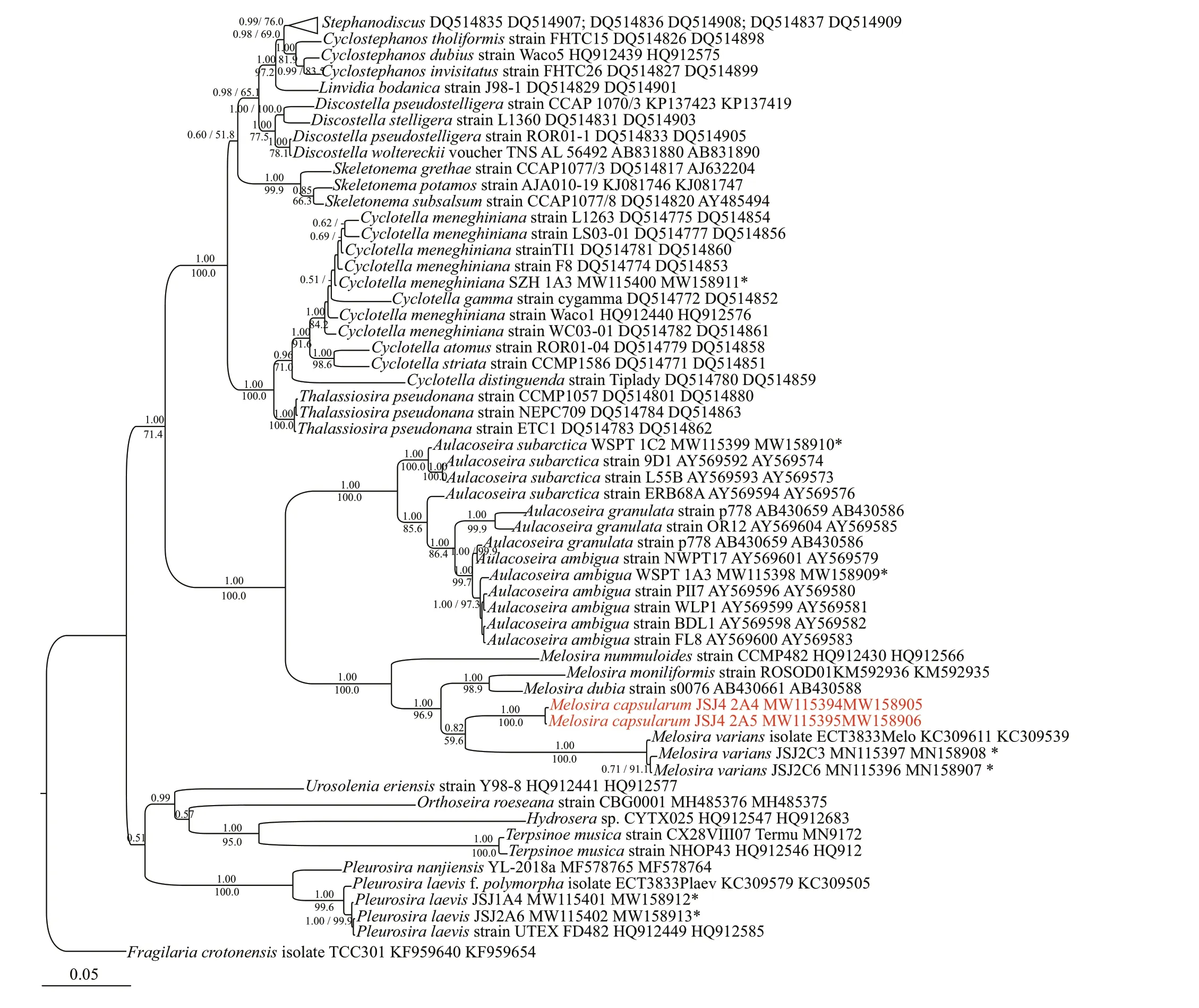
Fig.7 Bayesian tree of Melosira and Aulocoseira species constructed from a concatenated alignment of 59 partial rbc L and SSU rDNA sequences of 1 307 characters
4 DISCUSSION
4.1 Habitat analysis of the new species- Melosira capsularum L. Yang & Q. X. Wang
MostMelosiraspecies are planktonic and mass reproduction of certain species is through the formation of diatomite deposits. In this study, we combined morphological and molecular methods to analyze a putatively newMelosiraspecies that was collected from the surfaces ofCladophorasp. Because these diatoms grow in a given location, they accurately ref lect the water quality and changes in plankton populations in rivers, lakes, and oceans with higher f low rates. Therefore, the diatoms of the genus are good indicators for water environmental assessments.M.capsularumwas also observed in specimens collected in May and November, 2019. Therefore, we deduced that this species has existed in this location(i.e., it was not aff ected by water temperature).

Table 5 Taxonomic status revision of the freshwater Melosira species in China
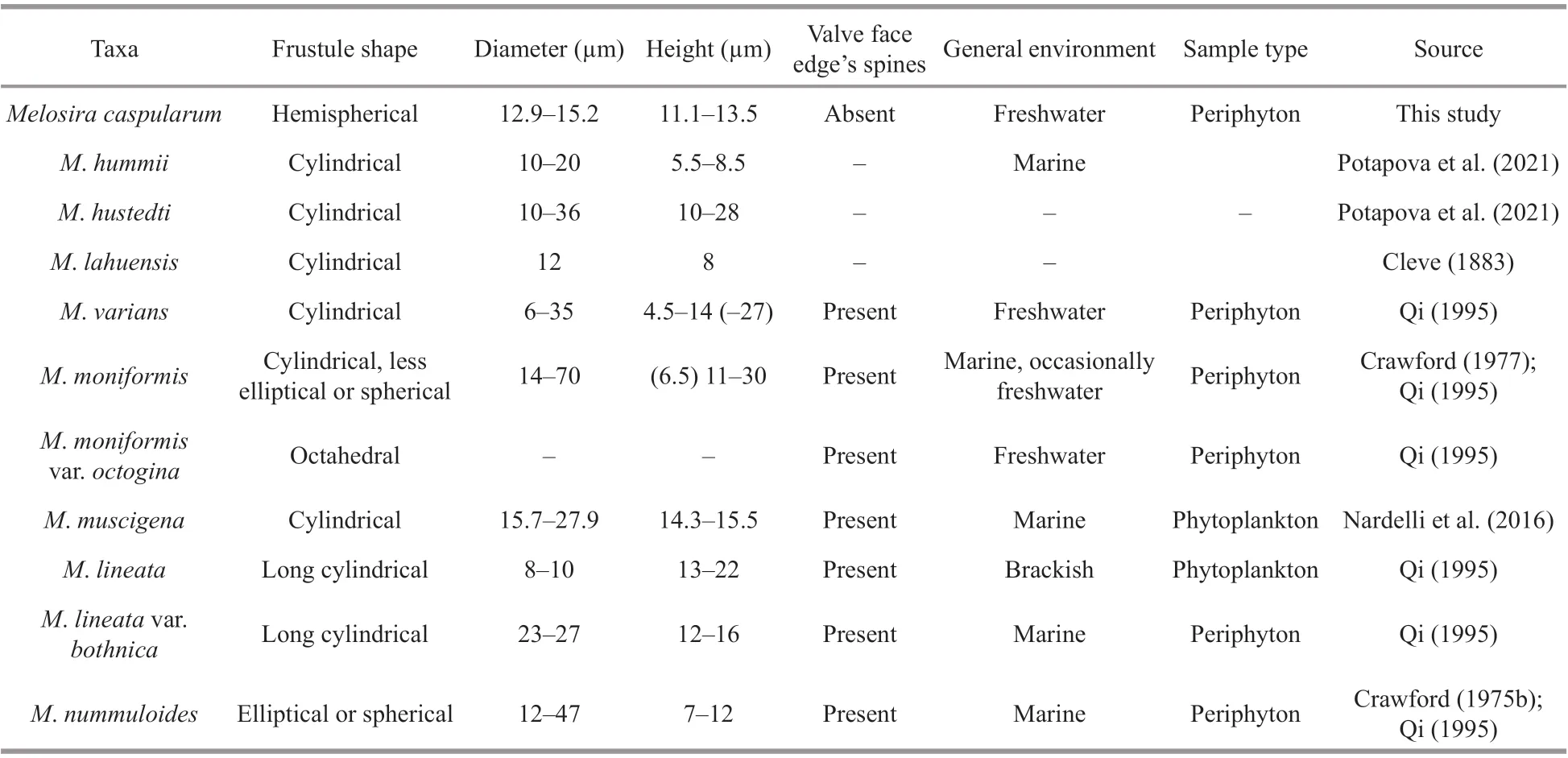
Table 6 Morphological characteristics of seven Melosira species
4.2 Morphological comparison between M. capsularum and other similar species
Based on the morphological characteristics and molecular results,M.capsularumis similar toM.varians,M.nummuloides,M.moniliformis,M.hummii,M.hustedti,M.lahuensis,M.muscigenaandM.lineata. All of these species exhibit the following characteristics: the position and arrangement of rimoportulae; the shape of girdle bands, and the arrangement of cell connections.However, they have obvious morphological diff erences, as summarized in Table 6.
4.3 Phylogenetic analysis of M. capsularum and other centric diatoms
In our phylogenetic analysis, most of the sequences for freshwater centric genera were obtained from GenBank. Nuclear SSU rDNA sequences and chloroplastrbcL sequences were applied in a previous phylogenetic analysis of centric diatoms of Thalassiosirales (Alverson et al., 2007), whereCyclotellaspecies formed a clade withThalassiosiraspecies, andDiscostella,Cyclostephanos, andStephanodiscusspecies clustered together on a large branch. This clustering result is consistent with our f indings (Fig.7). Our data support results by Gargas et al. (2018) thatOrthoseirais not closely related to other melosiroid diatoms, but it is closer to multipolar diatoms.Orthoseirais more closely related toTerpsinoeandHydroserathan toMelosiraorAulacoseira. Phylogenetic trees based on SSU rDNA andrbcL sequences showed thatM.capsularumwas distinct from otherMelosiraspecies. Diff erences in both morphological measurements and molecular analyses supportedM.caspularumas a new species.In our phylogenetic tree,Melosiraformed a lineage next toAulacoseiraspecies, indicating that these genera have a close relationship. This result was consistent with our morphological observations. In the phylogenetic tree,M.dubiaformed a branch withM.moniliformisand had higher statistical support(1.00/98.9). However, in AlgaeBase,M.dubiais regarded as a synonym ofPodosiradubia(Kützing)Grunow. Therefore, the phylogenetic position ofM.dubia/P.dubiarequires further investigation,including morphological observations and molecular analyses of available strains.
4.4 Current status in phylogeny of the genus Melosira
In the genusAulocoseira, 45 species have been phylogenetically analyzed using morphological and gene sequence data (SSU rDNA andrbcL) (Edgar and Theriot, 2004). Compared toAulocoseira, there is a lack of comparative systematic phylogenetic studies onMelosiraand the sequences ofMelosiraspecies are lacking as well. Thus, further work on additionalMelosirataxa, including both morphological and molecular analyses, is required in order to form a more complete picture of the evolutionary history of this genus.
5 CONCLUSION
A new speciesMelosiracapsularum, which was collected from the surfaces ofCladophorasp. during an investigation of periphytic diatoms in the Jinsha River, China, was identif ied in this study. The results of both the morphological comparisons and phylogenetic analysis support the f inding that this algal taxon is a new species. In addition, after a series of revisions of the 48 taxa of freshwaterMelosirathat have been reported in China, 18 taxa (13 species and 5 varieties) were retained the genusMelosiragenus;22 taxa (13 species, 7 varieties, and 2 forms) have been transferred into the genusAulacoseira; 1 species and 1 variety were transferred intoEllerbrckia; 4 taxa(1 species, 3 varieties, and 1 form) were transferred intoOrthoseira; 1 species was transferred intoParaliaand 1 species was transferred intoStephonodiscus.
6 DATA AVAILABILITY STATEMENT
All of the data obtained and/or analyzed in this study are available from the corresponding author upon request.
 Journal of Oceanology and Limnology2022年2期
Journal of Oceanology and Limnology2022年2期
- Journal of Oceanology and Limnology的其它文章
- Identif ication of Antarctic minke and killer whales with passive acoustic monitoring in Prydz Bay, Antarctica*
- Eff ects of dissolved oxygen and nutrients from the Kuroshio on hypoxia off the Changjiang River estuary*
- Methane in the Yellow Sea and East China Sea: dynamics,distribution, and production*
- Longitudinal genetic analysis of growth-related traits in red swamp crayf ish Procambarus clarkii (Girard)*
- Early life migration and population discrimination of the small yellow croaker Larimichthys polyactis from the Yellow Sea: inferences from otolith Sr/Ca ratios*
- A new oil spill detection algorithm based on Dempster-Shafer evidence theory*
